Abstract
In bacteria, stalled ribosomes are recycled by a hybrid transfer–messenger RNA (tmRNA). Like tRNA, tmRNA is aminoacylated with alanine and is delivered to the ribosome by EF-Tu, where it reacts with the growing polypeptide chain. tmRNA entry into stalled ribosomes poses a challenge to our understanding of ribosome function because it occurs in the absence of a codon–anticodon interaction. Instead, tmRNA entry is licensed by the binding of its protein partner, SmpB, to the ribosomal decoding center. We analyzed a series of SmpB mutants and found that its C-terminal tail is essential for tmRNA accommodation but not for EF-Tu activation. We obtained evidence that the tail likely functions as a helix on the ribosome to promote accommodation and identified key residues in the tail essential for this step. In addition, our mutational analysis points to a role for the conserved K131GKK tail residues in trans-translation after peptidyl transfer to tmRNA, presumably EF-G-mediated translocation or translation of the tmRNA template. Surprisingly, analysis of A1492, A1493, and G530 mutants reveals that while these ribosomal nucleotides are essential for normal tRNA selection, they play little to no role in peptidyl transfer to tmRNA. These studies clarify how SmpB interacts with the ribosomal decoding center to license tmRNA entry into stalled ribosomes.
Keywords: tmRNA, SmpB, decoding, EF-Tu, ribosome
INTRODUCTION
Eubacteria contain a conserved quality-control system that rescues ribosomes stalled on truncated mRNAs. Arising from premature termination of transcription or from mRNA decay, transcripts lacking a stop codon trap the ribosome at their 3′ ends for prolonged periods. Stalled ribosomes are rescued by transfer–messenger RNA (tmRNA), a stable RNA that acts both as a tRNA and an mRNA. tmRNA is aminoacylated with Ala by alanyl-tRNA synthetase. Together with its protein partner SmpB, tmRNA enters the empty aminoacyl-tRNA site of stalled ribosomes and adds Ala to the growing peptide chain. The ribosome then resumes translation on the tmRNA template, adding a 10-amino-acid tag to the nascent polypeptide and releasing at a stop codon on tmRNA. This process, known as trans-translation, results in the recycling of stalled ribosomes and the tagging of the aborted polypeptide for degradation by proteases (for review, see Moore and Sauer 2007).
One unsolved puzzle in the trans-translation model is how tmRNA is allowed to enter stalled ribosomes. Prior to undergoing peptidyl-transfer, an aminoacyl-tRNA must pass through robust decoding mechanisms that exclude tRNAs that cannot form correct codon–anticodon pairs. Because decoding is essential for accurate translation of the genetic code, it has been studied with a variety of tools for many years, and as a result, the selection of tRNAs during canonical translation is well understood (see below). During ribosome rescue, however, codon–anticodon pairing cannot occur because tmRNA lacks an anticodon and binds to ribosomes with little or no mRNA in the A-site (Ivanova et al. 2004). While tmRNA entry into stalled ribosomes is not decoding per se, as no genetic information is decoded, tmRNA must somehow trick the decoding machinery into licensing its entry, and do so without a codon–anticodon interaction. This poses a challenge to our understanding of the trans-translation mechanism and the canonical decoding process.
Canonical tRNA selection involves two kinetic discrimination steps that are separated by the essentially irreversible hydrolysis of GTP (Daviter et al. 2006). Aminoacyl-tRNAs are delivered to the ribosome by EF-Tu. In the first selection step, cognate tRNAs trigger GTP hydrolysis by EF-Tu at a faster rate than noncognate tRNAs (Pape et al. 1999; Gromadski and Rodnina 2004); in contrast, noncognate tRNAs dissociate from the ribosome more readily. Following GTP hydrolysis, the aminoacyl-tRNA is released from EF-Tu, allowing it to be fully accommodated within the ribosome. Accommodation is the movement of the 3′-aminoacylated end of a tRNA from EF-Tu to the peptidyl-transferase center ∼70 Å away (Stark et al. 2002; Valle et al. 2003). Cognate tRNAs are more rapidly and efficiently accommodated into the A-site than noncognate tRNAs (Pape et al. 1999); this is the second selection step.
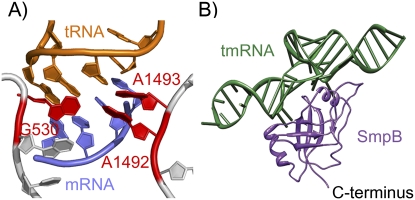
The faster rates of GTP hydrolysis and accommodation that allow cognate tRNAs to pass these two selection steps arise from conformational changes in the ribosome in response to proper codon–anticodon pairing. Conserved 16S nucleotides A1492 and A1493 flip out of a loop in helix 44 to bind the minor groove of the first and second base pairs in the duplex, as shown in Figure 1A (Ogle et al. 2001; Ogle and Ramakrishnan 2005). G530 also undergoes a conformational change to interact with the second and third base pairs. These local movements lead to global conformational changes, namely, a rotation of the head and shoulder of the 30S subunit toward the intersubunit space, effectively closing the 30S subunit over the codon–anticodon helix (Ogle et al. 2002). This change is then communicated to EF-Tu through its interactions with both of the ribosomal subunits and through distortion of the tRNA structure (Schmeing et al. 2009). Mutation of the conserved bases A1492, A1493, or G530 leads to dramatic reductions in the rates of EF-Tu activation and accommodation for cognate tRNAs, leading to error-prone protein synthesis (Cochella et al. 2007).
FIGURE 1.
(A) Structure of the decoding center showing mRNA (blue) paired with cognate tRNA (orange), and the recognition of this pairing by 16S rRNA nucleotides A1492, A1493, and G530 (red). Created with Pymol from PDB 2J00 (Selmer et al. 2006). (B) Cocrystal structure of the tRNA-like domain of tmRNA (green) and SmpB (purple) demonstrating the structural similarity between this complex and a canonical tRNA, where SmpB mimics the anticodon stem. The SmpB C-terminal tail was truncated and would add an additional 27 residues to the C terminus. Created with Pymol from PDB 2CZJ (Bessho et al. 2007).
During the rescue of stalled ribosomes, tmRNA is delivered to the ribosomal A-site by EF-Tu (Hallier et al. 2004; Kaur et al. 2006; Shimizu and Ueda 2006). Since it cannot participate in codon–anticodon pairing, tmRNA must use another means to effect EF-Tu activation and accommodation into the A-site. Several lines of evidence suggest that during ribosome rescue, the decoding center is engaged not by an RNA duplex but by the SmpB protein. SmpB and the tRNA-like domain of tmRNA form a structure that mimics tRNA (Fig. 1B); SmpB acts as the anticodon stem–loop (Bessho et al. 2007). Cryogenic electron microscopy studies of a pre-accommodation complex place SmpB so that the C-terminal tail of SmpB, roughly 30 amino acids long, could be bound in the decoding center (Kaur et al. 2006). Hydroxyl radical and chemical probing experiments have likewise detected interactions of SmpB with ribosomal RNA nucleotides in the 30S A-site (Kurita et al. 2007). Indeed, SmpB binding protects nucleotides A1492, A1493, and G530 from reacting with chemical probes (Nonin-Lecomte et al. 2009).
Taken together, these data support a model in which SmpB acts as a codon–anticodon mimic. By binding the decoding center nucleotides A1492, A1493, and G530, SmpB might trigger the conformational changes associated with canonical decoding, leading to EF-Tu activation and accommodation of tmRNA (Nonin-Lecomte et al. 2009). Currently, this model is supported exclusively by structural and equilibrium binding studies; it has not been determined if these conserved bases in the decoding center are essential for licensing tmRNA's entry into the ribosome. We have tested this model directly, measuring the rates of GTP hydrolysis by EF-Tu and the rate of peptidyl transfer with a series of ribosome and SmpB mutants. Our data show that mutations in the SmpB tail reduce rates of peptidyl transfer to tmRNA, but not GTP hydrolysis by EF-Tu, consistent with a role for the SmpB tail in tmRNA accommodation. We identify key residues in the tail required for this activity and provide evidence that the tail functions as a helix within the ribosome. Surprisingly, analysis of ribosomes containing mutations in A1492, A1493, or G530 reveals that although these nucleotides are essential for both EF-Tu activation and accommodation with normal tRNAs, they play little or no role in licensing tmRNA entry into stalled ribosomes.
RESULTS
Functional importance of conserved residues in the SmpB C-terminal tail
The C-terminal tail of SmpB (residues 131–160 in Escherichia coli) is predicted to bind the 30S A-site (Kaur et al. 2006; Kurita et al. 2007; Nonin-Lecomte et al. 2009), and deletion of the tail sequence prevents peptidyl-transfer to tmRNA (Sundermeier et al. 2005; Shimizu and Ueda 2006). To identify which residues in the tail sequence are essential for trans-translation, we mutated conserved residues to Ala and assayed SmpB and tmRNA activity in vivo. Ribosomes translating a glutathione-S-transferase (GST) construct ending in Glu-Pro-Opal (UGA) stall during termination (Hayes et al. 2002a). These stalled ribosomes were rescued by tmRNA encoding an altered tag, ANDHHHHHHD, that does not target the aborted GST protein for proteolysis (Hayes et al. 2002b). Addition of this tag was detected by anti-His6 antibodies, indicating completion of all the steps in the trans-translation process. The loss of the anti-His6 signal on the blot reports on the inhibition of the trans-translation process by the relevant SmpB mutations. GST levels were also monitored to control for protein expression, loading, and blotting. The GST levels are not expected to vary; in the absence of tmRNA activity, ribosomes stalled on the GST template are presumably released by an alternative rescue pathway involving ArfA, allowing GST synthesis by other ribosomes to continue (Chadani et al. 2010).
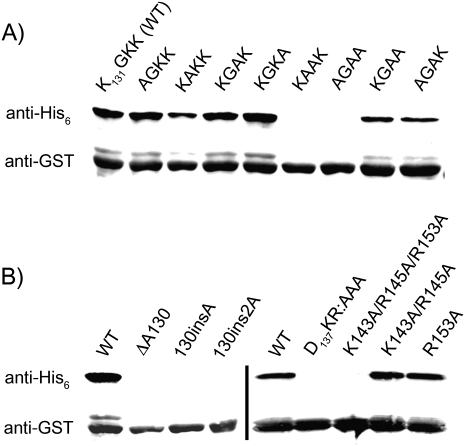
Analysis of the alignment of 470 SmpB genes (Fig. 2) identifies conserved residues that are possible sites of interaction with rRNA, including the D137KR sequence and four partially conserved positive charges at residues 143, 145, 149, and 153 (Andersen et al. 2006). Previously, Karzai and coworkers reported that the D137KR sequence was required for tagging in vivo and for peptidyl transfer to tmRNA (Sundermeier et al. 2005). We confirmed that tagging is lost in the DKR:AAA mutant in our assay (Fig. 3B, right). We also tested the relevance of the positively charged residues by replacing Lys143, Arg145, and Arg153 with Ala. (The basic residue at 149 is not conserved in E. coli, where it is Val.) The triple mutant abolishes tagging activity, while either the double mutant Lys143Ala/Arg145Ala or the single mutant Arg153Ala has no effect (Fig. 3B, right), consistent with the notion that at least one of these conserved positively charged residues is absolutely required for SmpB function.
FIGURE 2.
Alignment of the SmpB C-terminal tail. An alignment of 470 SmpB sequences (Andersen et al. 2006) is displayed as a sequence logo (Crooks et al. 2004). Positively charged residues are highlighted in black. The corresponding E. coli sequence (131–160) is shown below. Residues 137–157 are predicted to be helical by the Jpred algorithm (Cole et al. 2008), based on empirical structural propensities and analysis of the multiple sequence alignment.
FIGURE 3.
Mutations in the C-terminal tail impair SmpB function. Various SmpB tail mutants were analyzed for their ability to support tagging of a stalled protein by tmRNA. The complete GST protein with the stalling sequence Glu-Pro-Stop at the C terminus served as a substrate for tagging. tmRNA was altered to express an ANDHHHHHHD tag; tagging was detected with an anti-His6 antibody. A GST expression control was visualized on the same blot with anti-GST antibodies. (A) Single or multiple Ala mutations reveal essential residues in the conserved K131GKK sequence at the beginning of the tail. (B) To study the junction where the tail leaves the body of SmpB, Ala130 was deleted or one or two Ala residues were inserted between Ala130 and Lys131 (left). The roles of three conserved positive charges in the tail and the D137KR sequence were tested by mutagenesis (right).
The alignment also reveals that the K131GKK sequence is highly conserved. Substitution of Gly132 in the K131GKK sequence by Ala resulted in a threefold reduction in tagging (K131AKK), but no loss of function was observed when the surrounding Lys residues (131, 133, and 134) were substituted, one at a time, with Ala (Fig. 3A). When the two residues with the highest conservation, Gly132 and Lys133, were both mutated to Ala together, tagging was no longer detectable (K131AAK). Tagging was also abolished by replacing all three Lys residues with Ala (A131GAA). Mutation of two Lys residues (K131GAA or A131GAK) lowered tagging approximately threefold. These data point to an important role for Gly132 and suggest that at least one positively charged Lys residue is also essential.
The K131GKK sequence is at the beginning of the C-terminal tail. In the cocrystal structure of the Thermus thermophilus SmpB–tmRNA complex (Bessho et al. 2007), residues corresponding to 131–133 are the last ones seen (the rest of the tail was truncated for crystallization purposes). Residues 131–133 were seen to exit the body of SmpB at the bottom of the protein, on the opposite side from the tmRNA-binding site. To further examine this body/tail junction, we deleted Ala130 or inserted one or two Ala residues between Ala130 and Lys131. Immunoblot analysis revealed that all three of these mutations destroy tagging activity (Fig. 3B). These observations strongly suggest that the spacing or orientation of the tail is critical for SmpB function as it leaves the body of SmpB.
The helicity and function of the C-terminal tail
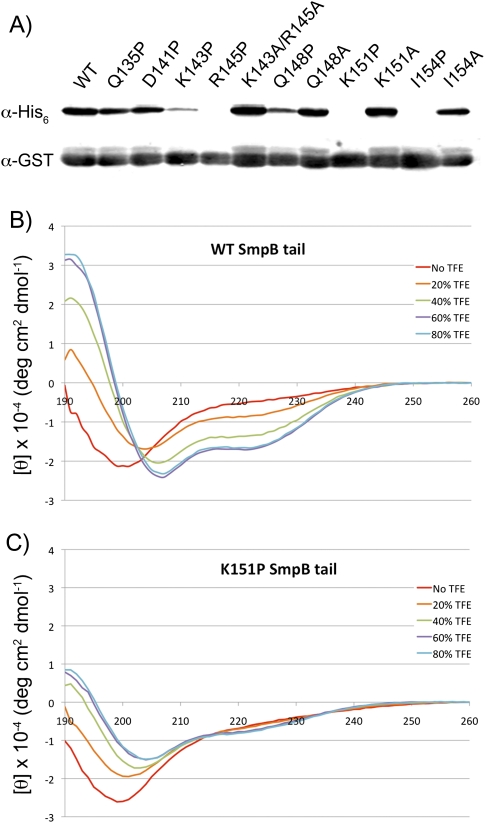
The periodicity of basic residues in the SmpB C-terminal tail suggest that the tail, although unstructured in solution, might form an amphipathic helix inside the ribosome. According to our analysis of the tail sequence with the JPred software (Cole et al. 2008), residues 137–157 are likely to form a helix (Fig. 2). While the helical propensity of the SmpB tail has been noted for some time (Jacob et al. 2005), it has never been determined whether helix formation plays a role in SmpB function. To address this question, we introduced Pro substitutions to destabilize helix formation in the tail. Several residues from 135–154 were mutated to Pro residues one at a time, and tagging was monitored in the immunoblot assay. As a control, corresponding Ala mutants were also tested to ensure that the observed effects result from helix destabilization and not the deletion of essential side chains. None of these Ala mutants reduced tagging (Fig. 4A; data not shown), so any observed effects are due to the introduction of Pro.
FIGURE 4.
Helicity of the SmpB C-terminal tail. (A) Several residues in the tail were mutated to Pro to destabilize helix formation. The same residues were separately mutated to Ala to control for loss of the original residue. (Only relevant Ala mutants are shown.) Addition of the tmRNA-encoded tag was monitored by immunoblot with anti-His6 antibodies. (B) CD spectra of a peptide corresponding to residues 137–157 of E. coli SmpB (a portion of the C-terminal tail). Trifluoroethanol (TFE) was added at various concentrations to induce secondary structural formation. (C) CD spectra of a similar peptide with the Lys151Pro mutation.
Loss of tagging activity in several Pro mutants supports the hypothesis that the SmpB tail forms a helix inside the ribosome to perform its function. Mutation of Lys143, Arg145, Lys151, or Ile154 to Pro reduced tagging to low or undetectable levels, indicating loss of SmpB activity (Fig. 4A). In contrast, replacing Gln135 or Asp141 with Pro had little or no effect, and the Gln148Pro mutation resulted in only a moderate reduction. These data are consistent with a functional requirement for a helix that spans at least residues 143–154, with a possible break surrounding residue 148.
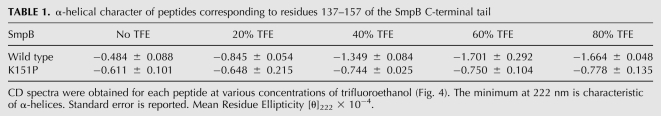
To further characterize the helicity of the tail sequence, we collected circular dichroism spectra of a short peptide corresponding to residues 137–157 (DKRSDIKEREWQVDKARIMKN). As expected, given that the tail is unstructured in solution, the CD spectrum of this peptide in water is consistent with a predominantly random-coil conformation (Fig. 4B). We added trifluoroethanol (TFE) at concentrations up to 80% to stabilize helix formation. TFE enhances the strength of hydrogen bonds between amides in the peptide backbone (Luo and Baldwin 1997), presumably by decreasing hydrogen-bonding with the solvent. In the presence of 40% TFE, the tail peptide exhibits a CD spectrum with α-helical characteristics: a maximum around 190 nm and minima of 208 and 222 nm (Fig. 4B). In contrast, the same peptide with a Lys151Pro mutation exhibits spectra characteristic of a random coil, with a minimum around 200 nm, even at 80% TFE (Fig. 4C). Quantification of the helical character by comparing the mean residue ellipticity at 222 nm confirms that the Pro mutant has little helical character at any TFE concentration (Table 1). These data demonstrate that the wild-type tail peptide has a helical propensity, consistent with earlier predictions. They also show that the Lys151Pro mutation prevents helix formation. Taken together with the finding that the Lys151Ala mutation is tolerated, these studies support our model that loss of helical propensity in the SmpB tail reduces its function in vivo.
TABLE 1.
α-helical character of peptides corresponding to residues 137–157 of the SmpB C-terminal tail
The SmpB tail is required for peptidyl transfer...…
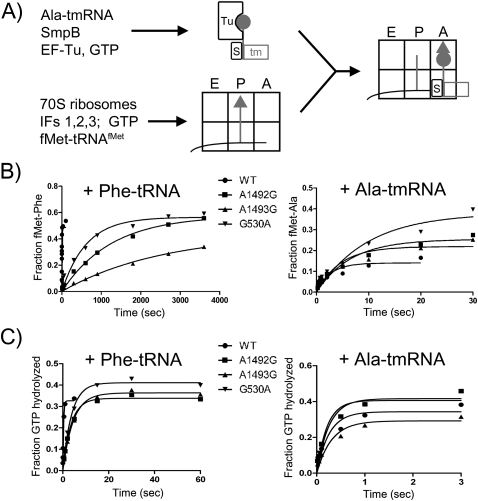
While our results show that certain conserved residues in the SmpB tail are essential for tmRNA function in vivo and that the tail may function as a helix, they do not reveal which step in the trans-translation process is inhibited by mutating the tail, either peptidyl transfer to tmRNA or engaging the tmRNA template sequence to resume translation. To determine which step is inhibited, we used an in vitro assay to further characterize SmpB tail mutants that are inactive in vivo. Because we expected that defects in tmRNA entry into the A-site were involved, we measured peptidyl-transfer rates to tmRNA using purified components (Fig. 5A). Ehrenberg and coworkers previously showed that ribosome complexes with fewer than 6 nt in the A-site are good substrates for ribosome rescue by tmRNA and SmpB in vitro (Ivanova et al. 2004).
FIGURE 5.
(A) Reaction scheme for determining GTP hydrolysis and peptidyl transfer rates. The quaternary complex (top) contains EF-Tu, GTP, SmpB, and Ala-tmRNA, and the initiation complex (bottom) contains mRNA, fMet-tRNAfMet, and 70S ribosomes. The mRNA has only a UUC codon in the A-site, allowing the initiation complex to react either with Phe-tRNAPhe or Ala-tmRNA. Peptidyl transfer rates are determined by monitoring the rate of formation of the dipeptide fMet-Ala or fMet-Phe using 35S-labeled fMet-tRNAfMet. GTP hydrolysis rates are measured by using [γ-32P]GTP in the quaternary EF-Tu complex and following the appearance of 32P-labeled phosphate upon hydrolysis. (B) Representative primary data for dipeptide bond formation rates with Phe-tRNAPhe (left) or Ala-tmRNA (right), with a series of 16S mutants (see also Table 3). (C) Representative primary data for GTP hydrolysis rates with Phe-tRNAPhe (left) or Ala-tmRNA (right).
We assembled ribosome initiation complexes containing formyl-[35S]Met-tRNAfMet bound to an AUG codon in the P-site. Downstream from this start codon, the mRNA sequence contains only a single phenylalanine codon (UUC). The rate of peptidyl transfer was determined by measuring the amount of fMet-Ala dipeptide at various time points after mixing the initiation complex with an excess of quaternary complex composed of Ala-tmRNA, SmpB, EF-Tu, and GTP. As the concentration of the quaternary EF-Tu complex was not saturating in the reaction, the reported rates reflect both binding and catalysis (kcat/Km).
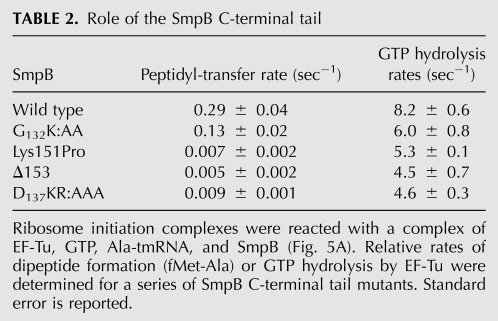
We measured the rate of dipeptide formation with wild-type SmpB and four SmpB mutants. The K131AAK, D137KR:AAA, and Lys151Pro mutants were shown above to abolish tagging in vivo. The Δ153 mutant is truncated at residue 153; Karzai and coworkers showed that deletion of the last seven residues in the tail inhibits tagging in vivo (Sundermeier et al. 2005). The peptidyl-transfer rates of D137KR:AAA, Lys151Pro, and Δ153 were far lower than that of wild-type SmpB (30-fold to 60-fold) (Table 2, left). These results show that conserved residues D137KR are essential for rapid peptidyl transfer to tmRNA and that helix formation is likewise required.
TABLE 2.
Role of the SmpB C-terminal tail
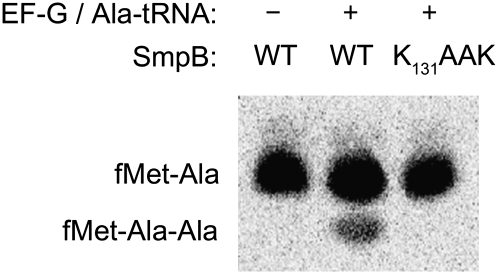
In contrast, we found that the K131AAK mutant supported peptidyl transfer at a rate only twofold slower than wild-type SmpB (Table 2). This suggests that the K131GKK sequence at the beginning of the C-terminal tail is not essential for tmRNA functions upstream of peptidyl transfer, including activation of EF-Tu or A-site accommodation of tmRNA. To test if these residues were required for tmRNA to serve as a template sequence, we performed a dipeptide reaction, waited until the endpoint was reached, and then added EF-G and Ala-tRNAAla (the first codon on tmRNA is Ala). In the presence of wild-type SmpB, the tripeptide fMet-Ala-Ala was formed as expected (12% yield) (Fig. 6). In contrast, no tripeptide was formed when the reaction was performed with the K131AAK mutant (<0.5% yield), suggesting that these residues are required for tmRNA to serve as a template.
FIGURE 6.
The K131GKK sequence in the SmpB tail is required for tmRNA to serve as a template. fMet-Ala dipeptide was formed by reacting initiation complexes with Ala-tmRNA complexes as in Figure 5A. After 5 min, EF-G and Ala-tRNAAla were added to the reaction and incubated for an additional 10 min. The tripeptide fMet-Ala-Ala is synthesized only if tmRNA is translocated to the P-site and the resume codon on tmRNA (Ala) is positioned properly in the A-site.
...…but not for EF-Tu activation
The peptidyl-transfer assays above do not allow us to distinguish between defects in accommodation and defects in the activation of EF-Tu. As discussed above, structural studies indicate that the C-terminal region of SmpB is positioned in the decoding center prior to the release of tmRNA by EF-Tu (Kaur et al. 2006). As a result, we wondered whether the C-terminal tail is capable of activating EF-Tu, presumably by altering the conformation of key decoding center nucleotides or EF-Tu itself.
Since the activation of EF-Tu is slower than the chemistry of GTP hydrolysis, GTP hydrolysis rates can be used to report on EF-Tu activation as the decoding signal is read in the 30S A-site (Pape et al. 1999). GTP hydrolysis rates were determined by mixing initiation complexes with substoichiometric amounts of the quaternary complex composed of Ala-tmRNA, SmpB, EF-Tu, and [γ-32P]GTP. The levels of free radioactive phosphate were monitored at various time points. As expected, no significant GTP hydrolysis was observed in the absence of SmpB (data not shown). Several SmpB mutants were used to test the role of the tail: wild-type, K131AAK, D137KR:AAA, Lys151Pro, and Δ153. Surprisingly, we found that all four SmpB mutants catalyzed GTP hydrolysis very efficiently, less than twofold slower than wild-type (Table 2, right). Since these SmpB mutants support efficient EF-Tu activation, they must inhibit peptidyl transfer by interfering with the accommodation of tmRNA.
The role of rRNA nucleotides in the decoding center
A1492, A1493, and G530 bind and recognize correctly paired codon–anticodon helices in the A-site (Fig. 1A) during the canonical decoding process (Ogle and Ramakrishnan 2005). Mutation of these nucleotides results in a dominant lethal phenotype in E. coli (Powers and Noller 1990). Anticipating that these nucleotides would also be involved in licensing tmRNA entry during ribosome rescue, we purified ribosomes containing the A1492G, A1493G, or G530A mutations and measured peptidyl-transfer rates to tmRNA in vitro. Mutant ribosomes were purified to homogeneity by an affinity-purification procedure from cells that also express wild-type ribosomes (Youngman and Green 2005; Cochella et al. 2007). The MS2 hairpin was inserted into the mutant 16S rRNA genes, providing a chemical handle for isolation of mutant 30S subunits. In addition to the three decoding center mutants, wild-type MS2-tagged ribosomes were isolated for the wild-type control.
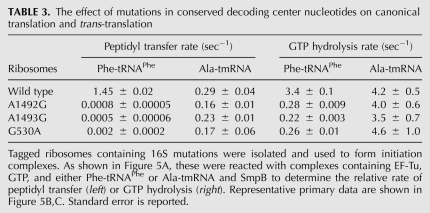
We formed initiation complexes with the MS2-tagged ribosomes and measured the rates of dipeptide formation for either Phe-tRNAPhe or wild-type tmRNA and SmpB. Representative primary data are shown in Figure 5B. Time courses of peptidyl transfer with Phe-tRNAPhe showed that the decoding center mutations led to an ∼1000-fold rate reduction (Table 3, left). This is consistent with the reductions in peptidyl-transfer rates reported previously for these mutants (Cochella et al. 2007). In contrast, peptidyl transfer to Ala-tmRNA was reduced by less than twofold in the A1492G, A1493G, or G530A mutants.
TABLE 3.
The effect of mutations in conserved decoding center nucleotides on canonical translation and trans-translation
The fact that mutation of these nucleotides does not significantly reduce peptidyl transfer rates suggests that they do not play an important role in tmRNA accommodation. We were concerned, however, that defects in EF-Tu activation could be masked by the slower, rate-limiting accommodation step. The fact that the SmpB tail is required for accommodation but not EF-Tu activation (as shown above) suggests that the two steps may occur via different mechanisms during ribosome rescue. We therefore analyzed the GTP hydrolysis rates for the decoding center mutants, using both Phe-tRNAPhe and the tmRNA–SmpB complex. Representative primary data are shown in Figure 5C. Time courses of GTP hydrolysis by EF-Tu with Phe-tRNAPhe, used to address canonical decoding, revealed an ∼15-fold rate reduction in the mutant ribosomes (Table 3, right), consistent with earlier studies (Cochella et al. 2007). In contrast, no rate reduction was seen for the trans-translation reaction with EF-Tu complexed with tmRNA and SmpB. Taken together, these data show that although A1492G, A1493G, and G530A play a critical role in the canonical decoding process, both in the EF-Tu activation and accommodation steps, they play little to no role in either step as tmRNA enters stalled ribosomes.
DISCUSSION
Stalled ribosomes accept tmRNA into their A-sites in the absence of a codon–anticodon interaction. Like canonical tRNAs, Ala-tmRNA is delivered to the ribosome by EF-Tu complexed with GTP. Somehow tmRNA must activate EF-Tu to hydrolyze GTP and release tmRNA into the ribosomal A-site, after which it has to swivel into the appropriate conformation for peptidyl transfer to occur. Structural and biochemical studies show that the SmpB protein binds the decoding center in the 30S A-site (Kaur et al. 2006; Kurita et al. 2007; Nonin-Lecomte et al. 2009) and that the SmpB C-terminal tail in particular is essential for peptidyl transfer to tmRNA (Sundermeier et al. 2005; Shimizu and Ueda 2006). This function of the tail is independent of SmpB's ability to bind the ribosome or to bind to tmRNA (Sundermeier et al. 2005; Nonin-Lecomte et al. 2009). We have extended these studies by determining the step at which the tail acts during ribosome rescue and identifying characteristics of the tail that are essential for its function.
Our data indicate that the SmpB C-terminal tail is not involved in activating EF-Tu. This is surprising because the tail is positioned such that it could easily interact with decoding center nucleotides or EF-Tu itself to activate GTP hydrolysis. While this work was under way, Himeno and coworkers reported that truncation of the tail does not inhibit GTP hydrolysis by EF-Tu (Kurita et al. 2010). While consistent with our findings, their conclusions were based on the analysis of reaction yields at very long time points (5 and 10 min) and not the comparison of rate constants. This is problematic because they may have overlooked important defects in activity. For example, we found that when reacted with Phe-tRNA, ribosomes carrying the A1493G mutation activated GTP hydrolysis 15-fold slower than wild-type ribosomes. In spite of this defect, the A1493G mutant reached the same endpoint as wild-type ribosomes after only 15 sec (data not shown). Because Himeno and coworkers did not obtain rate constants for their SmpB mutants, this kind of defect in GTPase activity cannot be ruled out. By determining rates that were physiologically relevant (on the order of 10 sec−1) with pre-steady-state kinetic methods that have been used extensively to study canonical decoding, we have ruled out defects in our SmpB tail mutants in activating GTP hydrolysis by EF-Tu.
Our findings support the conclusion that the SmpB tail plays an essential role in the accommodation of tmRNA into the ribosomal A-site. Accommodation of the tmRNA/SmpB complex requires both flexibility and significant motion. During the first selection step, canonical tRNAs are conformationally strained. The strain is relaxed as accommodation occurs and the 3′-CCA end moves into the peptidyl transferase center (Ogle and Ramakrishnan 2005). The necessary flexibility is associated with the elbow region of the tRNA (Valle et al. 2003; Cochella and Green 2005; Schmeing et al. 2009). Does the tmRNA/SmpB complex possess the same flexibility? Only the acceptor stem of tmRNA functions as a tRNA; SmpB acts as the anticodon stem–loop, with their interaction lying just below the elbow region (Fig. 1B). It makes sense that the junction between SmpB and tmRNA is right at the position where flexibility is likely to be important. Structural studies suggest that rotation of SmpB is also involved; the tmRNA–SmpB complex rotates 30° during accommodation while remaining bound in the decoding center (Weis et al. 2010). Perhaps the interaction of the SmpB tail with the ribosome is required for these motions to occur.
The SmpB tail, although unstructured in solution, may function as a helix inside the ribosomal A-site during accommodation. Our Pro-scanning data are consistent with a model in which residues 143–154 function as a helix. The Lys151Pro mutation was found to dramatically reduce the helical potential of the tail peptide and lower the peptidyl-transfer rate 40-fold. The hydroxyl-radical probing studies of Himeno and coworkers also imply a helical structure in the latter half of the C-terminal tail (Kurita et al. 2007). We have added to this work by defining the extent of the helix and demonstrating its functional importance. Helix formation may position key residues in the tail for interaction with elements in the ribosomal A-site. Conserved positively charged residues at positions 143, 145, 149, and 153 are likely sites of interaction with rRNA, and their loss inhibits SmpB function. Conserved residues D137KR are also essential for accommodation and may also be a ribosome-binding site.
While the SmpB residues discussed above are required for the accommodation of tmRNA into stalled ribosomes, the K131GKK sequence is essential for the translation of tmRNA but not for peptidyl transfer. Two steps occur following the transfer of the nascent peptide to Ala-tmRNA that could be inhibited by mutation of the K131GKK sequence. First, tmRNA and SmpB must be translocated into the P-site by EF-G. Hydroxyl radical probing studies by Himeno and coworkers show that when SmpB is bound to the A-site, the tail lies along the downstream mRNA path, but that the tail tucks under the body of SmpB when bound to the P-site (Kurita et al. 2007). This conformational change moves the tail out of the way for the tmRNA template to enter the A-site. The K131GKK sequence at the beginning of the tail could act as a hinge allowing this movement to occur. A second possibility is that K131GKK affects the placement of the tmRNA template within the decoding center. We previously reported that SmpB plays a role in selecting the reading frame on tmRNA (Watts et al. 2009), a finding later confirmed by structural studies (Fu et al. 2010; Weis et al. 2010). Residues Tyr24 and Ala130 were implicated in this process; mutation of these residues alters the reading frame on tmRNA. Tyr24 and Ala130 interact where the tail exits the body of the protein at the K131GKK site. Addition or deletion of residues between Ala130 and Lys131 obliterate SmpB function. Whether it is translocation or template placement that is affected, it seems that the angle of exit from the SmpB body and flexibility in the beginning of the tail are critical for SmpB function.
How does SmpB binding in the A-site trigger the decoding machinery? Using NMR and chemical probing experiments, Felden and coworkers showed that SmpB binding to ribosomes changes the conformation and reactivity of A1492, A1493, and G530 (Nonin-Lecomte et al. 2009). They concluded that SmpB mimics the codon–anticodon duplex, triggering the same response in the ribosome as cognate tRNA binding does. Our data, however, contradict this model; mutation of these nucleotides has no effect on GTP hydrolysis rates and only very minor effects on the rate of peptidyl transfer to Ala-tmRNA. It appears that A1492, A1493, and G530 do not play a significant role in promoting EF-Tu activation or accommodation of tmRNA. This is striking given their central role in these steps during canonical decoding. Note that our data do not contradict the findings of Felden and coworkers—SmpB binding to nearby nucleotides may alter the conformations of A1492 and A1493 as reported. But the conformational changes in A1492, A1493, and G530 are probably not a result of direct binding by SmpB, nor do they have the same functional significance as they do in canonical decoding.
We conclude that SmpB is not a codon–anticodon mimic, strictly speaking, and that SmpB binding to the 30S A-site must activate EF-Tu by some other mechanism. This may involve other SmpB–rRNA interactions that account for the majority of binding energy for SmpB in the A-site. Alternatively, the S12 protein is known to play an important role in the decoding process and may also influence tmRNA acceptance. S12 mutants can inhibit tmRNA tagging, although their mechanism of action is still unclear (Holberger and Hayes 2009; M Miller and A Buskirk, unpubl.). Experiments to determine the mechanisms by which SmpB activates the decoding machinery will likely yield more insight into trans-translation and perhaps canonical decoding as well.
MATERIALS AND METHODS
Circular dichroism
Peptides corresponding to residues 137–157 of E. coli SmpB were purchased from GenScript. The wild-type peptide has the sequence DKRSDIKEREWQVDKARIMKN; the Lys151Pro mutant was also synthesized. CD spectra were recorded on a Avic Model 420 CD spectrometer in a quartz cuvette with a path length of 0.1 cm. The peptides were dissolved at a concentration of 35 μM and in 10 mM Tris (pH 7.5) in the presence or absence of 2,2,2-trifluoroethanol (TFE). The concentrations of TFE varied from 0% to 80% in increments of 20%. Spectra were recorded from 260 nm to 190 nm with 1-nm step size and a time constant of 1.0 sec. Data from three or four replicates were averaged and are reported in mean residue ellipticity. In Table 1, the MRE at 222 nm is reported with the associated standard error.
Immunoblot assays
The pDH210 vector expresses glutathione S-transferase (GST) with the stall-inducing sequence Glu-Pro-Stop at the C terminus and also expresses tmRNA altered to encode ANDHHHHHHD. SmpB mutants were expressed from derivatives of the pDH113 vector (Watts et al. 2009). Tagging of the GST protein in the presence of the various SmpB mutants was assayed by immunoblotting as described (Tanner et al. 2009).
Expression and purification of MS2-tagged ribosomes
Wild-type and mutant MS2-tagged ribosomes were expressed and purified as described (Youngman and Green 2005; Cochella et al. 2007) with the following modifications. Crude MS2-tagged ribosome pellets were purified over a 15-mL FPLC amylose resin column to which the MBP-MS2-His protein was prebound. Elution was carried out with 10 mM maltose, and the eluted ribosomes were concentrated over Amicon Ultra filters (MWCO 100,000; Millipore). Purified ribosomes were depleted of 50S subunits, so purified MRE600 50S subunits were added back for the formation of initiation complexes.
Purification of translation components
IF1, IF2, IF3, and His-tagged EF-Tu, EF-G, PheRS, and AlaRS were purified as described (Shimizu et al. 2001; Cochella and Green 2005; Brunelle et al. 2006). Formyl-[35S]Met-tRNAfMet was prepared as described (Moazed and Noller 1991). mRNA (GGAAUUCGGGCCCUUGUUAACAAUUAAGGAGGUAUACUAUGUUC) and tRNAAla were synthesized by T7 transcription of a template assembled by annealing sense and antisense oligonucleotides.
Purification of SmpB
SmpB with an N-terminal His6-tag was expressed from a pET15b derivative in BL21/DE3 cells. Upon reaching an OD600 of 0.5, the cells were treated with 1 mM IPTG for 2 h to induce SmpB expression. The cells were pelleted and resuspended in lysis buffer (20 mM Tris-HCl at pH 7.5, 300 mM NaCl, 5 mM imidazole) and cracked using a French press. The lysate was clarified by centrifugation, and SmpB was purified on NiNTA agarose resin (QIAGEN). Purified SmpB was then dialyzed in SmpB storage buffer (50 mM Tris-HCl at pH 6.0, 150 mM NH4Cl, 200 mM KCl, 5 mM MgCl2, 6 mM β-mercaptoethanol, 50% glycerol).
tmRNA synthesis and aminoacylation
The tmRNA gene was amplified from pKW11 (Roche and Sauer 2001) by PCR, adding the T7 promoter sequence, using the forward primer GAAATTAATACGACTCACTATAGGGGCTGATTCTGGATTCGACGG, and the reverse primer TGGTGGAGCTGGCGGGAGTTGAACC. The PCR product was purified and transcribed using the Ambion MEGAshortscript Kit. tmRNA was purified from the reaction by phenol/chloroform extraction followed by ethanol precipitation. tmRNA (5 μM) was aminoacylated with purified AlaRS in buffer 101 (20 mM Tris-HCl at pH 7.5, 20 mM MgCl2, 1 mM DTT), 2 mM ATP, and 10 mM Ala. tmRNA was then purified by phenol/chloroform extraction followed by ethanol precipitation and resuspended in 20 mM KOAc (pH 5.1). The extent of tmRNA aminoacylation was 10%–20% as determined by a small parallel reaction with 50 μM [14C]Ala. Likewise, E. coli tRNAPhe (Sigma) was aminoacylated with purified PheRS, and tRNAAla synthesized by run-off transcription was aminoacylated by AlaRS.
Peptide-bond formation reactions
70S initiation complexes were formed by incubating 4 μM–tagged 70S ribosomes, 10 μM mRNA, 6 μM f[35S]Met-tRNAfMet, 5 μM each IF (1, 2, and 3), and 2 mM GTP in buffer A for 45 min at 37°C. Buffer A is 50 mM Tris-HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 1 mM dithiothreitol (Pape et al. 1999). The complex was purified by layering over a 1.3-mL sucrose cushion (1.1 M sucrose, 20 mM Tris-HCl at pH 7.5, 500 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA) and spun at 258,000g in a TLA100.3 rotor for 2 h. The resulting pellet was resuspended in buffer A, diluted to 100 nM, and aliquots were stored at −80°C.
The Phe-tRNAPhe ternary complex was prepared by incubating 2 μM charged Phe-tRNAPhe, 8 μM EF-Tu, and 1 mM GTP in buffer A. The tmRNA–SmpB quaternary complexes were prepared by incubating 2 μM charged tmRNA (20 μM total), 40 μM SmpB, and 1 mM GTP in buffer A for 5 min at 37°C; 20 μM EF-Tu was added, and the reaction mixture was incubated for another 5 min at 37°C.
Peptide-bond formation rate reactions were carried out at 37°C by mixing equal volumes of initiation complexes with either the ternary or quaternary complexes described above. The reactions were stopped at desired time points by addition of KOH to a final concentration of 0.3 M. Reactions with relatively fast rate constants (>0.05 sec−1) were performed on a KinTek RQF-3 quench-flow instrument. Reaction products were resolved using cellulose TLC plates in pyridine acetate (pH 2.8) as described (Youngman et al. 2004) and analyzed by autoradiography. The data were fit to a first-order exponential equation with GraphPad Prism5 software. All reported reactions were performed at least twice, and the standard error is given.
Tripeptide reactions were performed with an mRNA with a weaker Shine-Dalgarno sequence: GAAGCUGAACGAGAAACGUAAAAUGUAGUAC. Initiation complexes were formed as above and diluted to 100 nM. The Ala-tmRNA quaternary complex was prepared by incubating 5 μM total tmRNA, 15 μM SmpB, 1 mM GTP, and 8 μM EF-Tu for 5 min at 37°C. The quaternary complex was reacted with an equal volume of initiation complex for 5 min at 37°C. The resulting pre-translocation complex was then combined with an equal volume of a solution containing 600 nM Ala-tRNAAla, 1 mM GTP, and 10 μM EF-G in buffer A, reacted for 10 min at 37°C, and analyzed as above.
GTP hydrolysis reactions
70S initiation complexes were formed as above except nonradioactive fMet-tRNAfMet was used and the complexes were diluted to 500 nM prior to storage at −80°C. The Phe-tRNAPhe ternary complex was prepared by first incubating 20 μM EF-Tu, 17.5 μCi of [γ-32P]GTP (6000 Ci/mmol), 3 mM phosphoenol pyruvate (PEP), and 0.1 mg/mL pyruvate kinase (PK) in buffer A for 30 min at 37°C. Phe-tRNAPhe was then added to 2 μM and incubated for 30 min on ice. The tmRNA–SmpB quaternary complexes were prepared by incubating 5 μM tmRNA, 20 μM SmpB (wild type or mutant), 20 μM EF-Tu, 17.5 μCi of [γ-32P]GTP (6000 Ci/mmol), 3 mM PEP, 0.1 mg/mL PK, 20 mM L-alanine, 2 mM ATP, and 10 μM AlaRS in buffer A for 1 h at 37°C. The ternary and quaternary complex mixes were passed through two P30 columns to remove excess [γ-32P]GTP.
GTP hydrolysis rate reactions were carried out on a KinTek RQF-3 quench-flow instrument at 20°C, where equal volumes of initiation complexes and either the ternary or quaternary complexes described above were rapidly mixed and quenched with 40% formic acid at the desired times. Reaction products were resolved on PEI cellulose TLC plates in 0.5 M KH2PO4 (pH 3.5) and analyzed by autoradiography. The data were fit to a first-order exponential equation with GraphPad Prism5 software. All reported reactions were performed at least twice, and the standard error is given.
ACKNOWLEDGMENTS
This work was supported by grant GM77633 to A.B. and grant GM059425 to R.G. from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2821711.
REFERENCES
- Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C 2006. The tmRDB and SRPDB resources. Nucleic Acids Res 34: D163–D168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S 2007. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci 104: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle JL, Youngman EM, Sharma D, Green R 2006. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA 12: 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T 2010. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol 78: 796–808 [DOI] [PubMed] [Google Scholar]
- Cochella L, Green R 2005. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R 2007. Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36: W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Gromadski KB, Rodnina MV 2006. The ribosome's response to codon–anticodon mismatches. Biochimie 88: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Fu J, Hashem Y, Wower I, Lei J, Liao HY, Zwieb C, Wower J, Frank J 2010. Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J 29: 3819–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B 2004. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J Biol Chem 279: 25978–25985 [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT 2002a. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem 277: 33825–33832 [DOI] [PubMed] [Google Scholar]
- Hayes CS, Bose B, Sauer RT 2002b. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci 99: 3440–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberger LE, Hayes CS 2009. Ribosomal protein S12 and aminoglycoside antibiotics modulate A-site mRNA cleavage and transfer-messenger RNA activity in Escherichia coli. J Biol Chem 284: 32188–32200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M 2004. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol 338: 33–41 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP 2005. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J Biol Chem 280: 5503–5509 [DOI] [PubMed] [Google Scholar]
- Kaur S, Gillet R, Li W, Gursky R, Frank J 2006. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc Natl Acad Sci 103: 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Sasaki R, Muto A, Himeno H 2007. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res 35: 7248–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita D, Muto A, Himeno H 2010. Role of the C-terminal tail of SmpB in the early stage of trans-translation. RNA 16: 980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P, Baldwin RL 1997. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry 36: 8413–8421 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1991. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci 88: 3725–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124 [DOI] [PubMed] [Google Scholar]
- Nonin-Lecomte S, Germain-Amiot N, Gillet R, Hallier M, Ponchon L, Dardel F, Felden B 2009. Ribosome hijacking: a role for small protein B during trans-translation. EMBO Rep 10: 160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF 1990. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci 87: 1042–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem 276: 28509–28515 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV IV, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T 2006. SmpB triggers GTP hydrolysis of elongation factor Tu on ribosomes by compensating for the lack of codon-anticodon interaction during trans-translation initiation. J Biol Chem 281: 15987–15996 [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T 2001. Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751–755 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M 2002. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat Struct Biol 9: 849–854 [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW 2005. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc Natl Acad Sci 102: 2316–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR 2009. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem 284: 34809–34818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Watts T, Cazier D, Healey D, Buskirk A 2009. SmpB contributes to reading frame selection in the translation of transfer-messenger RNA. J Mol Biol 391: 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis F, Bron P, Giudice E, Rolland JP, Thomas D, Felden B, Gillet R 2010. tmRNA-SmpB: a journey to the centre of the bacterial ribosome. EMBO J 29: 3810–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Green R 2005. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods 36: 305–312 [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599 [DOI] [PubMed] [Google Scholar]