Abstract
The thyroid hormone receptors (TR) are members of the nuclear hormone receptor (NHR) superfamily that regulate development, growth, and metabolism. Upon ligand binding, TR releases bound corepressors and recruits coactivators to modulate target gene expression. Steroid Receptor Coactivator 2 (SRC2) is an important coregulator that interacts with TRβ to activate gene transcription. To identify novel inhibitors of the TRβ and SRC2 interaction, we performed a quantitative high throughput screen (qHTS) of a TRβ-SRC2 fluorescence polarization assay against more than 290,000 small molecules. The qHTS assayed compounds at six concentrations up to 92 uM to generate titration-response curves and determine the potency and efficacy of all compounds. The qHTS dataset enabled the characterization of actives for structure-activity relationships as well as for potential artifacts such as fluorescence interference. Selected qHTS actives were tested in the screening assay using fluoroprobes labeled with Texas Red or fluorescein. The retest identified 19 series and 4 singletons as active in both assays with 40% or greater efficacy, free of compound interference and not toxic to mammalian cells. Selected compounds were tested as independent samples and a methylsulfonylnitrobenzoate series inhibited the TRβ-SRC2 interaction with 5 uM IC50. This series represents a new class of thyroid hormone receptor-coactivator modulators.
Keywords: thyroid receptor, small molecule, HTS, coactivator, protein-protein interaction
Introduction
Thyroid hormone is instrumental in controlling aspects of metabolism, growth, and development (reviewed in 1; 34). The thyroid receptor (TR) is a member of the family of nuclear hormone receptors (NHR) that induce the expression of transcriptional targets. There are two TR genes, TRα and TRβ, both of which are alternatively spliced 29. TRα regulates key aspects of heart function, while TRβ controls lipid cholesterol levels as well as feedback regulation between the hypothalamus, pituitary and thyroid (reviewed in 5).
Like other NHRs, TR is composed of three domains: a ligand-independent transcriptional activation domain, a DNA-binding domain, and a ligand-binding transcriptional activation domain (LBD, 15). In the absence of ligand, TR is bound to corepressor proteins 4; 14 as a complex on responsive promoters that prevents transcriptional activation. Ligand binding to TR causes the dissociation of corepressors and the subsequent association of coactivators 21 that stimulate target gene expression.
A coactivator family that interacts strongly with TRs, as well as other NHRs, is the p160 protein family of steroid receptor coactivators (SRCs) 19; 32; 33 that include SRC1, SRC2 (GRIP1/TIF2), and SRC3 (AIB1/TRAM1/RAC3/ACTR). All SRCs contain a nuclear receptor interaction domain composed of three repeated consensus LXXLL motifs, termed NR boxes, and an activation domain that interacts with other coregulatory proteins. The LXXLL motif is necessary and sufficient for interaction with NRs 6; 12. Peptides comprised of LXXLL motifs from SRC2 family members bind to TR with affinities comparable to full-length SRC2 protein 6; 7. NR boxes interact with a hydrophobic groove located in AF-2 region of TR 28.
We have previously identified a pro-inhibitor β-aminoketone series (SJ1, Figure 1A) from a high throughput screen, which disrupts TR-SRC2-2 interaction in the presence of T3 2. SJ1 can undergo deaminiation to form an enone in situ, followed by a modification of a nucleophilic cysteine in AF-2 cleft. Mechanism of action studies suggest that SJ1 modifies Cys298, one of four accessible cysteines in the AF-2 region of TR, resulting in disruption of SRC binding 8. Preliminary evaluation of in vivo toxicology revealed significant dose-related cardiotoxicity, suspected to arise from ion channel inhibition. Although subsequent chemical optimization work improved the pharmacological properties of SJ1, including increased potency, reduced cytotoxicity, and elimination of hERG potassium channel activity 16, these analogs still exhibit ion channel binding. Therefore, new chemotypes of TR-coactivator inhibitors are desired. The goal of this project is to identify new small molecule inhibitors that are TR selective by disrupting the association of SRC2
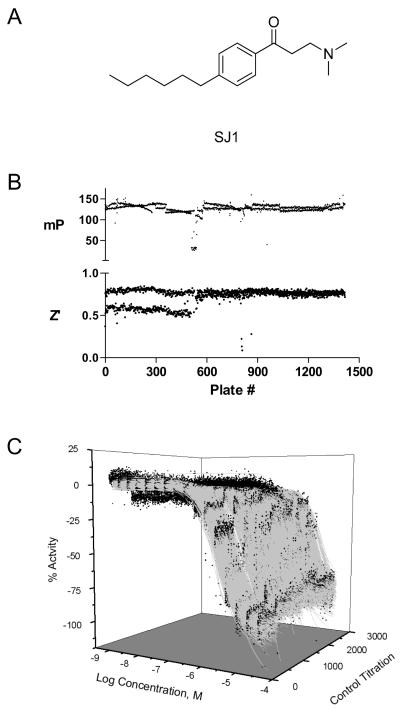
Figure 1. Performance of the quantitative high throughput screen.
A) The structure of the β-aminoketone control inhibitor, SJ1, is shown. B) mP values and Z’ factor of control wells are shown for each plate screened. The bifurcated Z’ scores of the first ~500 plates resulted from a software problem in one of the two EnVision detectors used in the screen. Following correction, the detector configuration was optimized and the Z’ scores aligned with those of the second detector. C) Concentration-response data (black circles) and curve fits (gray lines) of SJ1 titrations from each plate screened is shown. SJ1 was titrated as 16 two-fold titrations in duplicate beginning at 46 uM on each plate.
Methods
Protein Expression and Purification
hTRβ LBD (His6; residues T209-D461) was expressed in BL21(DE3) (Invitrogen) (10 × 1L culture) at 20 °C and 0.5 mM isopropyl-1-thio-b-D-galactopyranoside added at 0.6 Abs600 to induce protein expression. When the Abs600 reached 4, cells were harvested, resuspended in 20 ml of buffer/1 liter of culture (20 mM Tris, 300 mM NaCl, 0.025% Tween 20, 0.10 mM phenylmethylsulfonyl fluoride, 10 mg of lysozyme, pH 7.5), incubated for 30 min on ice, and then sonicated for 3 × 3 min on ice. The lysed cells were centrifuged at 100,000 × g for 1 h, and the supernatant was loaded onto Talon resin (20 ml, Clontech). Protein was eluted with 500 mM imidazole (3 × 5 ml) plus ligand (3,3′,5-triiodo-L-thyronine (Sigma)). Protein purity (>90%) was assessed by SDS-PAGE and high pressure size exclusion chromatography, and protein concentration was measured by the Bradford protein assay. The protein was dialyzed overnight against assay buffer (3 × 4 liters, 50 mM sodium phosphate, 150 mM NaCl, pH 7.2, 1 mM dithiothreitol, 1 mM EDTA, 0.01% Nonidet P-40, 10% glycerol).
Peptide synthesis and labeling
SRC2-2 peptide was synthesized and purified by reverse phase HPLC in the Hartwell Center (St. Jude Children’s Research Hospital). Texas Red- or fluorescein- maleimide (Molecular Probes) fluoroprobes were conjugated to the N-terminal cysteine of SRC2-2 peptide as described 10.
Cell toxicity assay
Human follicular thyroid carcinoma WRO cells (kindly provided by Dr. M. D. Ringel) were maintained in RPMI-1640 culture medium supplemented with 10% heat-inactivated FBS, 50 units/ml penicillin and 50 μg/ml streptomycin. Cells were harvested and suspended at 330,000 cells/mL in assay medium (50% DMEM, 50% F-12 medium without Phenol-Red (Cellgro #16-405-CV), L-glutamine, 10% heat inactivated and charcoal treated FBS, 100 μM non-essential amino acid, 50 units/ml penicillin, 50 μg/ml streptomycin). Approximately 10,000 cells were dispensed in 30 μL/well into 384-well plates (Corning 3917) and incubated 12 h at 37 °C and 5% CO2. Compounds and T3 (100 nM final concentration) were added using a pin tool and plates were incubated 16 h incubation at 37 °C and 5% CO2. Following incubation, plates were equilibrated at ambient temperature for 30 min, 20 ul of CellTiter-Glo (Promega) was added and luminescence measured using an EnVision (PerkinElmer) reader. The data were normalized to negative (DMSO) and positive (5 μM staurosporine) control wells on each plate. Compounds were tested in four independent runs and those displaying concentration response curves with 30% activity or greater (Class 1-3) in at least three runs were considered cytotoxic.
Quantitative high throughput screen
The screen was performed on an integrated robotic platform 20 using the protocol outlined in Table 1. In brief, 5 μL/well 0.6 uM TRβ and 20 nM SRC2-2 Texas Red in protein buffer (20mM Tris hydrochloride, pH 7.4, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 0.01% NP-40, 1 mM DTT, 1 μM T3 and 5% DMSO) was dispensed into black solid 1536-well plates (Greiner) using a solenoid-based dispenser. Following transfer of 23 nL compound or DMSO vehicle by a pin tool, the plates were centrifuged for 15 s at 1000 RPM and incubated for 5 h at ambient temperature. The plates were read using an EnVision (Perkin Elmer) reader to detect fluorescence polarization of SRC2-2 Texas Red (555 nm excitation and 632 nm emission). The screening collection included the following libraries with number of compounds in parentheses: Molecular Libraries Small Molecule Repository of diversity compounds (222,431), a NCGC diversity collection (44,000), natural product extracts from the University of Georgia and University of Michigan (9,243), diversity libraries from the Centers of Excellence in Chemical Methodology and Library Development at Boston University (2,401), Kansas University (1,033) and University of Pittsburgh (421), and bioactive compounds from Sigma Aldrich (3,533), Tocris (1,746), the MicroSource Spectrum collection (2,032) and BioMol (1,993). Chemical structures and qHTS data were deposited in PubChem (AID 1469, and 1479). While 296,587 samples were screened, curve-fits were performed on titration-response data for 292,732 samples because some assay wells failed quality control and were masked.
Table 1.
Protocol for the TRβ-SRC2-2 qHTS
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Reagent | 5 μL | 0.6 μM TRβ, 20 nM SRC2-2 |
| 2 | Controls | 23 nL | β-aminophenylketone inhibitor SJ1 |
| 3 | Library compounds | 23 nL | 92 μM to 2.9 nM dilution series |
| 4 | Centrifuge | 15 sec | 1000 RPM |
| 5 | Incubation time | 5 hr | Ambient temperature |
| 6 | Assay readout | FP polarization | EnVision |
Step Notes
Greiner 1536-well black, medium binding, solid bottom plates; 1 tip dispense to all columns except column 3 of protein buffer (20 mM Tris hydrochloride pH 7.4, 100 mM NaCl, 10% Glycerol, 1 mM EDTA, 0.01% NP-40, 1 mM DTT, 20 nM SRC2-2 Texas Red, 600 nM TRβ, 1 μM T3 and 5% DMSO). Column 3, 1 tip dispense of protein buffer without TRβ
Control compound plate Column 1, SJ1 titration starting at 46 uM, 16 points in duplicate 1:2 dilutions; Columns 2-4, DMSO
Pin tool transfer. The highest concentration library plates were pinned two times to achieve the highest tested concentrations. Most compounds were screened at 92, 46, 9.2, 1.8, 0.36, and 0.0029 μM final assay concentration.
Plates covered with stainless steel rubber gasket-lined lids containing pinholes for gas exchange.
Detector settings- 555 nm excitation, 632 nm emission, 75 flashes.
Follow up fluorescence polarization assays
For retest, the screening assay was performed as above using 20 nM SRC2-2 fluoroprobe labeled with either Texas Red or fluorescein. Samples were plated as 24 two-fold dilutions in duplicate beginning at 5 mM and 23 nL were transferred to the assay plate four times to achieve a top concentration of 92 μM.
For confirmation, 20 μL/well 0.6 uM TRβ and 20 nM SRC2-2 Texas Red or fluorescein fluoroprobe in protein buffer (20 mM Tris hydrochloride, pH 7.4, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 0.01% NP-40, 1 mM DTT, 1 μM T3 and 4% DMSO) was dispensed into black solid 384-well plates (Costar 3710) using a Biomek FX (Beckman Coulter) liquid handling system. Compounds were plated at 10 three-fold dilutions and 260 nL were transferred to the assay plate to achieve a top concentration of 130 μM. After incubation for 3 h at ambient temperature, the plates were read using an EnVision (Perkin Elmer) plate reader to detect fluorescence polarization of SRC2-2 Texas Red (555 nm excitation and 632 nm emission) or SRC2-2 Fluorescein (480 nm excitation and 535 nm emission). Five confirmation compounds were tested in the 1536-well plate format described above and yielded results similar to the 384-well plate format, indicating little or no difference in assay sensitivity for the two plate formats.
Data analysis and SAR derivation
The titration-response data was processed by normalizing mP and total fluorescence (TF) values to controls as follows: % Activity = ((Vcompound − Vpos)/(Vpos − Vneg)) × 100, where Vcompound denotes the compound well values, Vpos denotes the median value of the DMSO-treated control wells containing TRβ, and Vneg denotes the median values of the DMSO-treated control well without TRβ (free fluoroprobe). mP window was defined as the difference in mP values of the DMSO-treated control wells containing free and bound fluoroprobe. TF was calculated as follows: TF = S + 2P where S and P are fluorescence readings in the parallel and perpendicular channels, respectively. These normalized activity values were then corrected by applying a pattern correction algorithm to remove plate backgrounds such as dispense patterns and edge effects 26 using DMSO-only plates placed at 24-plate intervals in the screen as well as at the beginning and end.
Concentration-response curves were fit and classified as described 18. Four major curve classes (1-4) were created based on the completeness of curve, goodness of fit, and efficacy. The compounds with class 1.1, 1.2, and 2.1 curves are statistically the most reliable, while the compounds with class 2.2 and 3 curves are less reliable. Class 4 compounds are inactive, showing no concentration response. An in-house auto scaffold detection program was used to cluster 511 actives (curve classes 1.1, 1.2 and 2.1 as well as class 2.2 curves with >40% efficacy) yielding 730 structural series and 128 singletons. Each series contained at least three compounds of which at least one was active. Series were flagged for the following potential liabilities (see Table S2 for additional details): concentration dependent changes in total fluorescence in the FP assay or at 547 nm excitation and 618 emission 24, promiscuous aggregation (selectively active in the detergent free Cruzain assay, PubChem AID 2383), promiscuous redox activity (active in the caspase-1 or caspase-7 assay, PubChem AID 889 and 2389), low potency (> 20 uM) or low efficacy (< 50 %), significantly lower actives compared to mean actives in all series using a Fisher’s exact test (P < 0.05), and promiscuous activity in other assays (promiscuity score >0.5; Table S1). This process identified eight series and two singletons with no criteria liabilities.
Three-dimensional plots of concentration-response data and associated curve fits were made using OriginPro software. Screen and follow up data were deposited into PubChem (AID 1469, 1479, 1570-73)
Results
Quantitative high-throughput screen for TRβ-SRC2-2 inhibitors
Small molecules that prevent the interaction of TRβ with the steroid receptor coregulator 2 (SRC2) were detected using a quantitative high throughput screen (qHTS) of a previously described fluorescence polarization assay with a Texas Red labeled SRC2-2 peptide, corresponding to a 20 amino acid region of the nuclear receptor interaction domain 2. qHTS is a method where each compound is assayed at multiple concentrations to generate a titration-response curve for each tested sample 18. Small molecule inhibitors were identified by a decrease in fluorescence polarization.
For screening, the TR assay was scaled from 384-well to 1536-well format by reducing the assay volume by one quarter, from 20 μL to 5 μL per well. Assay performance was validated in this format using standard procedures 20. A collection of 292,732 small molecule samples was tested in the TR assay. The qHTS was conducted on an integrated robotic platform where the compounds were assayed at six concentrations, with over two million wells tested. The screen performed well; of the 1418 assay plates screened, 1380 (97%) plates passed quality control, giving a mean Z’ score of 0.73 and mean mP window of 128 (Figure 1B). The plates that failed quality control did so because of reagent dispense problems arising from mechanical problems. The affected rows or plates were masked to exclude this data. The control titration of the β-aminophenylketone inhibitor, SJ1 (Figure 1A, 8), present on each plate performed consistently, showing a mean IC50 of 8.2 ± 5.2 μM (Figure 1C).
Identification of TR inhibitors
Concentration response curves (CRC) were fit for the fluorescence polarization (FP, mP values) and total fluorescence (TF) data sets (Figure 2). As previously described, the CRCs were categorized into four general classes 18. Class 1 are curve fits having both upper and lower asymptotes and >0.9 r2. Class 2 are incomplete curve fits having only one asymptote and >0.9 r2. Class 3 are low confidence curve fits with <0.9 r2 or have activity at a single concentration. Class 1 and 2 curves are divided further into subclasses to indicate efficacies 80% or greater (Class 1.1 and 2.1) or between 30% and 80% (Class 1.2 and 2.2). Class 4 compounds are inactive having either no curve fit or an efficacy below threshold activity (three SD of the mean activity).
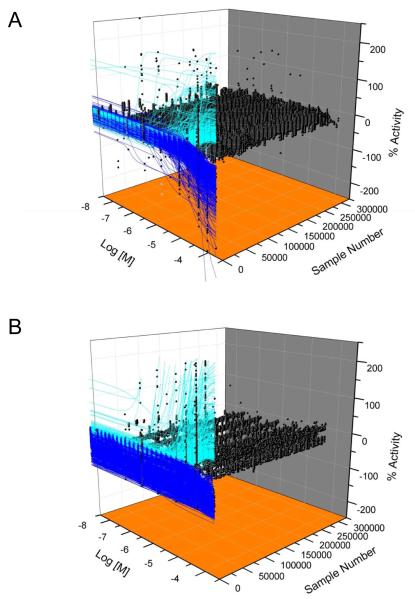
Figure 2. Compound activity of the quantitative high throughput screen.
A and B) Shown are concentration-response data (black circles) and curve fits of compounds with decreasing (blue lines) or increasing (cyan) FP (A) or TF (B) activity. Inactive compounds (black circles only) do not have curve fits.
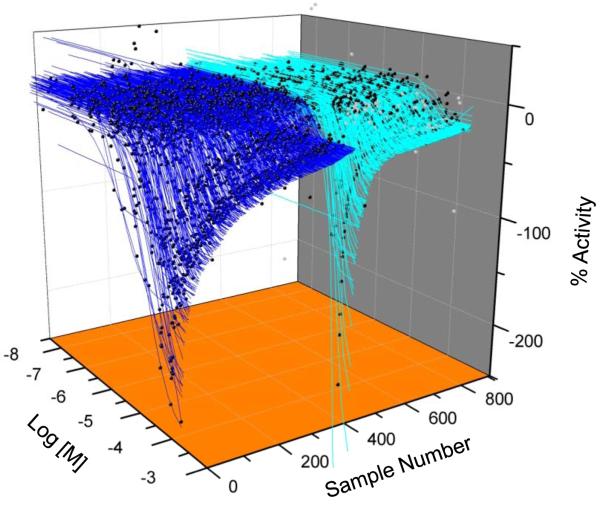
The FP qHTS data revealed 1222 compounds with activity (Classes 1-3), of which 312 were activators and 910 were inhibitors (Figure 2A, Table 2). Compounds showing concentration-dependent increases in mP values were not pursued further because their activity likely arose from compound interference, such as light scattering 22; 27. Of the 910 FP inhibitors, 511 were scored as active (Class 1.1, 1.2, 2.1 and Class 2.2 with >40% efficacy; Table 2) and 399 as inconclusive (Class 2.2 with <40% efficacy and Class 3; Figure 3).
Table 2.
Activity profile of TRβ-SRC2 qHTS
| Curve Class | |||||||
|---|---|---|---|---|---|---|---|
| 1.1 | 1.2 | 2.1 | 2.2 | 3 | 4 | ||
| FP | Inhibitor # % library |
48 0.02% |
159 0.05% |
65 0.02% |
332 0.11% |
306 0.10% |
291,510 99.58% |
|
|
|||||||
| Activator # % library |
21 0.01% |
39 0.01% |
23 0.01% |
114
|
115 0.04% |
||
|
| |||||||
| TF | Inhibitor # % library |
120 0.04% |
1,006 0.34% |
127 0.04% |
571 0.20% |
2,119 0.72% |
288,323 98.49% |
|
|
|||||||
| Activator # % library |
34 0.01% |
13 0.00% |
125 0.04% |
159 0.05% |
135 0.05% |
||
Figure 3. Concentration-response profile of TR qHTS actives.
Concentration-response data (black circles) and curve fits of the FP inhibitors scored as active (blue lines) or inconclusive (cyan).
The TF qHTS data were used to counterscreen the FP inhibitors as changes in TF are useful for identifying artifacts arising from compound interferences such as fluorescence, aggregation, or light scattering 22. From the entire screen, 466 samples showed concentration dependent increases in TF and 3,943 samples displayed decreases (Table 2, Figure 2B). This total of 4,409 samples comprised 1.5% of the screening collection. Of the 511 FP actives, 212 (41%) showed concentration dependent changes in TF (data not shown) suggesting the activity of these FP inhibitors arose from compound interference rather than inhibition of TRβ-SRC association. Over 80% of these interfering compounds showed increases of TF (data not shown).
The FP inhibitors showed a wide range of potencies. Examination of all 511 FP actives indicated 17 (4%) samples with IC50 <1 μM and 72 (14%) samples having IC50 in the range of 1-10 μM (Table 3). Most of these actives displayed Class 1 curves, indicating a saturated response. The majority of samples, 409 (80%), had IC50 values in the range between 10 and 100 μM. Almost 70% of these actives showed Class 2 curves, which are incomplete and have only an upper asymptote.
Table 3.
Potency profile of TRβ-SRC2 FP inhibitors
| FP inhibitors Curve Class |
Noninterference FP inhibitors Curve Class |
|||||||
|---|---|---|---|---|---|---|---|---|
| μM | 1.1 | 1.2 | 2.1 | 2.2 | 1.1 | 1.2 | 2.1 | 2.2 |
| <0.1 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | 0 |
| 0.1 - 1 | 2 | 11 | 0 | 0 | 0 | 5 | 0 | 0 |
| 1 - 10 | 14 | 52 | 0 | 6 | 0 | 49 | 0 | 1 |
| 10-100 | 32 | 92 | 63 | 222 | 1 | 79 | 2 | 152 |
| >100 | 0 | 0 | 2 | 11 | 0 | 0 | 1 | 6 |
|
|
||||||||
| Subtotal | 48 | 159 | 65 | 239 | 1 | 136 | 3 | 159 |
| Total | 511 | 299 | ||||||
|
|
||||||||
After removing the 212 interfering compounds that showed TF activity, 299 FP inhibitors remained (Table 3). Of the 299 inhibitors, 266 were recovered from the Molecular Libraries Small Molecule Repository, 33 from internal diversity and bioactive libraries and none from the natural product extract collections. Almost all of these inhibitors showed Class 1.2 and 2.2 curves, indicating efficacies below 80%. However, the general distribution of potencies of this subset was similar to the total set of FP inhibitors, where 8 (3%) samples displayed IC50 <1 μM, 50 (17%) with IC50 in the range of 1-10 μM IC50, and 234 (78%) with IC50 in the range of 10-100 μM. Clearly compound optical properties, as measured by TF activity, resulted in high efficacy FP false positives over a broad potency range. This is commonly observed in FP assays 25.
Identification of TR inhibitor series
To find structurally related compounds, the 511 validated inhibitors were analyzed using a fragment-based approach to yield 730 scaffolds and 128 singletons. Compounds containing a common scaffold formed a series. Each active could be part of more than one series depending on which part of the compound was used as a scaffold. Thus, the number of series was larger than the number of actives clustered. After the series of interest were defined, structurally related compounds that gave either inconclusive activity or no activity were added to each series, thus providing lists containing three or more compounds of which at least one was active. Each series and singleton was flagged for the following potential liabilities with the number of series indicated in parentheses: concentration-dependent changes in TF or solution fluorescence at 547 nm excitation and 618 emission (363), promiscuous aggregation (43), promiscuous redox activity (136), IC50 value greater than 20 μM or efficacy less than 50% (299), underrepresentation of actives compared to the library average (67), and promiscuous activity in other assays (509). Series and singletons flagged for potential artifacts (change of total fluorescence intensity, redox interference, and aggregation) or low potency (>20 μM IC50) were excluded from the follow-up study.
The remaining series were clustered further such that series with similar scaffolds were merged resulting in 42 series and 23 singletons. From this list, compounds were chosen for retest using the following method: all singletons were chosen and for each series, 2 actives and 1 inactive were chosen, when available. This process yielded a set of 103 available compounds containing 92 actives and 11 inactives.
The retest samples were characterized in two TR FP assays, one using a SRC2-2 fluoroprobe labeled with Texas Red and the other using the same probe labeled with fluorescein, as well as a cell viability assay. The samples were tested as concentration-response experiments using a range of 24 two-fold dilutions in duplicate beginning at 92 μM. Samples that displayed Class 1-3 curve-fits were scored as active. Compounds having efficacy below 40% were considered active, but the IC50 values were not considered reliable and therefore not used for subsequent work. The Texas Red FP assay identified as active 75 (83%) of 92 qHTS positives as well as 7 of 11 qHTS inactives (Table S2). The orthogonal fluorescein FP assay was used to confirm activity and eliminate compounds that interacted with the Texas Red labeled fluoroprobe. The fluorescein assay identified as active, 58 of 92 qHTS positives as well as 4 of 11 qHTS inactives. A viability assay using WRO cells showed eight compounds were toxic. Merging these results to select the compounds with a high probability of true activity indicated 38 compounds (19 series and 4 singletons) were active with 40% or greater efficacy in both FP assays, free of compound interference, and not toxic in cells (Table S2).
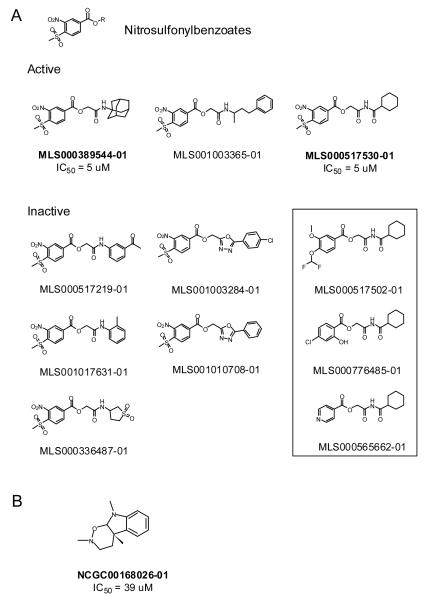
To prioritize compounds for subsequent work, the 38 retest actives were examined to eliminate compounds showing undesirable chemical or biological properties, such as low potency in one or both FP assays. In addition, one series (#38, Table S2) was excluded because of structural similarity to the β-aminoketone scaffold identified previously as a TRβ-coactivator inhibitor 2. This process identified four structurally distinct series and two singletons for further characterization. Independent samples of these actives as well as one analog were tested in the two FP assays to confirm activity (Table S2). One series containing a methylsulfonylnitrobenzoate (MSNB) core (#1) and one singleton (#S16), the plant alkaloid geneseroline 35, confirmed as active in both FP assays (Figure 4, Table S2). Two members of the MSNB series showed about 5 μM IC50 in both FP assays while geneseroline was weakly active with an IC50 above 30 μM in both assays (Table S2). The MSNB series was judged a validated screening hit and subsequent studies have shown this series is a specific and irreversible antagonist that inhibits a TR-mediated gene reporter in cells17.
Figure 4. Structures of confirmed TR actives and associated analogs from the qHTS.
A) The structures of active and inactive MSNB analogs identified from the qHTS are shown. The compounds within the box are inactive 2-(cyclohexanecarboxamido)-2-oxoethyl benzoate compounds structurally related to the MSNB series but lack the nitrosulfonylphenyl group. B) The structure of the geneseroline is shown. Compound names in bold indicate independent samples that were tested in confirmation studies.
Discussion
In this study, we have performed a qHTS to identify compounds that inhibit the interaction between TRβ and SRC2-2. The titration-response screen of over 290,000 samples enabled us to determine the activity of each compound as well as the potency, efficacy, and other pharmacological measures of the actives. Using only the primary qHTS data, actives were prioritized, interfering compounds were identified, and nascent structure-activity relationships were derived. This analysis allowed the selection of a small number of compounds for retest and the inclusion of analogs and inactives for series of interest. Confirmation studies of select actives with sufficient potency and chemical tractability identified a MSNB containing series as a new class of thyroid hormone receptor-coactivator antagonists.
While the goal of this qHTS was to recover compounds that decreased FP via fluoroprobe displacement, the screen recovered 312 compounds that showed FP increases. The TF dataset indicated that 41% of these samples showed concentration dependent decreases in TF and an additional 28% showed increases (data not shown). These increases in FP and changes in TF may reflect compound effects on light scattering 22; 27 or the emission and polarization of the fluoroprobe. In the latter case, compounds may cause the fluoroprobe to aggregate resulting in both increased fluorescence and anisotropy. A less likely effect is compound autofluorescence because spectroscopic profiling of over 70,000 library samples shows that fewer than ten samples fluoresce in the far red excitation and emission spectra used in this assay 24. There were a minority of compounds that showed increases in both FP and TF. Of the 312 compounds with increased FP and 466 compounds with increased TF, only 87 compounds showed increases in both.
Our assay used fluorescence polarization to detect the interaction of the TRβ LBD with a SRC2-2 fluoroprobe. However, other assay formats, like TR-FRET and AlphaScreen™, have been used to detect other NHR-coactivator interactions 11; 13; 31. While all of these methodologies have been implemented to identify validated small molecules inhibitors, each has advantages and liabilities (reviewed in 30). In general, FP formats are simpler, requiring the labeling of only one probe, which can result in lower assay costs and greater ease of implementation. However, FP assays typically require micromolar concentrations of target protein, resulting in lower sensitivity, and are susceptible to a variety of compound interferences 22. Methods based upon TR FRET determine a ratio of two fluorescent emissions and are less susceptible to compound interferences, though artifacts do occur 23. Antibody-based TR-FRET formats use nanomolar concentrations of target 11; 13, permitting greater sensitivity in detecting potent inhibitors. However, these formats are more complex where probe reagents may be difficult to label or expensive to generate and may make large-scale screening cost prohibitive30.
The TRβ-SRC2-2 qHTS was very selective in identifying actives. Only 299 FP inhibitors (0.1 % of screening collection) were recovered from the qHTS that were free of compound interference, as judged by total fluorescence activity. This percentage is similar to an earlier screen using this assay where 0.02% actives were identified by assaying compounds at a single concentration of 30 μM 2. Our five-fold higher recovery of actives reported here is due likely to the titration-based screening method used 18 combined with a higher starting concentration of 92 μM for most of the screened compounds. The low yield of TRβ-SRC2-2 inhibitors may be attributed to several factors: the difficulty in disrupting protein-protein interactions with small molecules 9, the relatively high concentration of target protein (0.6 μM), and the shallow pocket of TRβ that interacts with SRC2 8. Indeed, many of the compounds that retested as positive in both FP assays, had IC50 values greater than 20 μM (Table S2) indicating that most compounds weakly disrupted the interaction between TRβ and SRC2-2.
However, the qHTS identified one novel series that inhibited TRβ and SRC2-2 association with an IC50 value of 5 μM. This MSNB series represents a new chemotype inhibitor of this interaction. Eight MSNB analogs were tested in the qHTS, of which three were active. Because the qHTS assayed each compound at six concentrations, the determined potencies and efficacies allowed us to derive nascent structure-activity relationship for this series. All MSNB compounds containing either a hydrophobic imide (MLS000517530-01) or amide (MLS000389544-01 and MLS001003365-01) were active while those containing an aniline (MLS000517219-01 and MLS001017631-01), β-aminosulphone (MLS000336487-01), or oxadiazole (MLS001010708-01 and MLS001003284-01) were inactive (Figure 4A). These results suggest that a hydrophobic group is needed at this position for activity. Replacement of the MSNB group in MLS000517530-01 with other aromatic groups (MLS000517502-01, MLS000776485-01, and MLS000565662-01) also resulted in loss of the activity, implying that this group is essential. Further studies using a broader range of analogs will be needed to confirm these hypotheses.
The MSNB series is structurally distinct from the previously described β-aminoketones 2 and therefore represents a novel TR-SRC2 inhibitor scaffold. The β-aminoketone series of inhibitors has been successfully modified to improve potency, reduce cytotoxicity, and improve physiochemical determinants of bioavailability 3; 16. However, this class has an inherent cardiotoxicity due to the interaction of the requisite aminoketone moiety with cardiac ion channels. While optimization has lessened the impact of this issue, the liability has not been eliminated. The new MSNB series does not possess chemical moieties expected to cause this liability and thus presents advantages over the earlier series. Structural studies indicate that β-aminoketones series members alkylate cysteine 298 in the AF-2 pocket of TRβ, which normally forms the interface for binding coactivators 8. Initial characterization of the MSNB series indicates it binds irreversibly to the TRβ LBD and may inhibit in a similar manner 17. Future studies will elucidate the mechanism of MSNB inhibition more fully.
Supplementary Material
Acknowledgements
We thank Sam Michael for automation assistance, Paul Shinn and Danielle Van Leer for compound management, Bill Leister, Chris LeClair and Jeremy Smith for analytical chemistry, Rajarshi Guha for informatics, and Ryan MacArthur for preparation of three dimensional plots.
Funding acknowledgements This work was supported by the National Institutes of Health (grant number DK58080); the NIH Roadmap for Medical Research; the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health; the American Lebanese Syrian Associated Charities; and St. Jude Children’s Research Hospital.
References
- 1.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 2.Arnold LA, Estebanez-Perpina E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. Discovery of small molecule inhibitors of the interaction of the thyroid hormone receptor with transcriptional coregulators. J Biol Chem. 2005;280:43048–43055. doi: 10.1074/jbc.M506693200. [DOI] [PubMed] [Google Scholar]
- 3.Arnold LA, Kosinski A, Estebanez-Perpina E, Fletterick RJ, Guy RK. Inhibitors of the interaction of a thyroid hormone receptor and coactivators: preliminary structure-activity relationships. J Med Chem. 2007;50:5269–5280. doi: 10.1021/jm070556y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 8.Estebanez-Perpina E, Arnold LA, Jouravel N, Togashi M, Blethrow J, Mar E, Nguyen P, Phillips KJ, Baxter JD, Webb P, Guy RK, Fletterick RJ. Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor. Mol Endocrinol. 2007;21:2919–2928. doi: 10.1210/me.2007-0174. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher S, Hamilton AD. Protein-protein interaction inhibitors: small molecules from screening techniques. Curr Top Med Chem. 2007;7:922–927. doi: 10.2174/156802607780906735. [DOI] [PubMed] [Google Scholar]
- 10.Geistlinger TR, Guy RK. Steroid receptor coactivator peptidomimetics. Methods Enzymol. 2003;364:223–246. doi: 10.1016/s0076-6879(03)64013-9. [DOI] [PubMed] [Google Scholar]
- 11.Gunther JR, Du Y, Rhoden E, Lewis I, Revennaugh B, Moore TW, Kim SH, Dingledine R, Fu H, Katzenellenbogen JA. A set of time-resolved fluorescence resonance energy transfer assays for the discovery of inhibitors of estrogen receptor-coactivator binding. J Biomol Screen. 2009;14:181–193. doi: 10.1177/1087057108329349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 13.Hilal T, Puetter V, Otto C, Parczyk K, Bader B. A dual estrogen receptor TR-FRET assay for simultaneous measurement of steroid site binding and coactivator recruitment. J Biomol Screen. 15:268–278. doi: 10.1177/1087057109359196. [DOI] [PubMed] [Google Scholar]
- 14.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 15.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang JY, Arnold LA, Zhu F, Kosinski A, Mangano TJ, Setola V, Roth BL, Guy RK. Improvement of pharmacological properties of irreversible thyroid receptor coactivator binding inhibitors. J Med Chem. 2009;52:3892–3901. doi: 10.1021/jm9002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang JY, Huang W, Arnold LA, Huang R, Attia RR, Connelly M, Wichterman J, Zhu F, Augustinaite I, Austin CP, Inglese J, Johnson RL, Guy RK. Methylsulfonylnitrobenzoates, a new class of irreversible inhibitors of the interaction of the thyroid hormone receptor and its obligate coactivators that functionally antagonizes thyroid hormone. J Biol Chem. 2011;286:11895–11908. doi: 10.1074/jbc.M110.200436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 20.Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, Simeonov A. A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onate SA, Tsai SY, Tsai MJ, O′Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 22.Owicki JC. Fluorescence polarization and anisotropy in high throughput screening: perspectives and primer. J Biomol Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 23.Shukla SJ, Nguyen DT, Macarthur R, Simeonov A, Frazee WJ, Hallis TM, Marks BD, Singh U, Eliason HC, Printen J, Austin CP, Inglese J, Auld DS. Identification of pregnane X receptor ligands using time-resolved fluorescence resonance energy transfer and quantitative high-throughput screening. Assay Drug Dev Technol. 2009;7:143–169. doi: 10.1089/adt.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 25.Simeonov A, Yasgar A, Jadhav A, Lokesh GL, Klumpp C, Michael S, Austin CP, Natarajan A, Inglese J. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem. 2008;375:60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southall NT, Jadhav A, Huang R, Nguyen T, Wang Y. Enabling the large scale analysis of quantitative high throughput screening data. In: Seethala R, Zhang L, editors. Handbook of Drug Screening. Taylor and Francis; New York: 2009. pp. 442–462. [Google Scholar]
- 27.Turconi S, Shea K, Ashman S, Fantom K, Earnshaw DL, Bingham RP, Haupts UM, Brown MJ, Pope AJ. Real experiences of uHTS: a prototypic 1536-well fluorescence anisotropy-based uHTS screen and application of well-level quality control procedures. J Biomol Screen. 2001;6:275–290. doi: 10.1177/108705710100600502. [DOI] [PubMed] [Google Scholar]
- 28.Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 29.Williams GR. Cloning and characterization of two novel thyroid hormone receptor beta isoforms. Mol Cell Biol. 2000;20:8329–8342. doi: 10.1128/mcb.20.22.8329-8342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woelcke J, Hassiepen U. Fluorescence-based biochemical protease assay formats. In: Chen T, editor. A practical guide to assay development and high-throughput screening in drug discovery. Taylor & Francis Group, LLC; Boca Raton: 2010. pp. 25–48. [Google Scholar]
- 31.Wu X, Sills MA, Zhang JH. Further comparison of primary hit identification by different assay technologies and effects of assay measurement variability. J Biomol Screen. 2005;10:581–589. doi: 10.1177/1087057105275628. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Wu RC, O′Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen PM. Physiological and molecular basis of thyroid hormone action. Physiol Rev. 2001;81:1097–1142. doi: 10.1152/physrev.2001.81.3.1097. [DOI] [PubMed] [Google Scholar]
- 35.Yu QS, Yeh HJ, Brossi A, Flippen-Anderson JL. Geneserine and geneseroline revisited: acid-base catalyzed equilibria of hexahydropyrrolo-[2,3-b]-indole N-oxides with hexahydro-1,2-oxazino-[5,6-b]-indoles. J Nat Prod. 1989;52:332–336. doi: 10.1021/np50062a019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.