Abstract
The cellular factors involved in mRNA degradation and translation repression can aggregate into cytoplasmic domains known as GW bodies or mRNA processing bodies (P-bodies). However, current understanding of P-bodies, especially the regulatory aspect, remains relatively fragmentary. To provide a framework for studying the mechanisms and regulation of P-body formation, maintenance, and disassembly, we compiled a list of P-body proteins found in various species and further grouped both reported and predicted human P-body proteins according to their functions. By analyzing protein–protein interactions of human P-body components, we found that many P-body proteins form complex interaction networks with each other and with other cellular proteins that are not recognized as P-body components. The observation suggests that these other cellular proteins may play important roles in regulating P-body dynamics and functions. We further used siRNA-mediated gene knockdown and immunofluorescence microscopy to demonstrate the validity of our in silico analyses. Our combined approach identifies new P-body components and suggests that protein ubiquitination and protein phosphorylation involving 14-3-3 proteins may play critical roles for post-translational modifications of P-body components in regulating P-body dynamics. Our analyses provide not only a global view of human P-body components and their physical interactions but also a wealth of hypotheses to help guide future research on the regulation and function of human P-bodies.
Keywords: P-body, autophagy, ubiquitin, hnRNP, microRNA, translation, mRNA turnover, protein–protein interaction, deadenylation, bioinformatics, phosphorylation
INTRODUCTION
Conserved from yeast to human, mRNA processing bodies (P-bodies) are dynamic cytoplasmic foci in eukaryotic cells that contain nontranslating mRNAs as well as proteins involved in translational inhibition and mRNA degradation (Eulalio et al. 2007; Parker and Sheth 2007; Franks and Lykke-Andersen 2008; Anderson and Kedersha 2009). Initially identified through the use of an autoimmune serum targeting a glycine–tryptophan-rich protein, GW182, these membrane-free structures are also called GW bodies (Eystathioy et al. 2002). They are anchored to microtubules and move around in the cytoplasm (Aizer and Shav-Tal 2008; Lindsay and McCaffrey 2011). mRNA decay intermediates are found in P-bodies, and inhibition of 5′-to-3′ mRNA decay increases the size of P-bodies (Sheth and Parker 2003), suggesting that mRNAs can be degraded in P-bodies. P-bodies are free of ribosomes, thus mRNAs cannot be translated in P-bodies. However, it has been reported that mRNAs in P-bodies can be released from P-bodies and reenter polysomes (Brengues et al. 2005; Bhattacharyya et al. 2006). Moreover, in activated human bronchial epithelial cells, a decrease in P-bodies accompanies an increase in global translation, a decrease in mRNA turnover, and a decrease in miRNA function (Zhai et al. 2008). These observations support the notion that P-bodies may provide another layer of post-transcriptional regulation of gene expression by serving as a transient reservoir for nontranslating mRNAs.
Most of the proteins found in P-bodies have functions related to mRNA binding (e.g., eIF4E), translational inhibition (e.g., Rck/p54, eIF4ET, and CPEB1), mRNA deadenylation (e.g., Caf1 and Ccr4), mRNA decapping (e.g., Dcp1/2 and Lsm1-7), mRNA 5′-to-3′ degradation (e.g., Xrn1), or miRNA-mediated gene silencing (e.g., Ago1-4 and GW182) (Parker and Sheth 2007). Factors involved in nonsense-mediated decay (NMD) were also enriched in P-bodies when NMD was interrupted in yeast (Sheth and Parker 2006) and in mammalian cells (Durand et al. 2007). In contrast, most factors involved in mRNA translation (e.g., eIF4G and ribosome subunits) are normally absent from P-bodies (Kedersha et al. 2005). The functions of some P-body components (e.g., ZAR1l) are unknown (Hu et al. 2010). A recent cell-based siRNA screen identified proteins involved in O-linked N-acetylglucosamine (O-GlcNAc) modifications as being important for stress-granule and/or P-body formation, suggesting an important role for post-translational modifications in cytoplasmic granule formation (Ohn et al. 2008).
Although P-bodies have been studied in different species for nearly a decade, the mechanisms by which P-body formation, maintenance, or disassembly is accomplished and controlled remain poorly understood. The complete composition of cellular factors involved in P-body dynamics is not known either. Moreover, controversy still surrounds the function(s) and physiological significance of these dynamic aggregates of mRNA–protein complexes (mRNPs) in the cytoplasm. To better understand the functional significance and regulation of P-bodies, we began a comprehensive analysis of P-body components and their interacting factors. We used bioinformatics tools to retrieve and analyze information from different studies published since 2002, when the nature and importance of P-bodies began to be recognized (Eystathioy et al. 2002). Several important questions directly related to the biological significance and regulation of P-bodies were addressed. For example, how many P-body proteins have been identified? How do these proteins interact with each other and/or with other cellular proteins? How do these proteins and their interactions with each other affect the formation, dynamics, function, and regulation of P-bodies? Combining in silico and experimental approaches, our study provides not only a global view of human P-body components but also a framework for predicting genes involved in P-body regulation. In addition, based on the protein–protein interaction data, our analyses suggest new P-body components and help guide future investigations of P-body regulation by suggesting several hypotheses.
RESULTS
Compilation of information about P-body protein components found in different species
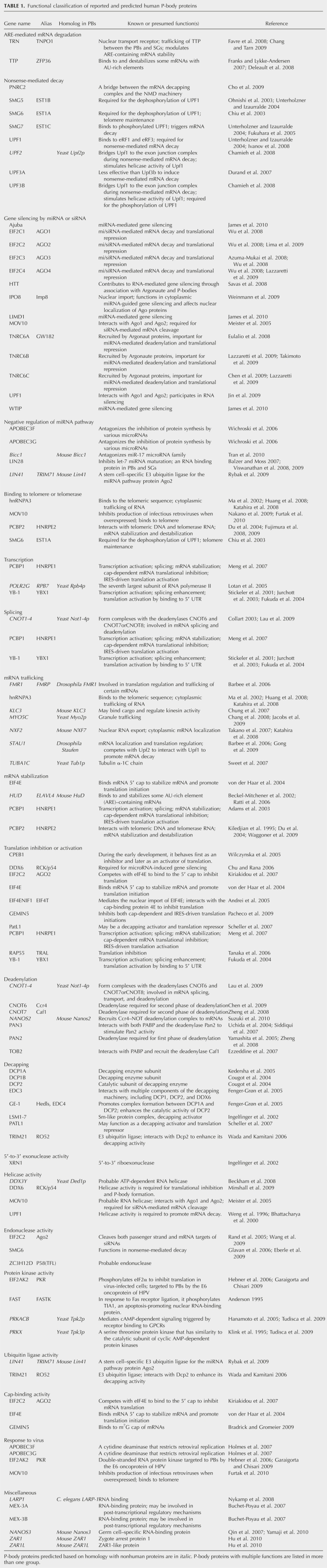
To have a comprehensive list of known P-body proteins, we retrieved PubMed articles published since 2002 that contain various synonyms of “P-bodies,” “GW-bodies,” or P-body or GW-body components in their title or abstract. Two hundred and ninety articles were retrieved. After careful examination of the evidence in each article for P-body localization of each candidate protein, we generated a list of P-body protein components reported in six different species (Supplemental Table 1). The list included 60 human proteins, 11 mouse proteins, 31 yeast proteins, and 17 proteins from plants, worms, or fruit fly.
Functional classification of human P-body protein components
To better understand the functions and regulations of P-bodies, we grouped the human P-body proteins based on their known or putative functions (Table 1). We also searched the NCBI HomoloGene database for human proteins whose orthologs were found to localize to P-bodies in other species but have not themselves been reported as P-body components. As most P-body components are conserved across species, it is likely that the human orthologs of P-body proteins in other species are also P-body components. Based on this homology search, we identified 23 potential human P-body components (Table 1, italics). Functional classification of these predicted human P-body components, along with the 60 previously reported human P-body proteins (Table 1), indicates that human P-body components are involved in many different aspects of mRNA metabolism. These include deadenylation, decapping, 5′-to-3′ degradation, AU-rich element (ARE)–mediated mRNA decay, nonsense-mediated decay (NMD), miRNA-mediated gene silencing, mRNA transport, and translational inhibition, etc. Moreover, some P-body components are involved in post-translational modifications, such as phosphorylation (e.g., PKR) and ubiquitination (e.g., RO52 or TRIM21) (Table 1), suggesting that these modifications play a role in regulating the dynamics and function of P-bodies.
TABLE 1.
Functional classification of reported and predicted human P-body proteins
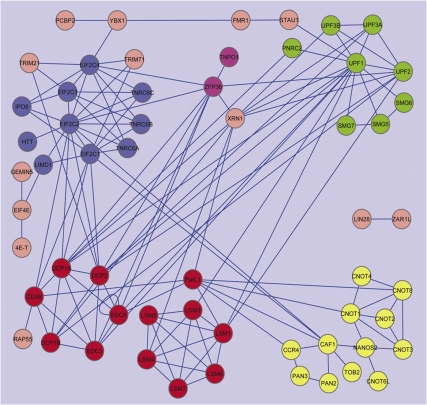
Protein–protein interactions among P-body components
Protein functions are often mediated and regulated through interacting with other proteins. To better understand how different P-body components may aggregate to form foci and how P-body components may be functionally linked, we analyzed protein–protein interactions (PPIs) among P-body components. Entry of the gene names of the 83 human P-body proteins listed in Table 1 to the STRING database version 8.3 (Jensen et al. 2009) returned 100 experimentally confirmed PPIs among these P-body components. An additional 39 experimentally confirmed PPIs among these proteins were found by manually checking recent publications. Together, our analysis included 139 PPIs among human P-body proteins. To better visualize this PPIs analysis, we imported the PPI data to Cytoscape (Fig. 1; Shannon et al. 2003). In this plot, proteins with similar or related functions are represented with similarly colored nodes. It is apparent that 58 out of 83 (i.e., more than two-thirds) of human P-body proteins are part of dense PPI networks, suggesting that the interactions among these proteins contribute to aggregation of mRNPs during P-body formation. Moreover, the plot in Figure 1 indicates that factors involved in ARE-mediated decay, miRNA-mediated gene silencing, and NMD are physically linked. Although deadenylation triggers mRNA degradation mediated by AREs, miRNAs, and nonsense codons (Shyu and Chen 2011), not many direct interactions between deadenylation factors and proteins involved in these mRNA decay pathways have been reported. This observation is consistent with a regulatory model in which deadenylation induces mRNP remodeling before nontranslatable mRNPs enter or form P-bodies (Zheng et al. 2008).
FIGURE 1.
Protein–protein interactions among P-body components. The protein–protein interactions (PPIs) among reported and predicted human P-body components are visualized using Cytoscape. Node colors: (green) NMD factors; (blue) miRNA-mediated gene silencing factors; (magenta) ARE-mediated decay factors; (red) decapping factors; (yellow) deadenylation factors. Lines between the nodes represent PPIs. Details are described in the Materials and Methods.
Interactions between P-body proteins and other cellular proteins
Currently, little is known about how P-body assembly, maintenance, and disassembly are regulated. As the function and localization of a protein may be changed by interactions with other proteins, we hypothesize that among proteins interacting with P-body components, some of them are likely to modulate P-body dynamics. Moreover, although some P-body-interacting proteins have not been reported or predicted as P-body components, they may actually localize to P-bodies. Thus, studying P-body-interacting proteins may reveal new P-body components. We retrieved experimentally confirmed PPIs between known or predicted human P-body components (Table 1) and other cellular proteins using PINA, which integrates PPI data from six databases (Wu et al. 2009). Proteins with obsolete Uniprot accession numbers and/or redundant interactions were culled. The data were then imported to Cytoscape for visualization (Supplemental Fig. 1), with proteins grouped based on the number of different P-body componentswith which they interact. The results show that among the 535 P-body interacting proteins, 400 interact with one P-body component, 84 interact with two P-body components, 31 interact with three P-body components, 10 interact with four P-body components, five interact with five P-body components, two interact with six P-body components, one interacts with seven P-body components, one interacts with eight P-body components, and one interacts with nine P-body components.
Supplemental Table 2 lists proteins that interact with three or more P-body components. This table includes some proteins that have already been reported to affect P-body dynamics, lending support to the validity of our in silico approach for identifying P-body-related factors. For example, HSP90AA1 and HSP90AB1, two major cytosolic HSP90 proteins, appear in Supplemental Table 2. Inhibition of Hsp90 activity was reported to block association of some P-body components (Pare et al. 2009; Johnston et al. 2010). Another example involves YWHAG, YWHAB, YWHAH, YWHAQ, and YWHAZ. These proteins belong to the highly conserved 14-3-3 family of proteins, which bind to phosphoserine- or phosphothreonine-containing proteins (Morrison 2009). It was reported that binding of 14-3-3 proteins to EDC3, an enhancer of mRNA decapping complex, altered P-body morphology, inhibited miRNA-mediated gene silencing, and changed protein–protein interactions of EDC3 (Larance et al. 2010). Our analysis further revealed interactions between 14-3-3 proteins and several other reported or predicted P-body components, including LARP1, KLC3, TRIM21 (RO52), TTP (ZFP36), and RCK/P54 (DDX6). Collectively, these observations suggest that 14-3-3 proteins play an important role in regulating P-bodies.
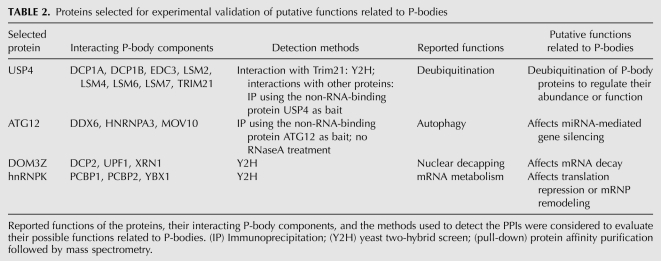
Evaluation of proteins that may have impact on P-bodies
As an initial test of the validity of our in silico approach, we evaluated the P-body interactants listed in Supplemental Table 2 by examining their reported functions as well as the methods used to detect their interactions with P-body components and selected USP4, DOM3Z, ATG12, and hnRNP K (Table 2) for experimental validation. These proteins have not been reported to link to P-bodies; however, each interacts with at least three P-body components whose functions appear to directly impact P-body dynamics. Moreover, they are all ubiquitously expressed in mammalian cells. USP4 is a deubiquitinating enzyme with isopeptidease activity, and it interacts strongly with RO52, an E3 ubiquitin ligase (Wada and Kamitani 2006). The two enzymes appear to have antagonistic effects (Wada and Kamitani 2006). A recent report found that RO52 associates with Dcp2 and that ectopically expressed RO52 colocalizes with P-bodies (Yamochi et al. 2008). We therefore also targeted RO52 for experimental validation.
TABLE 2.
Proteins selected for experimental validation of putative functions related to P-bodies
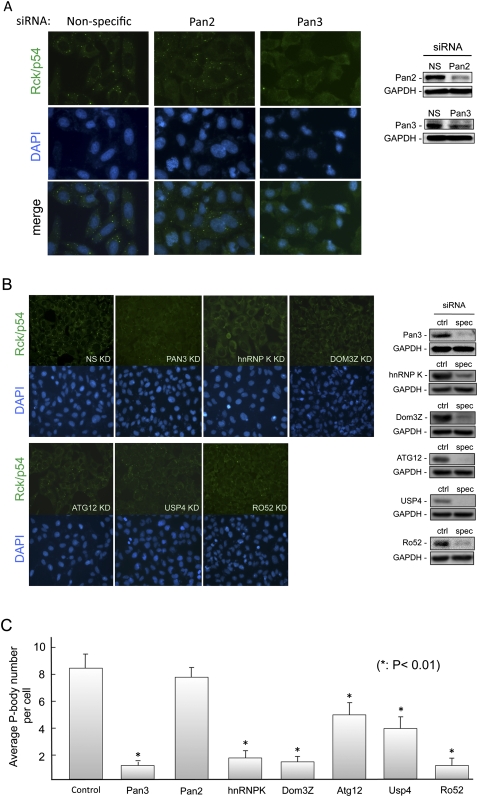
Human U2OS cells were used as the experimental model. Immunofluorescence microscopy, using endogenous Rck/p54 (a well-characterized P-body component) as a marker, showed, as in mouse NIH3T3 cells (Fig. 2A; Zheng et al. 2008), a nice staining of P-bodies in human U2OS cells treated with nonspecific siRNAs (Fig. 2B). Moreover, Pan3 knockdown in U2OS cells decreased P-body numbers as observed in NIH3T3 cells (Zheng et al. 2008), whereas knocking down Pan2 deadenylase had little effect in both cell lines (Fig. 2A,B). We then examined the effects of knocking down USP4, RO52, DOM3Z, ATG12, or hnRNP K on P-bodies in human U2OS cells. The results showed a significant reduction of P-bodies in cells treated with siRNAs specific for any one of these five candidates or Pan3 (Fig. 2B,C). Western blot analysis confirmed an effective knockdown of each protein using specific siRNAs but not control siRNAs (Fig. 2B). It should be noted that among the five genes tested, knocking down hnRNP K has the most dramatic effect on P-body number. While knocking down >90% of ATG12 or USP4 led to 50%–60% reduction of P-bodies, P-bodies could hardly be detected when only ∼60% of hnRNP K was knocked down. Collectively, these experiments demonstrated that USP4, RO52, DOM3Z, ATG12, and hnRNP K all impact the dynamics of P-bodies.
FIGURE 2.
Effects of siRNA knockdown of selected P-body interacting proteins. (A) Immunofluorescence microscopy and Western blot results showing a significant loss of P-bodies in mouse NIH3T3 cells when Pan3 (and not Pan2) was knocked down by specific siRNAs. P-bodies were visualized using rabbit anti-Rck/p54 antibody followed by secondary goat anti-rabbit conjugated to Alexa 488 (green). Cell nuclei were stained by DAPI. (B) Immunofluorescence microscopy results showing effects on P-bodies when the indicated endogenous proteins were knocked down by specific siRNAs in human U2OS cells. P-bodies were visualized with rabbit anti-Rck/p54 antibody followed by secondary goat anti-rabbit conjugated to Alexa 488 (green). Cell nuclei were stained with DAPI. Western blots showing effective knockdown of the targeted endogenous proteins. (C) The changes in P-body number following knockdown of target proteins were analyzed. The quantification of P-bodies was determined by counting the number of P-bodies of a group of cells per experiment. Data are presented as the mean P-body number per cell ± standard error of the mean (n > 4). The asterisk denotes a significant difference analyzed by a paired t-test.
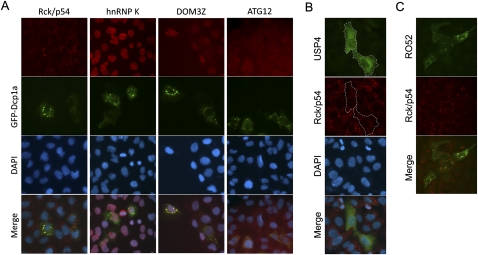
To evaluate whether any of the five proteins tested here are found in P-bodies, we performed indirect immunofluorescence microscopy, using ectopically expressed GFP-Dcp1a as a P-body marker (Eystathioy et al. 2003; Zheng et al. 2008). GFP-Dcp1a forms clear P-body foci and colocalized well with P-bodies marked by Rck/p54 in U2OS cells (Fig. 3A). HnRNP K showed strong nuclear staining without discernable foci in the cytoplasm. Although DOM3Z exhibited strong nuclear staining, it also formed a few clear foci in the cytoplasm that colocalized with P-bodies marked by GFP-Dcp1a (Fig. 3A). In contrast, endogenous ATG12 did not appear to form any obvious foci that colocalized with P-bodies in U2OS cells expressing GFP-Dcp1a. Due to the lack of anti-USP4 antibody that is suitable for indirect immunofluorescence study, it is not clear whether endogenous USP4 can be found in P-bodies. However, ectopic expression of Flag-tagged USP4 either abolished or appreciably reduced P-body formation (Fig. 3B). In most cases, Flag-USP4 gave a fairly even staining of the cytoplasm. In a few cases, even though Flag-USP4 was observed to form some cytoplasmic foci, they did not colocalize with P-bodies. In contrast to USP4, ectopically expressed RH-tagged RO52 forms many foci that colocalize well with P-bodies (Fig. 3C), consistent with the observation by a previous study (Yamochi et al. 2008). Taken together, our data identified DOM3Z as a new P-body component and further suggest a role for ubiquitination–deubiquitnation in regulating P-body dynamics.
FIGURE 3.
Subcellular localization of selected P-body-interacting proteins. Immunofluorescence microscopy of (A) endogenous Rck/p54, hnRNP K, Dom3Z, and ATG12; (B) ectopically expressed USP4; or (C) ectopically expressed RO52 in proliferating U2OS cells. U2OS cells were transfected with a plasmid encoding GFP-Dcp1a to mark P-bodies (green foci) in panel A. Endogenous proteins were visualized using the cognate rabbit antibodies followed by secondary goat anti-rabbit conjugated to Alexa 555 (red staining). Flag-tagged USP4 or RH-tagged RO52 was visualized using the corresponding anti-epitope monoclonal antibody followed by secondary goat anti-mouse conjugated to Alexa 488 (green staining). P-bodies were visualized using rabbit anti-Rck/p54 antibody followed by secondary goat anti-rabbit conjugated to Alexa 555 (red).
DISCUSSION
Our study combining in silico analyses and experimental data led to new observations and also lends support to several notions. First, P-body components are not limited to proteins involved in mRNA decay and translation repression in the cytoplasm (Eulalio et al. 2007; Parker and Sheth 2007; Franks and Lykke-Andersen 2008; Anderson and Kedersha 2009); they also contain proteins involved in various other aspects of mRNA metabolism in the nucleus as well, including transcription, splicing, or mRNA transport. Second, the present analysis reveals that at least two-thirds of human P-body proteins form dense PPI networks (Fig. 1), supporting the notion that PPIs among P-body components contribute to the aggregation of mRNPs during P-body formation. Third, in addition to deadenylases and the 5′-to-3′ exonuclease Xrn1, two endonucleases (SMG6 and \ZC3H12D) can also be found in P-bodies (Table 1). This suggests that degradation of some mRNAs in P-bodies may involve endonucleolytic cleavage. Fourth, although many mRNA degradation factors are found in P-bodies, some proteins identified as P-body components are actually mRNA stabilizers (e.g., eIF4E, HuD, PCBP1, and PCBP2). This suggests that some mRNAs in P-bodies may be protected from degradation, consistent with the notion that P-bodies can serve as temporary mRNP-storage sites (Eulalio et al. 2007; Parker and Sheth 2007; Franks and Lykke-Andersen 2008; Anderson and Kedersha 2009). Finally, localization of protein modification factors—including 14-3-3 protein family members, kinases, E3 ubiquitin ligases, and deubiquitinating enzymes—to P-bodies (Table 1) suggests that phosphorylation and ubiquitination may occur in P-bodies to modulate P-body dynamics (also see discussion below).
While more about the effectiveness of the in silico strategy can be further determined through additional experimentation in the future, our initial test (Fig. 2B,C) with individual knockdown of five genes selected based on the analyses supports the validity of our in silico approach. It is worth noting that P-body loss or reduction as a result of gene knockdown should not be extrapolated to mean that the corresponding gene product is physically required for P-body integrity. It is also possible that a gene knockdown may indirectly impact translation or mRNA decay, which, in turn, could influence P-body dynamics. Our results have stimulated several hypotheses that help to guide future research on P-bodies. Some examples of these hypotheses are discussed below.
The role of 14-3-3 family proteins in regulating P-body dynamics
Although proteins of the 14-3-3 families were not reported as P-body components, our analysis reveals that 14-3-3 proteins interact with several reported or predicted P-body components (Supplemental Table 2). The putative roles of these proteins in regulating P-body dynamics are also supported by several recent studies showing that binding of 14-3-3 proteins to EDC3, a P-body component, altered P-body morphology, inhibited miRNA-mediated gene silencing, and changed EDC3's PPIs (Larance et al. 2010). Furthermore, phosphorylated 14-3-3γ protein (YWHAG) translocates to P-bodies, and knocking down 14-3-3γ protein blocks P-body formation after UV damage (Okada et al. 2011). It is known that 14-3-3 proteins bind to protein ligands with phosphorylated serine or threonine residues, which, in turn, physically prevent molecular interactions or modulate the accessibility of a target protein to modifying enzymes such as kinases, phosphatases, and proteases (Mhawech 2005; Johnson et al. 2010). In addition, 14-3-3 proteins can act as scaffolds to bring target proteins in close proximity to one another. We hypothesize that these activities of 14-3-3 proteins and the involved signaling pathways play a pivotal role in regulating P-body assembly, maintenance, and/or disassembly.
P-body regulation via protein ubiquitination and deubiquitination
Our results show that knocking down USP4, a deubiquitinating enzyme with isopeptidease activity that interacts strongly with RO52 E3 ubiquitin ligase (Wada and Kamitani 2006), caused a significant reduction of P-bodies (Fig. 2B,C). We also show that ectopically overexpressing USP4 results in loss of P-bodies (Fig. 3B). One possibility is that the isopeptidease activity of USP4 is necessary for it to impact P-body dynamics. Given that mLin41 and RO52 (Trim21), two E3 ubiquitin ligases, are also found in P-bodies (Wada and Kamitani 2006; Rybak et al. 2009), it is possible that USP4-mediated deubiquitination of P-body components, which are ubiquitinated by these ubiquitin ligases, may influence P-body dynamics. It is of particular interest that RO52 colocalizes with P-bodies, and knocking down RO52 also impedes P-body formation (Figs. 2B, 3C). Moreover, RO52 can ubiquitinate itself and USP4, and USP4 can deubiquitinate itself and RO52 (Wada and Kamitani 2006). Taken together, it is tempting to postulate that a balance of the USP4–RO52 yin–yang actions on P-body components and on each other is important for P-body formation or maintenance, which may represent a new regulatory mechanism controlling P-body dynamics and/or functions.
A new role for the nuclear decapping enzyme DOM3Z in cytoplasmic P-body formation
The yeast ortholog of human Dom3Z, Rai1p, possesses both pyrophosphohydrolase activity toward 5′-triphosphorylated mRNA and a decapping endonuclease activity that removes the entire unmethylated cap dinucleotide from mRNA (Xiang et al. 2009; Jiao et al. 2010). Moreover, Rai1p was reported to be involved in a novel surveillance mechanism that ensures the integrity of mRNA's 5′ end by removing aberrant caps of mRNA in the nucleus (Jiao et al. 2010). Given the sequence conservation between Rai1p and Dom3Z, our finding that Dom3Z knockdown resulted in loss of P-bodies in human U2OS cells suggests that Dom3Z also plays an important role in mammalian cytoplasmic mRNA degradation. Moreover, our observation that Dom3Z colocalizes with P-bodies also supports the possibility that mammalian Dom3Z may have a surveillance role in P-bodies, e.g., by removing aberrant caps of mRNPs in P-bodies.
A potential role of autophagy in P-body formation
Our finding that knocking down ATG12 reduced P-bodies (Fig. 2B) raises an intriguing question as to how ATG12, a ubiquitin-like protein required for autophagy in mammals (Geng and Klionsky 2008), is involved in P-body formation. Autophagy plays a vital role in maintaining cellular homeostasis in differentiated mammalian cells (Glick et al. 2010). One possibility is that P-bodies help maintain cytoplasmic mRNA homeostasis in a way that coordinates with autophagy. The observation that ribosomes can be selectively degraded by autophagy (Glick et al. 2010), which could, in turn, affect the translation status of the mRNA pool, supports this possibility. Alternatively, given that ATG12 is a ubiquitin-like modifier that can be covalently attached to multiple protein substrates (Geng and Klionsky 2008), our finding also supports the notion that modifications of certain P-body component proteins may be necessary for P-body assembly.
A general role of hnRNP proteins in P-body formation
HnRNP K belongs to a large family of RNA-binding proteins that associate with mRNAs during mRNA biogenesis in the nucleus (Bomsztyk et al. 2004). They participate in various aspects of mRNA metabolism both in the nucleus and in the cytoplasm (Bomsztyk et al. 2004), e.g., pre-mRNA splicing, mRNA decay, translation, virus replication, and axon development. Therefore, hnRNP K may be important for general mRNP remodeling, and thus knocking down hnRNP K impairs formation of the proper mRNP structure required for P-body formation. However, it is equally possible that the effect of hnRNP K knockdown is indirect. It may alter the general integrity of mRNPs and thus their translation and decay, which, in turn, impacts P-body dynamics. The involvement of hnRNP K in P-body formation also raises a question as to whether and how many other hnRNP proteins also directly participate in or indirectly influence P-body assembly.
In summary, our study combines the identification of protein–protein interactions via in silico approach with experimental testing of some key protein targets. The analyses suggest several new pathways for controlling P-body assembly, maintenance, and disassembly and also provide several hypotheses that help to guide future research on human P-bodies.
MATERIALS AND METHODS
Compiling a comprehensive list of reported P-body protein components
To compile a comprehensive list of known P-body protein components in eukaryotes, we searched articles published since 2002 in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) that contain various synonyms of “P-bodies” in their titles or abstracts {(mRNA processing bod* [TIAB] OR P bod* [TIAB] OR (Dcp1* [TIAB] AND (foci [TIAB] or bodies [TIAB])) OR ((GW [TIAB] OR GW182 [TIAB]) AND (bodies [TIAB] OR foci [TIAB])) AND “last 8 years” [DP])}.
Prediction of new human P-body components and functional classification of human P-body proteins
To identify potential new human P-body components, the gene names of P-body components found in nonhuman species were submitted to the NCBI HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene) to retrieve their human homologs. Finally, the reported and predicted human P-body proteins were grouped according to their reported functions.
Bioinformatic analysis of protein–protein interactions (PPIs) among human P-body components
Gene names of the reported and predicted human P-body components were searched in the STRING database version 8.3 (http://string-db.org/newstring_cgi/show_input_page.pl?UserId=bP_o9PRBhSK_&sessionId=ZaAJSCDzZs5j&input_page_type=multiple_identifiers), with “organism” set to Homo sapiens. PPIs were limited to those experimentally detected (e.g., by coimmunoprecipitation, pull-down, or yeast two-hybrid assays). Additional PPIs were identified manually via a PubMed literature search. All PPIs were assembled in an Excel file that was imported to Cytoscape version 2.6.3 (Shannon et al. 2003) for visualization. The nodes were colored to reflect the presumed functions of the P-body proteins.
Bioinformatic analysis of protein–protein interactions (PPIs) between human P-body components and other cellular proteins
A list of Uniprot accession numbers for both reported and predicted human P-body components was submitted to the Protein Interaction Networks Analysis (PINA) platform (http://csbi.ltdk.helsinki.fi/pina/interactome.batchForm.do), which integrates PPI data from six databases. PPIs detected by affinity technology (e.g., co-IP, pull-down, or tandem affinity purification) or transcriptional complementation assay (e.g., two-hybrid or two-hybrid array) were selected and saved as new networks on the PINA server. These networks were then combined and downloaded as an MITAB file in TSV format, which was processed with a PHP script to remove records that were duplicates or contained obsolete accession numbers.
The resulting network data were then further imported to Cytoscape (version 2.6.3). Nodes representing P-body components were selected by opening a text file containing all reported and predicted P-body components and organized into a circle. Next, the non-P-body nodes were grouped based on the number of P-body nodes they interact with. The color of each group of nodes was set via VizMapper. To create a list of cellular proteins reported to interact with three or more P-body components, we gathered the gene names of the selected non-P-body nodes and their interacting P-body nodes from the Node Attribute Browser.
Cell culture, transfection, gene knockdown, and Western blotting
NIH3T3 cells and U2OS cells were transfected using Lipofectamine 2000 (Invitrogen) and FuGENE 6 (Roche), respectively, using the manufacturers' protocols. A pool of siRNAs (SMARTpool; DHARMACON) was used to knock down expression of each target gene expression.
Cell lysates were prepared, resolved on a 7% or 10% SDS-polyacrylamide gel, transferred to PVDF membranes, and immunoblotted using an ECL Western blotting kit (Amersham). The primary antibodies are indicated in the figure legends; bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce). The primary antibodies were rabbit anti-Rck/p54 (Bethyl; 1:2000 dilution); mouse monoclonal anti-GAPDH (Research Diagnostics; 1:10,000 dilution); rabbit anti-USP4 (Bethyl; 1:2000 dilution); rabbit anti-RO52 (Lifespan Biosciences; 1:800); rabbit anti-ATG12 (Cell Signaling; 1:2000 dilution); rabbit anti-Dom3Z (ProteinTech Group; 1:400 dilution); and rabbit anti-hnRNP K (Bethyl; 1:2000 dilution). Horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham; 1:4000 dilution) or goat anti-mouse IgG (Bio-Rad; 1:5000 dilution) was used as the secondary antibody.
Immunofluorescence assays
NIH3T3 or U2OS cells were seeded in six-well plates (0.4 × 106 cells per well) 24 h before transfection. At 22–26 h after transfection, cells were reseeded to slide chambers (BD Falcon) and incubated overnight. Cells in the slide chambers were fixed for 10 min each with 3.7% (w/v) paraformaldehyde (Sigma, Cat. P6148) in PBS, with cold methanol, and then with 0.2% (v/v) Triton X-100 in PBS (Zheng et al. 2008). For indirect immunofluorescence microscopy, the primary and secondary antibodies were diluted 1:1000 with 1% (w/v) BSA in PBS. Endogenous Rck/p54 was detected using rabbit anti-Rck/p54 and Alexa Flour 488 (green) or Alexa Flour 555 (red) conjugated to goat anti-rabbit IgG. Endogenous USP4, RO52, DOM3Z, ATG12, and hnRNP K were detected in U2OS cells, transiently expressing GFP-Dcp1a, using antibodies raised in rabbits and Alexa Flour 555 (red) conjugated to goat anti-rabbit IgG. After incubation with primary antibody, cells were washed three times in PBS for 5 min and then incubated with the fluorescent-labeled secondary antibody. After washing with PBS, fluorescence mounting medium with DAPI was added. Slides were examined with a Leica 40× objective lens with a Leica DM IL light microscope, or with a Deltavision deconvolution microscope and a 100× objective lens. Optical z-sectioning was set at 18 sections in total (0.2-μm space between sections), and images were deconvoluted using the SoftWorx Suite (Applied Precision). To analyze the changes in P-body number following gene knockdown, cells transfected with either control nonspecific (NS) or gene-specific siRNAs were analyzed using Image J software. Particles with values greater than 50 square pixels were considered above background staining and were selected for P-body analysis.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank R. Kulmacz for critical reading of the manuscript; T. Kamitani (Medical College of Georgia) for plasmids; and A. Chadee, K. Rogers, and Z. Xia for technical assistance. This work was supported by the National Institutes of Health (RO1 GM046454 to A.-B.S.) and in part by the Houston Endowment, Inc. to A.-B.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2789611.
REFERENCES
- Adams DJ, Beveridge DJ, van der Weyden L, Mangs H, Leedman PJ, Morris BJ 2003. HADHB, HuR, and CP1 bind to the distal 3′-untranslated region of human renin mRNA and differentially modulate renin expression. J Biol Chem 278: 44894–44903 [DOI] [PubMed] [Google Scholar]
- Aizer A, Shav-Tal Y 2008. Intracellular trafficking and dynamics of P bodies. Prion 2: 131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PTQ 1995. Fas-activated serine/threonine kinase (FAST) phosphorylates TIA-1 during Fas-mediated apoptosis. J Exp Med 182: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Lührmann R 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC 2008. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci 105: 7964–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E, Moss EG 2007. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol 4: 16–25 [DOI] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckel-Mitchener AC, Miera A, Keller R, Perrone-Bizzozero NI 2002. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J Biol Chem 277: 27996–28002 [DOI] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko A-M, Noueiry A, Ramaswami M, Parker R 2008. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol Biol Cell 19: 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 71: 513–521 [DOI] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J 2004. hnRNP K: One protein multiple processes. Bioessays 26: 629–638 [DOI] [PubMed] [Google Scholar]
- Bradrick SS, Gromeier M 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS ONE 4: e7030 doi: 10.1371/journal.pone.0007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchet-Poyau K, Courchet J, Le Hir H, Séraphin B, Scoazec J-Y, Duret L, Domon-Dell C, Freund J-N, Billaud M 2007. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res 35: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93 [DOI] [PubMed] [Google Scholar]
- Chang W-L, Tarn W-Y 2009. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res 37: 6600–6612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Zaarour RF, Reck-Peterson S, Rinn J, Singer RH, Snyder M, Novick P, Mooseker MS 2008. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid–protein complex that contains mRNAs and subunits of the RNA-processing body. RNA 14: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 16: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S-Y, Serin G, Ohara O, Maquat LE 2003. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Chu C-Y, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210 doi: 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Zhang Y, Van Der Hoorn F, Hawkes R 2007. The anatomy of the cerebellar nuclei in the normal and scrambler mouse as revealed by the expression of the microtubule-associated protein kinesin light chain 3. Brain Res 1140: 120–131 [DOI] [PubMed] [Google Scholar]
- Collart MA 2003. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Séraphin B 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego Otero Y, Severijnen L-A, van Cappellen G, Schrier M, Oostra B, Willemsen R 2002. Transport of fragile X mental retardation protein via granules in neurites of PC12 cells. Mol Cell Biol 22: 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault KM, Skinner SJ, Brooks SA 2008. Tristetraprolin regulates TNF TNF-α mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45: 13–24 [DOI] [PubMed] [Google Scholar]
- Du Z, Yu J, Chen Y, Andino R, James TL 2004. Specific recognition of the C-rich strand of human telomeric DNA and the RNA template of human telomerase by the first KH domain of human poly(C)-binding protein-2. J Biol Chem 279: 48126–48134 [DOI] [PubMed] [Google Scholar]
- Durand S, Cougot N, Mahuteau-Betzer F, Nguyen C-H, Grierson DS, Bertrand E, Tazi J, Lejeune F 2007. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 178: 1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EKL, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell 13: 1338–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EKL, Séraphin B, Cougot N, Fritzler MJ 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chang T-C, Zhu W, Yamashita A, Chen C-YA, Zhong Z, Yamashita Y, Zheng D, Shyu A-B 2007. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 27: 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre N, Camps M, Arod C, Chabert C, Rommel C, Pasquali C 2008. Chemokine receptor CCR2 undergoes transportin1-dependent nuclear translocation. Proteomics 8: 4560–4576 [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M 2008. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M 2009. Selective localization of PCBP2 to cytoplasmic processing bodies. Biochim Biophys Acta 1793: 878–887 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Ashizuka M, Nakamura T, Shibahara K, Maeda K, Izumi H, Kohno K, Kuwano M, Uchiumi T 2004. Characterization of the 5′-untranslated region of YB-1 mRNA and autoregulation of translation by YB-1 protein. Nucleic Acids Res 32: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, KewalRamani VN, Unutmaz D 2010. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS ONE 5: e9081 doi: 10.1371/journal.pone.0009081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U, Chisari FV 2009. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 6: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ 2008. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavan F, Behm-Ansmant I, Izaurralde E, Conti E 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25: 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE 2009. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 23: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamoto T, Ozaki T, Furuya K, Hosoda M, Hayashi S, Nakanishi M, Yamamoto H, Kikuchi H, Todo S, Nakagawara A 2005. Identification of protein kinase A catalytic subunit β as a novel binding partner of p73 and regulation of p73 function. J Biol Chem 280: 16665–16675 [DOI] [PubMed] [Google Scholar]
- Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA 2006. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J Gen Virol 87: 3183–3193 [DOI] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem 282: 2587–2595 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang F, Zhu X, Yuan Y, Ding M, Gao S 2010. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev Dyn 239: 407–424 [DOI] [PubMed] [Google Scholar]
- Huang P-R, Tsai S-T, Hsieh K-H, Wang T-CV 2008. Heterogeneous nuclear ribonucleoprotein A3 binds single-stranded telomeric DNA and inhibits telomerase extension in vitro. Biochim Biophys Acta 1783: 193–202 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DT, Weigert R, Grode KD, Donaldson JG, Cheney RE 2009. Myosin Vc is a molecular motor that functions in secretory granule trafficking. Mol Biol Cell 20: 4471–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James V, Zhang Y, Foxler DE, de Moor CH, Kong YW, Webb TM, Self TJ, Feng Y, Lagos D, Chu CY et al. 2010. LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc Natl Acad Sci 107: 12499–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Suh MR, Han J, Yeom K-H, Lee Y, Heo I, Ha M, Hyun S, Kim VN 2009. Human UPF1 participates in small RNA-induced mRNA downregulation. Mol Cell Biol 29: 5789–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C 2010. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J 427: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Geoffroy M-C, Sobala A, Hay R, Hutvagner G 2010. HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer H-D 2003. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem 278: 27988–27996 [DOI] [PubMed] [Google Scholar]
- Katahira J, Miki T, Takano K, Maruhashi M, Uchikawa M, Tachibana T, Yoneda Y 2008. Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs. Nucleic Acids Res 36: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J 14: 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z 2007. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129: 1141–1151 [DOI] [PubMed] [Google Scholar]
- Klink A, Schiebel K, Winkelmann M, Rappold G 1995. The human protein kinase gene PKX1 on Xp22.3 displays Xp/Yp homology and is a site of chromosomal instability. Hum Mol Genet 4: 869–878 [DOI] [PubMed] [Google Scholar]
- Larance M, Rowland AF, Hoehn K, Humphreys DT, Preiss T, Guilhaus M, James DE 2010. Global phosphoproteomics identifies a major role for akt and 14-3-3 in regulating Edc3. Mol Cell Proteomics 9: 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N-C, Kolkman A, van Schaik FMA, Mulder KW, Pijnappel WWMP, Heck AJR, Timmers HTM 2009. Human Ccr4–Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453 [DOI] [PubMed] [Google Scholar]
- Lazzaretti D, Tournier I, Izaurralde E 2009. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST 2009. Binding and cleavage specificities of human Argonaute2. J Biol Chem 284: 26017–26028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW 2011. Myosin Va is required for P body but not stress granule formation. J Biol Chem 286: 11519–11528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ASW, Moran-Jones K, Shan J, Munro TP, Snee MJ, Hoek KS, Smith R 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem 277: 18010–18020 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T 2005. Identification of novel Argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW, Kumar R 2007. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci 104: 5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhawech P 2005. 14-3-3 proteins—an update. Cell Res 15: 228–236 [DOI] [PubMed] [Google Scholar]
- Minshall N, Kress M, Weil D, Standart N 2009. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell 20: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK 2009. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 19: 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Kakiuchi Y, Shimada Y, Ohyama M, Ogiwara Y, Sasaki-Higashiyama N, Yano N, Ikeda F, Yamada E, Iwamatsu A, et al. 2009. MOV10 as a novel telomerase-associated protein. Biochem Biophys Res Commun 388: 328–332 [DOI] [PubMed] [Google Scholar]
- Nykamp K, Lee M-H, Kimble J 2008. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 14: 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200 [DOI] [PubMed] [Google Scholar]
- Okada N, Yabuta N, Suzuki H, Aylon Y, Oren M, Nojima H 2011. A novel Chk1/2-Lats2-14-3-3 signaling pathway regulates P-body formation in response to UV damage. J Cell Sci 124: 57–67 [DOI] [PubMed] [Google Scholar]
- Pacheco A, de Quinto SL, Ramajo J, Fernández N, Martínez-Salas E 2009. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 37: 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, López-Orozco J, LaPointe P, Lasko P, Hobman TC 2009. Hsp90 regulates the function of Argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 20: 3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhao H, Kovanci E, Simpson JL, Chen Z-J, Rajkovic A 2007. Mutation analysis of NANOS3 in 80 Chinese and 88 Caucasian women with premature ovarian failure. Fertil Steril 88: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Ratti A, Fallini C, Cova L, Fantozzi R, Calzarossa C, Zennaro E, Pascale A, Quattrone A, Silani V 2006. A role for the ELAV RNA-binding proteins in neural stem cells: stabilization of Msi1 mRNA. J Cell Sci 119: 1442–1452 [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11: 1411–1420 [DOI] [PubMed] [Google Scholar]
- Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N 2008. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci 105: 10820–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, Díez J 2007. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim Biophys Acta 1773: 1786–1792 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Chen C-YA 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2: 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N, Mangus DA, Chang T-C, Palermino J-M, Shyu A-B, Gehring K 2007. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem 282: 25067–25075 [DOI] [PubMed] [Google Scholar]
- Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J 20: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Igarashi K, Aisaki K-I, Kanno J, Saga Y 2010. NANOS2 interacts with the CCR4–NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci 107: 3954–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet TJ, Boyer B, Hu W, Baker KE, Coller J 2007. Microtubule disruption stimulates P-body formation. RNA 13: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Miki T, Katahira J, Yoneda Y 2007. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res 35: 2513–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto K, Wakiyama M, Yokoyama S 2009. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA 15: 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem 281: 40096–40106 [DOI] [PubMed] [Google Scholar]
- Tran U, Zakin L, Schweickert A, Agrawal R, Döger R, Blum M, De Robertis EM, Wessely O 2010. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development 137: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hoshino S-I, Katada T 2004. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem 279: 1383–1391 [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596 [DOI] [PubMed] [Google Scholar]
- Villacé P, Marión RM, Ortín J 2004. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucl Acids Res 32: 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng T, Toffanin S, O'Sullivan M, Lu J, Philips LA, Lockhart VL, et al. 2009. Lin28 enhances tumorigenesis and is associated with advanced human malignancies. Nat Genet 41: 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JEG 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 11: 503–511 [DOI] [PubMed] [Google Scholar]
- Wada K, Kamitani T 2006. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem Biophys Res Commun 339: 415–421 [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Johannes GJ, Liebhaber SA 2009. Depletion of the poly(C)-binding proteins αCP1 and αCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem 284: 9039–9049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Wein G, Rössler M, Klug R, Herget T 2003. The 3′-UTR of the mRNA coding for the major protein kinase C substrate MARCKS contains a novel CU-rich element interacting with the mRNA stabilizing factors HuD and HuR. Eur J Biochem 270: 350–365 [DOI] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski MJ, Robb GB, Rana TM 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog 2: e41 doi: 10.1371/journal.ppat.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 118: 981–992 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2008. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol 18: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Vallenius T, Ovaska K, Westermarck J, Mäkelä TP, Hautaniemi S 2009. Integrated network analysis platform for protein–protein interactions. Nat Methods 6: 75–77 [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L 2009. Structure and function of the 5′ → 3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Tanaka T, Shigeta M, Chuma S, Saga Y, Saitou M 2010. Functional reconstruction of NANOS3 expression in the germ cell lineage by a novel transgenic reporter reveals distinct subcellular localizations of NANOS3. Reproduction 139: 381–393 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Yamochi T, Ohnuma K, Hosono O, Tanaka H, Kanai Y, Morimoto C 2008. SSA/Ro52 autoantigen interacts with Dcp2 to enhance its decapping activity. Biochem Biophys Res Commun 370: 195–199 [DOI] [PubMed] [Google Scholar]
- Zhai Y, Zhong Z, Chen CY, Xia Z, Song L, Blackburn MR, Shyu AB 2008. Coordinated changes in mRNA turnover, translation, and RNA processing bodies in bronchial epithelial cells following inflammatory stimulation. Mol Cell Biol 28: 7414–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]