Abstract
Apart from the ribosome, the crystal structure of the bacterial RNase P in complex with a tRNA, reported by Reiter and colleagues recently, constitutes the first example of a multiple turnover RNA enzyme. Except in rare exceptions, RNase P is ubiquitous and, like the ribosome, is older than the initial branch point of the phylogenetic tree. Importantly, the structure shows how the RNA and the protein moieties cooperate to process the pre-tRNA substrates. The catalytic site comprises some critical RNA residues spread over the secondary structure but gathered in a compact volume next to the protein, which helps recognize and orient the substrate. The discussion here outlines some important aspects of that crystal structure, some of which could apply to RNA molecules in general.
Keywords: RNase P, multiple-turnover ribozyme, RNA–RNA interactions, molecular evolution, crystallography
Despite the fact that it is one of the earliest known ribozymes, >30 yr have been necessary to solve the structure of the bacterial RNase P (Reiter et al. 2010). This latency period has mostly been due to bottlenecks inherent to crystallography, such as the identification of the RNA constructs capable of forming well-diffracting crystals and the phasing of the data. Despite overcoming these difficulties, the crystals from which the structure of the bacterial RNase P was extracted were still diffracting at a fairly low resolution and, moreover, in an anisotropic manner. Historically, RNase P has been the first RNA catalyst characterized as a multiple turnover ribozyme (Guerrier-Takada and Altman 1984), many years before the ribosome (Nissen et al. 2000). By comparison to group I ribozyme structures, the RNase P structure sheds light on the characteristics of a true RNA enzyme in terms of substrate binding, discrimination between the substrate and the product—by a protein “cofactor,” and organization of the catalytic site. The structure supports a view of the holoenzyme that is finally very close to what was deduced qualitatively from the accumulated biochemical and theoretical data that led to molecular models published >5 yr ago (Tsai et al. 2003; Buck et al. 2005). Yet, the fairly low resolution of the structure together with the fact that the tRNA product is actually bound to the holoenzyme instead of the pre-tRNA substrate still leaves open some questions related to the precise molecular interactions taking place between the components of the system and related to the mechanism and the dynamics of the catalysis.
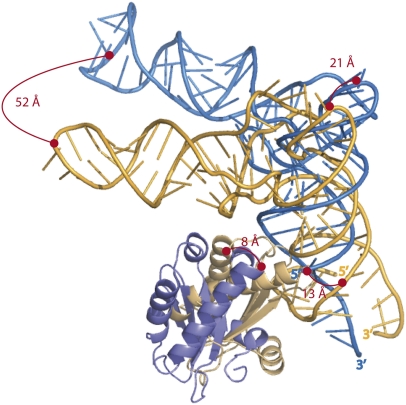
A striking feature of the structure is that the individual components do not undergo significant conformational changes upon formation of the holoenzyme, during pre-tRNA substrate or tRNA product release. This constitutes one of the main reasons why modeling the holoenzyme in the presence of the substrate was indeed possible. A superimposition of the RNase P RNAs from the crystal structure and from the model of the holoenzyme results in meaningful superimpositions between the tRNAs and the RNase P proteins (Fig. 1) and, importantly, with an almost correct orientation. The mode of recognition between the components of the bacterial RNase P seems to be quite ancient. All bacterial RNase Ps have the same overall organization with only minor differences within RNA peripheral domains. Moreover, there is only one major evolutionary step that led to the appearance of the Bacillus subtilis type B RNA from the ancestral type A RNA represented by the Thermotoga maritima or Escherichia coli enzymes. The type B has lost the P6 pseudoknot that has been replaced by a loop–loop interaction. A recent model of RNA evolution based on neutral sequence drift and alternative base-pairing leading to formation of transient specific structural elements like pseudoknots provides clues to explain how type B RNase P may have emerged from type A (Beckert et al. 2008). Thermotogales are located deeply in the phylogenetic tree and represent one of the oldest bacterial life forms, with very slow evolutionary dynamics compared with that of modern phyla (Woese 1987). It sounds reasonable that the common ancestor of all life forms may have processed pre-tRNAs using a similarly organized ribozyme. This idea is moreover supported by the observation that the ancestral ribosome was already relying on tRNAs (Cech 2000) and that the RNase P RNAs from all kingdoms are strongly related in structure, arguing for the appearance of the RNA moiety from RNase P well before the three kingdoms of life have diverged (Evans et al. 2006). Intriguingly, this view is complicated by the identification of an RNase P fully built from proteins that have evolved in the cellular lineage that gave rise to the animal mitochondria (Holzmann et al. 2008). This tRNA processing ability has seemingly appeared as an additional function for proteins already in charge of RNA-related functions such as RNA recognition or their chemical modification and should be seen as a genuine evolutionary exploration.
FIGURE 1.
Relative positions and orientations of the tRNA and of the RNase P protein resulting from the superimposition of the RNase P RNA from the crystal structure (blue) (Reiter et al. 2010) and the three-dimensional ab initio model (gold) (Tsai et al. 2003). The normalized root mean square deviation (nrmsd) by means of superimposition of phosphorus atoms from the RNase P RNA is 11.6 Å. Distances between elements from the tRNA and the protein are indicated on the picture. The large distance between the anticodon loops from the tRNAs results from a 35° rotation between the acceptor stems. The protein moieties have a 39° differential orientation and an offset of 16 Å (calculated with lsqman) (Kleywegt and Jones 1994).
Up to recently, crystal structures of tRNA-containing complexes were mainly represented by synthetases (Giegé et al. 2008) and tRNA modifying enzymes (Ishitani et al. 2008) where the partner of the tRNA substrate is made only of protein(s). Interactions between tRNAs and other RNAs could only be observed for the first time in the ribosome structures (Demeshkina et al. 2010)—except through symmetry-related interactions in crystal structures of tRNA, e.g., tRNAAsp (Westhof et al. 1985). Strikingly, the present crystal structure of the bacterial RNase P (Reiter et al. 2010) shows how the tRNA is actually recognized by means of the CCA 3′ end and the T loop, which are determinants also used by the 50S subunit. In more details, the tRNA that cocrystallized with the RNase P holoenzyme is recognized by means of a limited number of Watson-Crick base pairs taking place between the CCA 3′ end of the tRNA and the L15 loop of the P RNA (Fig. 2). However, as usually observed, such interactions are not sufficient to constrain the tRNA in the proper conformation. Recognition between the tRNA and the P RNA takes place through interactions involving non–Watson-Crick pairs typical of RNA folding (Fig. 2A). The specificity domain of the P RNA clamps the D and T loops of the substrate by means of stacking and A-minor interactions and regions of the backbone of the P RNA dock within the deep or shallow grooves of the tRNA. Interestingly, the molecular complex is experimentally formed by mixing and heating to 95°C the two RNAs and the protein together followed by snap cooling to 4°C. This is actually a very classical way of renaturing RNA molecules. The success of this very simple strategy could indicate that the tRNA actually helps stabilize the folding of the P RNA and points to a very close evolutionary relationship between those components. These features place the RNase P at a level different from single turnover ribozymes that, in general, recognize their substrate through extensive base-pairing interactions. Indeed, guide sequences constitute a very common mechanism of recognition between RNAs (see group I and II, ribosome A-site, and P-site on the 23S). Additionally, in many bacteria, unlike in E. coli, the CCA 3′ end is not directly encoded at the gene level (Shi et al. 1998). Rather, it is added by the CCA adding enzyme tRNA nucleotidyl transferase. Experimental data show that the bacterial protein actually helps processing substrates that do not bear a CCA 3′ end (Guerrier-Takada et al. 1984), and suggest that contacts between the 5′ leader of the substrate and the protein could compensate for the absence of the CCA 3′ end. Accordingly, it is seemingly not clear which enzyme could intervene before the other in vivo. Obtaining a crystal structure of a complex formed between the holoenzyme and a noncleavable pre-tRNA substrate lacking the CCA 3′ end would be in this respect very interesting.
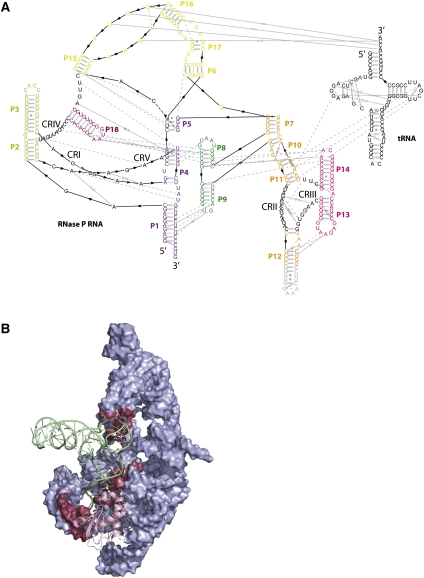
FIGURE 2.
(A) Interactions within the RNase P RNA and between the RNase P RNA and the tRNA represented on an expended secondary structure diagram, permitting a better view of the tertiary interactions (gray symbols). Colors are identical to the schemes described by Reiter et al. (2010). Conserved regions (CRs) are numbered (from I–V), and nucleotides are black type set. (B) Crystal structure of the complex highlighting residues (raspberry surface) from the RNase P RNA (blue surface) that interact with the tRNA (green ribbon). The protein is represented as a pink ribbon.
The crystal structure of the RNase P also exemplifies once again the predominant role of RNA engineering over the diversity of crystallization conditions in order to favor packing interactions promoting crystal growth. Crystals could be obtained only by creating artificial tertiary interactions based on the use of the tetraloop/tetraloop receptor motif that was proposed a decade ago as a general module for RNA crystallization (Ferré-D'Amaré et al. 1998). The RNase P crystal structure indeed relies on this tertiary structure motif as well as the structure of the U4 snRNP (Leung et al. 2010, 2011). In the case of RNase P, the designed interaction took place only in rare cases when two tRNAs with tetraloop receptor-containing anticodon stems of different length were combined with four distinct forms of P RNAs presenting P12 stems with varying length moreover capped by a GAAA tetraloop. Among those four combinations, one yielded crystals suitable for diffraction studies. An overall of 42 combinations involving six different tRNAs and nine different P RNAs were tested to obtain these results.
Nonetheless, the critical interaction resulting from the engineered motifs between P12 of the P RNA and the tRNA anticodon stem is outnumbered by the unpredicted and thus random packing interactions. The 3′ dangling ends of two P RNA base pair to form a 4-bp helix, and the tip of P18 interacts with the bulge separating P16 from P17 from a symmetry-related complex. The L3 loop from the P RNA contacts the D-loop from a neighboring tRNA, and the A residue from the engineered tRNA anti-codon loop contacts a distant P RNA at the kink between L15 and P16. Noteworthy, the latter contact was not expected, although it ensues from the engineering of the tRNA. Three symmetry-related molecules are brought in contact by a single tRNA by means of this unexpected interaction. The resulting trimer of trimers may constitute an initial crystalline nucleus, allowing for calling roll of the other ones up to completion of the unit cell and explaining why crystals can hardly be obtained without them. The protein moiety does not account for any packing interaction but is a pure P RNA binder in spite of its size, which is comparable to the U1A RNA binding domain that has been used many times to promote crystal packing interactions in RNA crystals (Ferre-D'Amare 2010).
One of the most important features is that the structure puts together for the first time all the components responsible for the catalysis. As underlined by Reiter et al. (2010), the 5′ end of the tRNA points out the residues of the P RNA that should carry the catalytic activity and thus the region that should contain the magnesium ions involved in catalysis. However, the fairly low resolution of the structure prevents the deduction of a firm catalytic mechanism, although it suffices to suggest a working hypothesis. The structure reveals the presence of heavy-atom derivatives (europium and samarium derivatives) in the vicinity of the 5′ end of the tRNA. These ionic sites may constitute an indirect observation of magnesium ions but cannot be confirmed at a resolution of 3.8 Å. Magnesium ions are as light as water molecules and can be identified with confidence at a resolution around 1.5 Å, which permits them to be distinguished with certainty from water molecules by analysis of their octahedral coordination geometry (Auffinger et al. 2003). With respect to other ribozymes, the RNase P context is quite original in the sense where (1) the nucleophile is a water molecule instead of a ribose hydroxyl group and (2) the cleavage products consist in two strands bearing a 5′ phosphate and a 3′ hydroxyl. To be activated presumably by proton abstraction, the nucleophilic water molecule should be presented to a specific chemical group with acidic properties. This water molecule may belong to the coordination inner sphere of a magnesium ion in the vicinity of the 5′ end of the tRNA observed in the structure. An outer-sphere RNA ligand that is capable of abstracting one of the protons could play the role of the acid. Yet the crystal structure does not suggest any candidate that could play this role. Whether the proton stabilizing the leaving group is provided by a water molecule or a nucleotide also stands as an important question. The crystal structure attempts to answer this question by suggesting that a water molecule bound to a second ion interacting with specific P4 residues could play the role of the acidic group stabilizing the tRNA product. Unfortunately, this proposal remains a working hypothesis since the potential ligands of the putative ions are not observable.
The fact that specific acid/base groups belonging to the RNA or to its ligands still need to be identified leaves open the possibility of drawing a parallel with the situation occurring in self-cleaving ribozymes. Self-cleaving ribozymes like the hairpin or the VS ribozymes need magnesium to acquire their native structure but can be active in a lesser extent under high monovalent salt concentration. This is due to the fact that their catalytic cycle relies purely on the acid/base properties of some nucleotides that could be identified by mutational analysis (Wilson and Lilley 2011). On the contrary, large ribozymes have co-evolved with magnesium and maybe other metal ions to participate in catalytic mechanisms (Steitz and Steitz 1993). In the case of RNase P, the magnesium ion may also allow placing the nucleophilic water molecule at the ideal place to promote the appearance of the right cleavage products. To promote the right in-line attack, the activated water molecule should attack the phosphorus atom of the scissile bond from the side opposite to the O3′ atom of the upstream residue. Noteworthy, among the two ions observed in the structure, the one supposedly bound to the nucleophilic water lies at a place compatible with this constraint, while the second one lies at a distance that would allow protonation of the leaving group via a ligand that has not been clearly identified yet. The architectural role of magnesium ions is thus not restricted to promoting RNA folding and stabilizing RNA structure but expands to anchoring the nucleophilic water molecule that is critical for RNase P activity to ensure its proper activation in order to obtain the right cleavage products (a 3′ hydroxyl and a 5′ phosphate tRNA).
The necessary role of magnesium for the folding of large RNAs may have resulted in enrolling them in active participation to catalysis by helping anchoring the chemical groups involved in the transesterification reactions that lead to the various products resulting from group I or group II introns, or RNase P catalysis. However, activation of reactive species or neutralization of leaving groups seem to rely on acid/base catalyzed processes. Identification of the precise catalytic mechanism of each ribozyme family may lead to deeper understanding on how ribozymes achieve the diversity of products by orchestrating variations around the transesterification reaction.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2841511.
REFERENCES
- Auffinger P, Bielecki L, Westhof E 2003. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem Biol 10: 551–561 [DOI] [PubMed] [Google Scholar]
- Beckert B, Nielsen H, Einvik C, Johansen SD, Westhof E, Masquida B 2008. Molecular modelling of the GIR1 branching ribozyme gives new insight into evolution of structurally related ribozymes. EMBO J 27: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Kazantsev AV, Dalby AB, Pace NR 2005. Structural perspective on the activation of RNase P RNA by protein. Nat Struct Mol Biol 12: 958–964 [DOI] [PubMed] [Google Scholar]
- Cech TR 2000. Structural biology. The ribosome is a ribozyme. Science 289: 878–879 [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Yusupova G, Yusupov M 2010. Interactions of the ribosome with mRNA and tRNA. Curr Opin Struct Biol 20: 325–332 [DOI] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR 2006. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341 [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare AR 2010. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods 52: 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Zhou KH, Doudna JA 1998. A general module for RNA crystallization. J Mol Biol 279: 621–631 [DOI] [PubMed] [Google Scholar]
- Giegé R, Touzé E, Lorber B, Théobald-Dietrich A, Sauter C 2008. Crystallogenesis trends of free and liganded aminoacyl-tRNA synthetases. Cryst Growth Des 8: 4297–4306 [Google Scholar]
- Guerrier-Takada C, Altman S 1984. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science 223: 285–286 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, McClain WH, Altman S 1984. Cleavage of tRNA precursors by the RNA subunit of E. coli ribonuclease P (M1 RNA) is influenced by 3′-proximal CCA in the substrates. Cell 38: 219–224 [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W 2008. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474 [DOI] [PubMed] [Google Scholar]
- Ishitani R, Yokoyama S, Nureki O 2008. Structure, dynamics, and function of RNA modification enzymes. Curr Opin Struct Biol 18: 330–339 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA 1994. A super position. CCP4/ESF-EACBM Newsletter on Protein Crystallography 31: 9–14 [Google Scholar]
- Leung AK, Kambach C, Kondo Y, Kampmann M, Jinek M, Nagai K 2010. Use of RNA tertiary interaction modules for the crystallisation of the spliceosomal snRNP core domain. J Mol Biol 402: 154–164 [DOI] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J 2011. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A 2010. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Maizels N, Weiner AM 1998. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J 17: 3197–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V 2003. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J Mol Biol 325: 661–675 [DOI] [PubMed] [Google Scholar]
- Westhof E, Dumas P, Moras D 1985. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol 184: 119–145 [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Lilley DM 2011. Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR 1987. Bacterial evolution. Microbiol Rev 51: 221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]