Abstract
Ribosomal proteins (RPs) are essential components of ribosomes, but several RPs are also present at transcription sites of eukaryotic chromosomes. Here, we report a genome-wide ChIP-on-chip analysis of the association of three representative 60S RPs with sites in the Schizosaccharomyces pombe chromosomes. All three proteins tend to bind at the same subset of coding and noncoding loci. The data demonstrate selective RNA-dependent interactions between RPs and many transcription sites and suggest that the RPs bind as components of a preassembled multiprotein complex, perhaps 60S or pre-60S subunits. These findings further indicate that the presence of RPs complexes at transcription sites might be a general feature of eukaryotic cells and functionally important. Unexpectedly, the RPs' chromosomal association is highest at centromeres and tRNA genes—the RPs were found at 167 of the 171 tRNA genes assayed. These findings raise the intriguing possibility that RP complexes are involved in tRNA biogenesis and possibly centromere functions.
Keywords: ribosomal proteins, nuclear function, tRNA genes, centromere
INTRODUCTION
It is generally expected that most ribosomal protein molecules (RPs) will be found in ribosomes (Warner 1989; Perry 2007), so the finding that at least 20 RPs—and rRNA—are present at transcription sites in Drosophila polytene chromosomes was an unexpected indication that ribosomal subunits associate with nascent mRNAs (Brogna et al. 2002). However, a later study showed that RPs bind to noncoding RNA genes in Saccharomyces cerevisiae: This suggested that the association might be independent of the translatability of the transcript and might involve free RPs that are not assembled into ribosomes (Schroder and Moore 2005). Several examples of RPs with extra ribosomal functions at transcription sites have been reported. Some RPs bind their own mRNA, pre-mRNA, or promoter and autoregulate their own expression by affecting translation, splicing, or transcription (Wool 1996; Lindstrom 2009; Warner and McIntosh 2009; De and Brogna 2010). There is also evidence of RPs binding with transcription factors at the promoters of other genes: RpL11 binds the oncoprotein c-MYC at the promoter of c-MYC target genes and inhibits transcription in human cells (Dai et al. 2007, 2010); RpS3 is a subunit of the NF-κB DNA-binding complex involved in chromatin binding and transcription regulation of specific genes (Wan et al. 2007), and RpL22, and possibly other RPs, bind histone protein H1 and suppress transcription in Drosophila (Ni et al. 2006).
Clearly, individual RPs can have specific functions at particular genes. The issue, however, is why multiple RPs are found together at transcription sites of a number of unrelated genes. If each RP binds individually, it should only associate with sites that match its individual RNA-binding or protein-binding affinities, and this would make it hard to explain why several RPs are found together at the same sites. It is possible that the presence of RPs at transcription sites is not functionally significant—they might be synthesized in excess of what is incorporated into ribosomes, with the “excess” proteins interacting nonspecifically with other proteins and/or nucleic acids while transiting the nucleoplasm (Lam et al. 2007). Most RPs are very basic (pI > 10), so at high concentrations they might bind nonspecifically to chromatin. Previous studies, however, have indicated that mechanisms that rapidly degrade excess RPs tend to keep the cellular concentrations of free RPs low (Warner 1977, 1989; Maicas et al. 1988; Lam et al. 2007; Perry 2007).
Here, we have investigated how widespread the association of three representative 60S RPs to chromosomal sites is in Schizosaccharomyces pombe. We have used chromatin immunoprecipitation (ChIP) assays to assess their association with some individual genes and ChIP-on-chip to identify sites to which RPs bind across the whole genome. We found that these three RPs tend to be most highly associated with a common set of at least 122 transcription loci—including 49 protein-coding and 65 noncoding RNA genes. The similar distributions of the three RPs suggest that they are bound as components of complexes consisting of multiple RPs, perhaps 60S or pre-60S subunits.
RESULTS
RPs associate with actively transcribed genes
To investigate whether RPs associate with actively transcribing genes in S. pombe we used the ChIP assay. We analyzed three 60S RPs: RpL7, RpL11, and RpL25. These evolutionarily conserved RPs were previously reported to associate with chromosomal sites in Drosophila and budding yeast (Brogna et al. 2002; Schroder and Moore 2005; Ni et al. 2006). Like other RPs in yeasts—both S. pombe and S. cerevisiae—these are encoded by duplicated genes called a or b isoform in S. cerevisiae or identified by a numeral suffix, typically, 01, 02, or 03 in S. pombe, which encode identical or very similar proteins (Komili et al. 2007).
We genetically tagged one of the paralogs of these proteins with three copies of the hemagglutinin epitope (3HA) by homologous recombination of the endogenous S. pombe genes (SPAC664.06, rpl7/rpl703; SPBC17G9.10, rpl11/rpl1102, and SPBC4F6.04, rpl25/rpl2502) (Fig. 1A). Western blot analysis of total protein extracts shows stable tagged proteins of the expected sizes (Fig. 1B). The tagged proteins appear to be functional, as they are incorporated into polysomes with only trace amounts of the proteins running into lighter fractions (Fig. 1C), and the recombinant strains grow as well as the wild type in serial dilution spot assays (Fig. 1D).
FIGURE 1.
HA-tagged RPs are functional. (A) Agarose gel showing that the PCR products expected from correct HA tagging in all integration strains; the primers were a gene-specific forward primer corresponding to the 3′ end of the gene and a common reverse primer corresponding to the kanMX6 tagging cassette (primers sequences in Supplemental Table S2). (B) Western blotting analysis of whole-cell protein extracts of cells expressing the HA-tagged RPs indicated. (C) Polysome fractionation and Western blot visualization of the HA-tagged RPs. (D) Serial dilution colony spotting assay of the tagged strains (from left to right, ∼105, 104, 103, and 102 cells/mL, 4 μL were spotted onto YES plate).
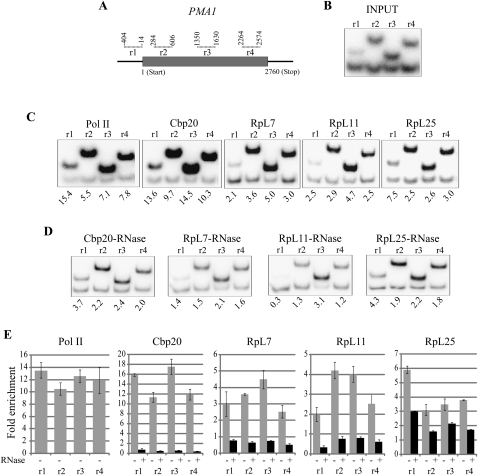
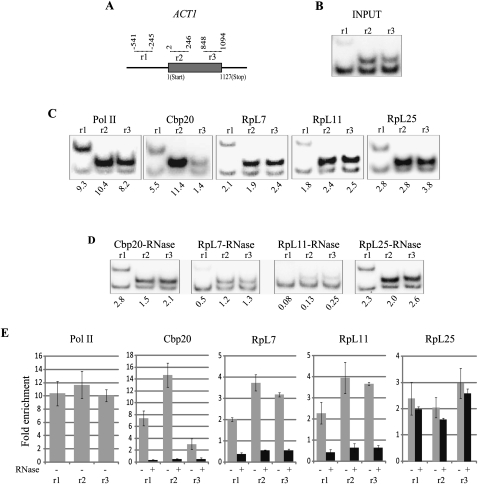
Initially, we tested for association of RPs with chromatin by examining the PMA1 (Fig. 2A) and ACT1 (Fig. 3A) genes, which are constitutively transcribed at a high level and have been used as models in many previous ChIP-based studies (Holstege et al. 1998; Komarnitsky et al. 2000; Abruzzi et al. 2004; Wilhelm et al. 2008). PMA1 codes for plasma membrane ATPase 1 and ACT1 for cytoplasmic actin.
FIGURE 2.
RPs are present at the PMA1 gene locus. (A) Schematic diagram of the PMA1 gene; gray bar represents the gene ORF; the PCR amplicons used for the ChIP assay are indicated by dotted lines above (numbers correspond to the primers positions relative to start codon). (B) Polyacrylamide gels showing radiolabeled PCR products produced by the PMA1-specific primer pairs (top bands) and by the pair specific for the intergenic region (bottom bands); using input DNA before ChIP. (C) PCR products as in B using ChIP-enriched DNA from the strains indicated. The relative enrichment of PMA1 DNA relative to intergenic sequence is expressed as ratio of the intensitity of the same fragments produced with the input DNA. (D) PCR analysis as in C of samples treated with RNases A and T1 prior ChIP. (E) Real-time PCR quantification using the same set of primers as above (indicated in the x-axis). Fold enrichments (y-axis) are relative to the intergenic control fragment and are calculated as ratio of ratios as in B. Gray bars show ChIP experiments not treated with RNase, black bars with RNase. Error bars represent standard deviation of three repeats.
FIGURE 3.
RPs are present at the ACT1 gene locus. (A) Schematic diagram of the ACT1 gene, gene ORF is represented by gray bar; dotted lines indicate PCR amplicons as in Figure 2. (B) Polyacrylamide gel with radiolabeled PCR products produced by the ACT1-specific primer pairs (top bands) and the pair corresponding to the intergenic region (bottom bands), using input DNA before ChIP. (C) PCR as in B of ChIP-enriched DNA. The relative enrichment of ACT1 fragments over the intergenic region was calculated as in Figure 2. (D) PCR of samples treated with RNases A and T1 prior ChIP as in C. (E) Quantification of ChIP-enriched DNA using real-time PCR with the primers indicated in the x-axis.
First, we optimized the ChIP assay with a monoclonal antibody against Pol II (8WG16). As expected, Pol II is at both genes and, relative to the control intergenic region, it was highly enriched throughout these two genes (promoter, beginning, middle, and end of coding region) (Fig. 2B,C) PMA1; (Fig. 3B,C) ACT1. The ChIP enrichment was estimated with two assays—by quantification of radioactive PCR bands from gels (panels C) and by quantitative real-time PCR (panels E)—these assays gave similar results.
As an additional positive control, we also tagged the Cbp20 subunit of the cap-binding complex (CBC), which is expected to bind the cap of all nascent pre-mRNAs. Similarly to Pol II, we found Cbp20 throughout the locus in both genes (Figs. 2C–E, 3C–E). The Cbp20 ChIP enrichment is RNA dependent: Treatment of the chromatin with RNase destroys the association (Figs. 2D,E, 3D,E)—and the high RNase sensitivity suggests that the association is with nascent RNA. Many previous studies have reported that digestion of RNA within cross-linked chromatin is not efficient even with high RNase concentrations (Abruzzi et al. 2004). Reasoning that ionic detergents in the chromatin preparations probably inhibited the RNase activity in those studies, we introduced an extra step into our assay, in which the sonicated chromatin is column purified before incubation with RNase: This modification dramatically improves the reliability of the RNase sensitivity assay (see Supplemental Fig. S1; Materials and Methods).
After validation of the ChIP technique, we assayed chromatin association of the HA-tagged RPs. We found that all three proteins associate with the two test genes, clearly more than with the intergenic region. The level of association is lower than that of Pol II or Cbp20—the caveat of this conclusion, however, is that even for proteins tagged with the same tag, epitope availability could affect the quantification, thereby giving misleading information on the relative abundance of the proteins. For RpL7 and RpL11, the highest enrichment is in the coding regions (Figs. 2C–E, 3C–E), but RpL25 is highest at the promoter (Figs. 2C–E, 3C–E). The enrichment was quantified by both radioactive PCR and real-time PCR, and again, the two assays gave similar results, with the gene-associated RPs showing two- to sevenfold enrichment relative to the intergenic region. The association of RpL7 and RpL11 with chromatin is very sensitive to RNase treatment, greatly reducing the ChIP signal, but the association of RpL25 is much less sensitive to RNase digestion (particularly with ACT1) (Fig. 3D,E). Perhaps some of the RpL25 molecules can also associate with chromatin via an RNA-independent mechanism, maybe by binding DNA directly (see Discussion).
In summary, the three RPs we have tested associate with active genes, the association is RNA dependent (this is apparent for RpL7 and RpL11, but only partially for RpL25). The association is highest with coding regions, but also apparent with the promoter, especially for RpL25.
RPs associate both with protein-coding genes and with other type of genes
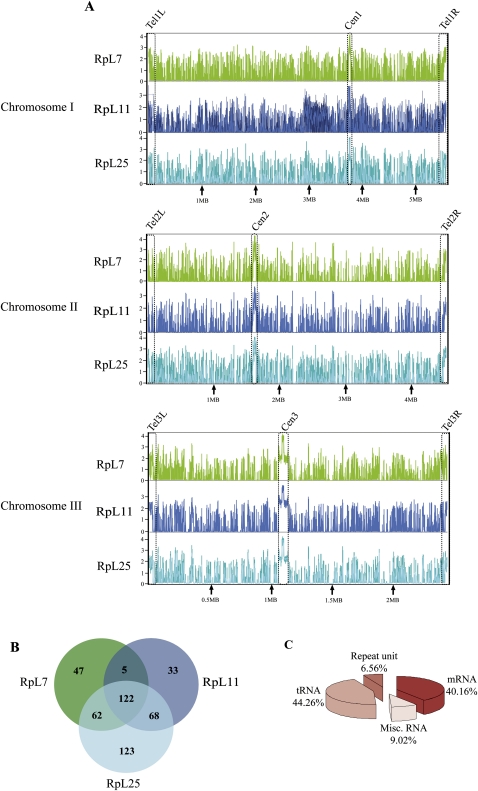
Finding RPs at both the PMA1 and ACT1 genes prompted us to investigate what other genes these proteins associate with. To do this we used ChIP-on-chip assays: Chromatin-immunoprecipitated DNA was hybridized to S. pombe genomic tiling arrays (Affymetrix GeneChip, see Materials and Methods). We analyzed the same three strains described above; for RpL7, RpL11, and RpL25, two independent biological replicas of the experiment were performed for each protein by assaying chromatin samples prepared at two different times from independent cultures. We analyzed the raw data and identified statistically significant binding sites with the MAT software, which is a program specifically designed for the analysis of ChIP-on-chip data produced using tiling arrays (Materials and Methods; Johnson et al. 2006). Between the replicas there is a high probe-to-probe signal correlation (Pearson correlation ≥0.76; see Supplemental Fig. S2), yielding almost identical gene enrichment profiles.
The analysis revealed that the three RPs associate with many loci throughout the three chromosomes: We identified 460 genes/genomic regions that were highly enriched with at least one of the proteins (Fig. 4A). RpL7 associated with 236, RpL11 with 228, and RpL25 with 375, and all three RPs associated with 122 of these genes/genomic regions (Fig. 4B; Supplemental Table S2, the 122 hits with enrichment score). The bioinformatical analysis indicates that RpL25 binds extra loci compared with the other two proteins. Visual inspection of the data, however, indicates that the binding profile is very similar between the three proteins (see below). Therefore, RpL25 might not be binding many extra loci, but it simply binds more strongly, so that at the stringent cutoff we used in the analysis it appears that there are more enriched loci than with the other two proteins.
FIGURE 4.
Genome-wide association of RPs. (A) Chromosomal binding profiles of the RPs on all three chromosomes, analyzed by the MAT software and visualized with the IGB software. Each RP is shown with a different color on the graph: green (RpL7), navy blue (RpL11), and sky blue (RpL25); x-axis shows the distance from the left chromosome end in megabases (Mb); the y-axis log2 MAT enrichment score (0–4). Each plot is based on two ChIP–chip biological replicas and two control arrays hybridized with input DNA, used as standard in all experiments. Position of centromeres (cen) and telomeres (tel) are highlighted with dotted boxes. (B) Venn diagram showing the number of genes/genomic regions associated with the three RPs. (C) Pie-chart showing the proportions of the genes that are associated with all three RPs that fall within various gene classes.
The hits are clearly shared between the three proteins. This tendency for the three RPs to bind to the same genes is highly statistically significant (P < 10−6, see Materials and Methods). The hits that are not shared have lower enrichment scores; however, with a less-stringent cutoff (P-value of 1 × 10−3 instead of 1 × 10−4, see Materials and Methods) they are also enriched above background with all three proteins. At the 1 × 10−3 cutoff, there are twice as many enriched regions (data not shown). Although many regions were not included in the hits list because their score was below the stringent 1 × 10−4 cutoff, enrichment peaks are apparent in close-up views of genomic regions with all three proteins. For example, a visual inspection of the enrichment profile over the 200-kb region around the PMA1 locus shows several enrichment peaks over flanking loci; yet, at the stringent cutoff that we have selected, the MAT software only considers the RNA-coding gene SPAPB15E9.02c enriched in all experiments, while PMA1, which the gene-specific ChIP experiments clearly indicated as enriched with all three RPs, was considered enriched only in the RpL11 and RpL25 experiments (see Supplemental Fig. S3). For these reasons, the number of regions that are associated with RPs is probably a conservative underestimate.
To gain further insights, the shared 122 hits were classified according to whether the genes encode proteins or nonprotein-coding RNAs. Surprisingly, only 40.16% (49 loci) are protein-coding genes. Of the others, 44.26% (54 loci) are tRNA genes, 6.56% are in repeat regions of the genome and 9.02% correspond to miscellaneous RNA genes, including one snoRNA, two snRNAs, one 5S rRNA, and seven noncoding RNAs (Fig. 4C). As detailed below, large plateaus of enrichment encompass all centromeric regions (Fig. 4A, centromeres are highlighted).
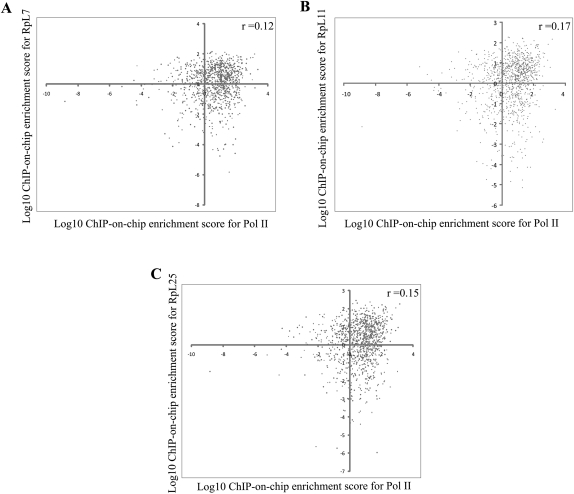
We also compared the enrichment of RPs on individual genes with published values for the enrichment of Pol II at the same genes (Wilhelm et al. 2008); surprisingly, we only found a small degree of correlation (Fig. 5). A second correlation analysis between ChIP-on-chip RPs enrichment and transcript levels—as a proxy for the transcription rates of the corresponding genes (Neil et al. 2009; Xu et al. 2009)—also showed little correlation between these processes (data not shown). For example, there is no sign of enrichment at some highly expressed Pol II genes like RpS17 (SPCC24B10.09) and Sec26 (SPBC146.14c); we also assayed the histone H2A gene (SPAC19G12.06c) by gene-specific ChIP during S phase and found no enrichment (data not shown). In summary, these data indicate that RPs bind specific genes rather than any transcribed locus (see Discussion).
FIGURE 5.
RPs' chromosomal association shows little correlation to Pol II occupancy. Scatter plot showing RPs versus Pol II occupancy at those regions that were found associated with Rps, based on published genome-wide enrichment scores for Pol II (x-axis) (Wilhelm et al. 2008) and the RPs enrichement (y-axis). A shows correlation with RpL7, B with RpL11, and C with RpL25. Pearson correlation is shown at the top right of each panel.
RPs are enriched on centromeric regions
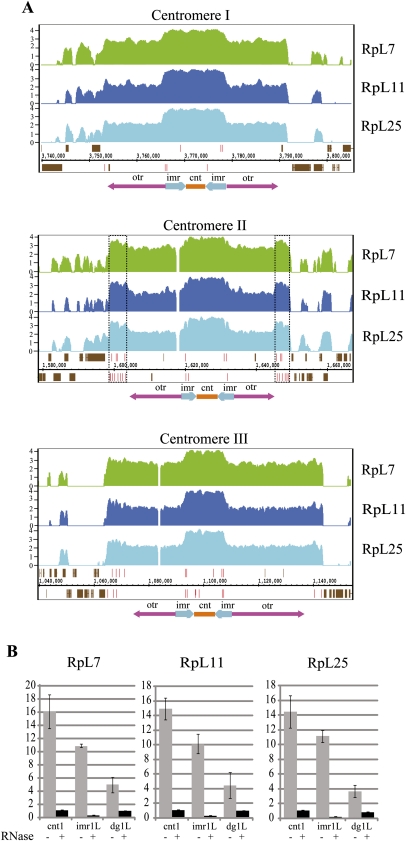
Unexpectedly, the nonprotein-coding loci with which the RPs associate are most abundant in the telomeric and centromeric regions (Fig. 4). The most enriched class of genes are tRNA genes (Fig. 4C). The enrichment is highest at tRNA genes found in dense clusters in the centromeres (Fig. 4A). Fission yeast centromeres span 35–110 kb. Each has a central core of nonrepetitive DNA (cnt), which is flanked by two repeat regions, the innermost repeats (imr) and the outer repeats (otr); the outer regions contain multiple copies of dh/dg repeats (Fig. 6A; Pidoux and Allshire 2004). The ChIP-on-chip analysis indicates that RPs associate with all three centromeres, and that the association is highest at the cnt and imr regions (Fig. 6A). To investigate this association further, we examined segments of the cnt, imr, and dg domains by sequence-specific ChIP and real-time PCR. The results confirm that the RPs associate with all three regions and that the association is sensitive to RNase treatment (Fig. 6B). Surprisingly, we found that the RpL25 association with centromeric regions, unlike at the ACT1 and PMA1, is also very sensitive to RNase treatment (Fig. 6B). The otr regions are transiently transcribed into small interfering RNAs (siRNA), which drive heterochromatin formation and transcription silencing (Volpe et al. 2002; Chen et al. 2008). Therefore, the association at these regions might be with nascent transcripts of the RNA-coding loci. Surprisingly, the association at the cnt region is also RNase sensitive. In this region there are only a few RNA-coding genes annotated in GeneDB, including some tRNAs, and until recently was believed not to be transcribed to any detectable extent (Wilhelm et al. 2008); a recent study, however, indicates that a large proportion of it is, in fact, transcribed by Pol II, yet RNAs fail to accumulate because they are rapidly destroyed by the exosome (Choi et al. 2011).
FIGURE 6.
Centromeric regions bind RPs. (A) ChIP-on-chip enrichment graphs for RPs at the centromeric region of each chromosome, generated with the IGB software. The map below each panel shows a schematic of fission yeast centromeres, with the three major domains labeled otr, imr, and cnt (see text for more details). Centromeric tRNA gene loci are indicated by red lines, and the ORFs of protein-coding genes by brown boxes. Two tDNA clusters in centromere 2 are highlighted by dotted boxes. Values on the y-axis show log2 enrichment scores. (B) Real-time PCR quantification of RPs enrichment at three specific centromeric repeats regions, with or without RNase digestion prior to ChIP.
The association of RPs with tRNA genes requires their transcription
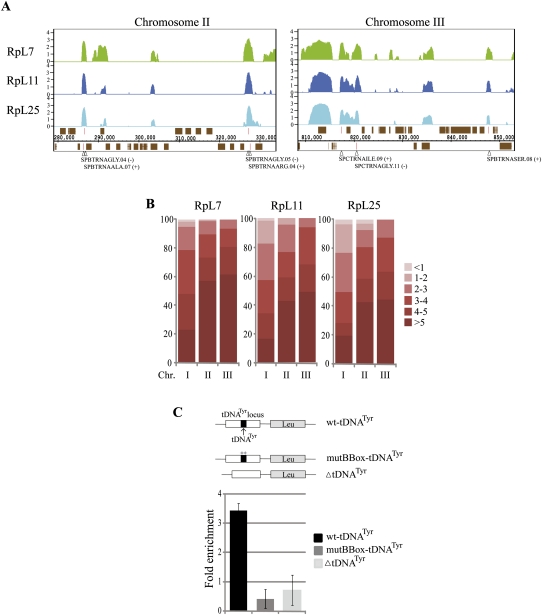
As mentioned above, within the centromeric regions there is a tendency for the RPs' association to be highest in correspondence of tRNA genes loci (Fig. 6A, tRNA genes are indicated by vertical red lines). Multiple tRNA genes are present at all three centromeres and are also present at the border between otr regions and the chromosome arms (Wood et al. 2002). For example, two apparent peaks of RPs enrichment in chromosome II are at the borders with the centromere and coincide with two dense clusters of tRNA genes (Fig. 6A, centromere II, tRNA clusters highlighted). RPs also clearly associate with tRNA genes dispersed in different chromosomal regions (Fig. 7A, specific tRNA genes are indicated by arrowheads). As reported above, at the stringent cutoff (P-value 1× 10−4) the MAT software indicate 54 tRNA genes as highly enriched (Fig. 4C). To evaluate the enrichment at all tRNA loci, we classified all known tRNA loci in a given chromosome into six classes depending on the RP enrichment scores, ranging from less than onefold to more than fivefold enrichment above background (the top 54 tRNA genes are on average 9.36-fold enriched with all three RPs, see Supplemental Table S2). The classification shows that as many as 170 of the annotated 171 tRNA genes in the array (there are 196 tRNA genes in the genome, of which 53 are in centromeric regions) (http://www.sanger.ac.uk/Projects/S_pombe/genome_stats.shtml) might associate with RPs, RpL7 (170), RpL11 (170), and RpL25 (167) (Fig. 7B).
FIGURE 7.
RPs associate with most tRNA genes. (A) Example of RPs' association at noncentromeric tRNA genes. The peaks of enrichment at tRNA loci are indicated by red vertical lines and by arrowheads at the bottom; individual tRNA are labeled (+ and − refer to genes in the upper and lower DNA strands, respectively). Values on the y-axis show log2 enrichment scores. (B) Histogram displaying the association of the RPs with all known 171 tRNA genes: All tRNA genes were classified in six classes based on the increasing degree to which they were RPs enriched (from less than onefold to more than fivefold, indicated by the color legend on the right). The heights of the bars represent cumulative percentages of the tRNAs encoded by each chromosome. (C) Schematic of the tRNATyr constructs. The top panel shows the wild-type construct; below that, a derivative carrying a mutation in the promoter (B Box deletion); and at the bottom, a derivative carrying a deletion of the entire tRNA coding sequence. Graph on the bottom shows real-time PCR quantification of ChIP enrichment for RpL7-HA at the different tRNA constructs.
To further investigate the association of RPs with tRNA genes, we assessed the recruitment of RpL7 to an ectopic tDNATyr construct integrated at the leu1+ locus (Pebernard et al. 2008). We found that RpL7 associates with a wild-type copy of the gene construct, but not with two mutant derivatives (Fig. 7C). One mutant (mutB Box-tDNATyr) carries a C→G mutation in the B Box of the Pol III promoter, inhibiting TFIIIC binding and transcription (Kurjan and Hall 1982; Baker et al. 1986; Pebernard et al. 2008), and the other (ΔtDNATyr) lacks the tRNA-coding sequence (Pebernard et al. 2008). It seems that RpL7 only binds to this tRNA gene if it is being actively transcribed.
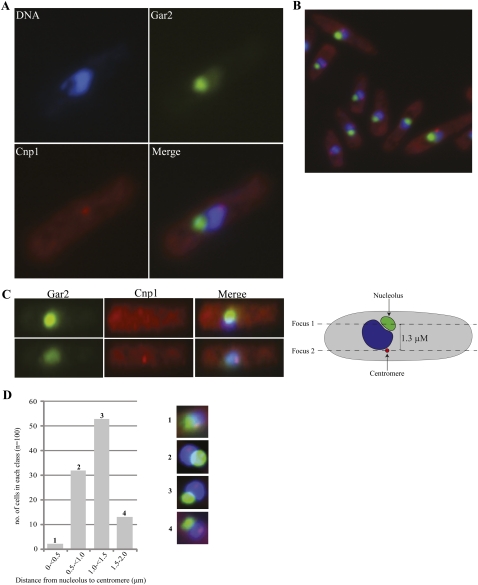
The centromeres are not adjacent to the nucleolus
In S. cerevisiae, tRNA genes are dispersed throughout the chromosome arms, yet within the intact nucleus they appear to cluster in the vicinity of the nucleolus (Thompson et al. 2003). It is conceivable that a similar phenomenon occurs in S. pombe, and that the prominent association of RPs with the centromere is simply the passive consequence of the region being spatially adjacent to the nucleolus. To evaluate this possibility we examined the spatial distribution of the nucleolus and centromere in cells using an antibody against Cnp1 (Kniola et al. 2001) in a strain carrying GFP-tagged Gar2, a nucleolar marker (Win et al. 2005). The results indicate that centromeres and the nucleolus are unconnected in S. pombe (Fig. 8A). The analysis was done with asynchronous culture and does not show any colocalization in cells at any stage of the cell cycle (Fig. 8B). In a few cells, the centromeric and nucleolar markers seemed to be nearby; however, in all cases a more detailed analysis showed that the two markers are in distant planes of focus (Fig. 8C); the seeming proximity is simply due to the 2D projection of the signals, and when only the nucleolar signal is visible, the weaker centromeric signal is simply out of focus (Fig. 8C, cf. the top and bottom pictures, which were taken at different focal planes). A measurement of the distance between the centers of the nucleolar and centromeric signals in pictures of 100 unsynchronized cells with both signals visible shows that, essentially in all cells, the flat projection distance between the two structures is >0.5 μm (Fig. 8D). Therefore, the data appear to exclude that the centromere and associated tRNA genes can contact the nucleolus at any stage of the cell cycle.
FIGURE 8.
Centromere and nucleolus are spatially detached. (A) Micrographs showing the subnuclear localization of the nucleolar protein Gar2-GFP (green) and the centromeric protein Cnp1 (red) in interphase S. pombe cells. 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI, blue) was used to stain the DNA. (B) Micrograph of Gar2 and Cnp1 proteins at various cell cycle stages. (C) Micrographs of a cell taken at two different focal planes (at the nucleolus or centromere). On the right is a schematic of the cell with the nucleolus in green, the DNA in blue and the centromere in red. (D) Histogram showing the distribution of cells within four different nucleolus-centromere distance classes (in microns [μm]). The quantification is based on images of 100 unsynchronized cells acquired with a Hamammatsu Orca R2 CCD camera using a 100× objective. Distances were measured with the Nikon NIS-element software from the center of the nucleolar to the center of the centromeric signal.
DISCUSSION
The results we have presented here indicate that RpL7, RpL11, and RpL25—chosen as representative of all RPs—are present at many transcription sites on S. pombe chromosomes. This finding confirms previous observations with the polytene chromosomes of Drosophila (Brogna et al. 2002) and indicates that the physical association of RPs with transcription sites might be a general feature of eukaryote cells. As in the previous study in S. cerevisiae (Schroder and Moore 2005), we found RPs both at protein-coding and at nonprotein-coding genes in S. pombe. The chromosomal association of RPs is RNA dependent: RNase treatment eliminates the ChIP signal for RpL7 and RpL11 and reduces that of RpL25. Perhaps RpL25 binding is less sensitive to RNase because it can bind DNA directly—this protein belongs to the RpL23 family of RPs, which in higher eukaryotes contains a histone-H1-like domain in the N terminus that could bind DNA (Ross et al. 2007). The observed RNase sensitivity suggests that RPs associate with RNAs at both proteins- and RNA-coding loci. However, we detect little correlation between RP enrichment at genes and either Pol II occupancy or the steady-state level of the transcripts. Highly transcribed genes typically have more Pol II molecules engaged at the locus (Wilhelm et al. 2008), so their DNA should be more accessible and replete with nascent RNA (Jackson et al. 1993; Wansink et al. 1996). Furtheremore, RPs are not at some highly transcribed genes. The observations are consistent, with their recruitment being selective and not primarily driven by their passive electrostatic affinity for RNA or DNA or the hyperphosphorylated Pol II C-terminal domain. The observation that the three RPs are typically found simultaneously at the same sites suggests that the proteins may be recruited together, most likely as parts of preassembled complexes, perhaps even as entire ribosomal subunits. In line with this interpretation, the RNase sensitivity of the interaction might in part be due, particularly at promoter regions, to the RPs being bound to the rRNA.
An unexpected finding of this work is that the RPs are mostly enriched at centromeres, and that their association is also RNase sensitive. Unlike in the euchromatic sites, the association of RpL25 with centromeric regions is also very sensitive to RNase treatment; perhaps the protein cannot bind DNA directly in heterochromatin and the specialized CENPACnp1 chromatin that assemble on the centromere core, and thus, the interaction is only with nascent RNAs (Choi et al. 2011).
Within the centromeres, the association is most apparent at tRNA genes, but tRNA genes located in other chromosomal regions also associate with the RPs. Strikingly, tRNA genes make up ∼0.1% of the S. pombe genome, yet represent >44% of the RPs' binding sites. Similar to other regions, statistical calculation and visual inspection of the enrichment profiles strongly indicate that all three RPs associate with the same centromeric and tRNA loci, suggesting also that, here, the proteins are recruited together as preassembled complexes. As mentioned above, in S. cerevisiae, tRNA genes physically associate with the nucleolus (Thompson et al. 2003) and dispersed tRNA and other Pol III genes appear to relocate at the centromeres in S. pombe (Iwasaki et al. 2010). Therefore, it can be argued that centromeres and tRNA genes preferentially associate with RPs simply because these regions are physically adjacent to the nucleolus. Our data argue against this explanation. We found no indication of the centromeres being proximal to the nucleolus at any stage of the cell cycle, consistent with our current understanding of how chromosomes are organized in the yeast nucleus (Zimmer and Fabre 2011). Furthermore, in S. pombe the rDNA repeats are at the telomeres of chromosome III (Wood et al. 2002); yet, telomeres and centromeres appear to stay separated throughout the cell cycle (Funabiki et al. 1993). In addition, the regions near the rDNA repeats are not any more enriched than those flanking the telomeres of the other two chromosomes. The rDNA repeat regions, as expected, bind the RPs (Supplemental Fig. S4); however, the level of enrichment is lower than at other sites—perhaps due to the microarray carrying probes for only one to two rDNA repeats rather than the 100–120 of the genomic regions (Wood et al. 2002; Materials and Methods). In summary, at this stage our conclusion is that the RPs' chromatin association is not a passive consequence of spatial proximity to the nucleolus. We speculate that the RP complexes might be involved in centromere functions and tRNAs biogenesis. This is in agreement with the reports that three other RPs—RpL6, RpL26, and RpL14—copurify with TFIIIE in S. cerevisiae (Dieci et al. 2009) and that RpL11 represses Pol III transcription in mammalian cells (Dai et al. 2010).
It is communally understood that to carry out the extra ribosomal functions RPs need to be detached from the ribosomal subunits—future studies might change this view (De and Brogna 2010); in E. coli, for example, ribosomal protein S10 (a classic example of RP moonlighting, see Warner and McIntosh 2009) can bind the transcription factor NusG while still associated to the small ribosomal subunit (Burmann et al. 2010)—this interaction couples bacterial transcription to translation (Proshkin et al. 2010). Here, many of the genes with which RPs associate do not encode proteins. While this argues against translation occurring at these nonprotein-coding loci, it leaves open the possibility that tRNAs, which readily bind to the ribosome in vitro nonenzymatically (Prince et al. 1982; Fahlman and Uhlenbeck 2004), might first associate to ribosomes or preribosomes at tRNA transcription sites. Of course, the interaction with protein-coding genes might have a different explanation than that of tRNA and other RNA-coding loci. Future work shall focus on unveiling the functional significance of RP complexes at transcription sites and address the important issue of whether bona-fide ribosomal subunits are present.
MATERIALS AND METHODS
Yeast strains and methods
The complete list of S. pombe strains used in this study is shown in Table 1. Fission yeast transformation was as described earlier, with minor modifications (Gietz and Woods 2006). Typically, a 50-mL culture grown to an OD600 of 0.7–0.8 was pelleted, washed with water, and resuspended in 100 μL of lithium acetate buffer. Then, 1 μg of linearized plasmids or 4 μg of PCR product was mixed with 4 μg of ssDNA, added to the cells, and incubated at room temperature for 10 min, followed by the addition of 260 μL of 50% PEG4000, incubation for a further 60 min at 30°C, followed by heat shock at 42°C for 15 min. The target proteins were HA-tagged by homologous recombination using a PCR-amplified fragment containing the kanMX6 cassette flanked by targeting sequences (Bahler et al. 1998); all of the PCR primers are listed in Supplemental Table S1.
TABLE 1.
List of strains used in the study
Western blot analysis and polysome fractionation
Protein extraction for Western blot analysis was done as described previously (Matsuo et al. 2006). For detection of HA-tagged proteins, blots were incubated with a mouse monoclonal anti-HA (12CA5, CRUK). The secondary antibody was a rabbit HRP-conjugated anti-mouse IgG, which was detected with chemiluminescent HRP substrate (Immobilon Western, Millipore). The chemiluminescent signal was analyzed with Quantity One (Bio-Rad). For polysome analysis, yeast cultures were grown in 50 mL of YES to an OD600 of 0.3–0.4 and treated with 100 μg/mL cycloheximide for 15 min prior to harvesting. Cell pellets were washed with 10 mL of lysis buffer (20 mM HEPES at pH 7.4, 2 mM magnesium acetate, 100 mM potassium acetate, 100 μg/mL cycloheximide, 0.5 mM dithiothreitol). The cells were pelleted again and resuspended in 600 μL of lysis buffer with 40 U/mL of RNase inhibitor (RNaseOUT, Invitrogen) and EDTA-free protease inhibitor cocktail (Roche). Whole-cell extracts were prepared by glass-bead grinding using the Precellys 24 (Bertin Technologies) beads shaker—typically three rounds of 15 sec at 6300 rpm. The whole-cell extracts containing polysome were adjusted to 10 OD260 U/mL and gently loaded onto the top of the 10%–50% sucrose gradients and centrifuged for 2.5 h at 38000 rpm in a Beckman SW41 rotor. Positions of ribosomal species in the gradient were determined by monitoring OD254 absorbance with a UV monitor (Pharmacia LKB-Optical Unit UV-1). Fractions, 1.0 mL each, were collected and precipitated with trichloroacetic acid and analyzed by SDS-PAGE (10% gel) and Western blotting.
Immunostaining and imaging
Cells were fixed for 8 min by adding freshly prepared formaldehyde solution (3.7% final) directly to the liquid culture immediately after removing from the incubator, and processed as previously described (Kniola et al. 2001). The primary antibody used was sheep anti-Cnp1 (provided by Robin Allshire), the secondary antibody was an Alexa Fluor555 conjugated (Molecular Probes). Imaging was performed using the Eclipse Ti Nikon Microscope.
ChIP
ChIP was performed according to Abruzzi et al. (2004). Cleared cell extracts were incubated with Protein-A Sepharose beads (Sigma) prebound to anti-HA (12CA5, Roche) or anti-Pol II (8WG16, Covance) antibodies. The amount of DNA in the “input” and “IP” samples was quantified using both radioactive (PCR fragments were labeled with 0.03 μCi [α32P]dCTP with 24 cycles of amplification) and real-time PCR (SYBR Green, QIAGEN). Radioactive PCR signals were quantified with a PhosphorImager (Bio-Rad). Normalization and calculation of the ChIP enrichment was done as described previously: The enrichment level was expressed as a ratio of the signal of the gene-specific PCR fragment to that of the intergenic control in IP DNA, divided by the same fragments ratio in input DNA (Komarnitsky et al. 2000; Abruzzi et al. 2004). For the RNase sensitivity test, samples were treated with 7.5 U of RNase A (Sigma) and 300 U of RNase T1 (Sigma) for 30 min (Abruzzi et al. 2004). To improve RNase activity, the sheered chromatin sample was purified by centrifugal filtration before RNase treatment using an YM-10 Microcon cartridge (Millipore); this step removes SDS and other chemicals that block RNase enzymes.
ChIP–chip
Immunoprecipitated DNA was first linearly labeled using Sequenase (USB Corporation) with a random primer flanked by an adaptor that was used for PCR amplification (Robyr and Grunstein 2003). Amplified DNA was fragmented and labeled using the GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix). The labeled DNA was hybridized to S. pombe Tiling 1.0FR Arrays (Affimetrix), (probe labeling, hybridization, and scanning were performed by the Dr. Andy Hayes, COGEME facility, Manchester University, except for the second RpL11 experiment, which was done by Dr. John Arrand at the Affimetrix facility of the School of Cancer Sciences, University of Birmingham). We used the Model-based Analysis of Tiling Array (MAT) software (Johnson et al. 2006) for analysis of the Affymetrix hybridization data. ChIP input DNA was used as a control for the analysis. Enrichment scores were assigned to genomic positions using the S. pombe genome coordinates and a bpmap file for the Affymetrix array (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/GFF; and S. pombe 8/23/07 library). Enriched regions were initially defined at different P-value thresholds; the P-value of 1 × 10−4 was chosen because this was the lowest P-value at which both the experimentally validated PMA1 (all but one experiment) and ACT1 genes were flagged as enriched. Given genes were classified as positive hits only if the enrichment was at least 50% or more of the gene sequence, therefore excluding regions with minimal levels of enrichment. Other than the P-value, default parameters were used with the MAT software. The results of the MAT software were visualized with the Affymetrix Integrated Genome Browser (IGB). In order to test the statistical significance of the overlap between the enriched regions identified with the three proteins, a program was written that randomly sampled the observed number of enriched regions from the total number of unique features of the S. pombe genome (total size of 10694—including all genes and unknown repeat regions) for each of the proteins, and determines what the overlap is between the three samples. We never observed an overlap larger than 6 in 4.5 × 106 simulations, implying a P-value <10−6.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Laura O'Neill, Katy Abruzzi, and Michael Rosbash for help in setting up the ChIP procedure, and Robin Allshire for the anti-Cnp1 antibody. We also thank Andrew Hayes and John Arrand for doing the DNA microarray hybridization and Jurg Bahler for providing the homologus recombination vectors. We thank Tony Carr and Shao-Win Wang for providing strains, Michael N. Boddy for the tDNA vectors, and Kim Piechocki for help with the polysome fractionation. We also thank the people in charge of the BBSRC-COGEME facility (Manchester University) for providing the pilot grant to carry out the microarray experiments, and Marc Buehler, Bob Michell, Oliver Muhlemann, and Michael Rosbash for critically reading the manuscript. S.B. is supported by a Royal Society URF fellowship, and S.D. by a Darwin Trust PhD scholarship. This work was also in part funded by a Wellcome Trust project grant to S.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2808411.
REFERENCES
- Abruzzi KC, Lacadie S, Rosbash M 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Baker RE, Gabrielsen O, Hall BD 1986. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem 261: 5275–5282 [PubMed] [Google Scholar]
- Brogna S, Sato TA, Rosbash M 2002. Ribosome components are associated with sites of transcription. Mol Cell 10: 93–104 [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P 2010. A NusE:NusG complex links transcription and translation. Science 328: 501–504 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI 2008. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC 2011. Identification of non-coding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem doi: 10.1074/jbc.M111.228510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Arnold H, Sun XX, Sears R, Lu H 2007. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J 26: 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H 2010. Ribosomal protein L11 associates with c-Myc at 5 S rRNA and tRNA genes and regulates their expression. J Biol Chem 285: 12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Brogna S 2010. Are ribosomal proteins present at transcription sites on or off ribosomal subunits? Biochem Soc Trans 38: 1543–1547 [DOI] [PubMed] [Google Scholar]
- Dieci G, Ruotolo R, Braglia P, Carles C, Carpentieri A, Amoresano A, Ottonello S 2009. Positive modulation of RNA polymerase III transcription by ribosomal proteins. Biochem Biophys Res Commun 379: 489–493 [DOI] [PubMed] [Google Scholar]
- Fahlman RP, Uhlenbeck OC 2004. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43: 7575–7583 [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA 2006. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol 313: 107–120 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol Biol Cell 21: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR 1993. Visualization of focal sites of transcription within human nuclei. EMBO J 12: 1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniola B, O'Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K 2001. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell 12: 2767–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J, Hall BD 1982. Mutations at the Saccharomyces cerevisiae SUP4 tRNA(Tyr) locus: isolation, genetic fine-structure mapping, and correlation with physical structure. Mol Cell Biol 2: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS 2007. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS 2009. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun 379: 167–170 [DOI] [PubMed] [Google Scholar]
- Maicas E, Pluthero FG, Friesen JD 1988. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol 8: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Asakawa K, Toda T, Katayama S 2006. A rapid method for protein extraction from fission yeast. Biosci Biotechnol Biochem 70: 1992–1994 [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL 2006. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev 20: 1959–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN 2008. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J 27: 3011–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP 2007. Balanced production of ribosomal proteins. Gene 401: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC 2004. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534 [DOI] [PubMed] [Google Scholar]
- Prince JB, Taylor BH, Thurlow DL, Ofengand J, Zimmermann RA 1982. Covalent crosslinking of tRNA1Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc Natl Acad Sci 79: 5450–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Grunstein M 2003. Genomewide histone acetylation microarrays. Methods 31: 83–89 [DOI] [PubMed] [Google Scholar]
- Ross CL, Patel RR, Mendelson TC, Ware VC 2007. Functional conservation between structurally diverse ribosomal proteins from Drosophila melanogaster and Saccharomyces cerevisiae: fly L23a can substitute for yeast L25 in ribosome assembly and function. Nucleic Acids Res 35: 4503–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder PA, Moore MJ 2005. Association of ribosomal proteins with nascent transcripts in S. cerevisiae. RNA 11: 1521–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. 2007. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131: 927–939 [DOI] [PubMed] [Google Scholar]
- Wansink DG, Sibon OC, Cremers FF, van Driel R, de Jong L 1996. Ultrastructural localization of active genes in nuclei of A431 cells. J Cell Biochem 62: 10–18 [DOI] [PubMed] [Google Scholar]
- Warner JR 1977. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol 115: 315–333 [DOI] [PubMed] [Google Scholar]
- Warner JR 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev 53: 256–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB 2009. How common are extraribosomal functions of ribosomal proteins? Mol Cell 34: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Win TZ, Mankouri HW, Hickson ID, Wang SW 2005. A role for the fission yeast Rqh1 helicase in chromosome segregation. J Cell Sci 118: 5777–5784 [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Wool IG 1996. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci 21: 164–165 [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Fabre E 2011. Principles of chromosomal organization: lessons from yeast. J Cell Biol 192: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]