Abstract
Angiogenesis is of major interest because of its involvement in numerous pathologies or for promoting tissue repair. It is often assessed by the ability of endothelial cells to sprout, migrate, and form vascular tubules in Matrigel in vitro. Matrigel contains a mixture of basement membrane components, which stimulate endothelial cells to form capillary-like hexagonal structures, and is often preferred over other in vitro assays because of its ease of use, rapidity and the ability to measure key steps in angiogenesis, including migration, protease activity, and tubule formation. Various methods have been used to quantitate tubule formation, yet no consensus has been reached regarding the best quantification method for evaluating the efficacy of angiogenic stimulants or inhibitors in this Matrigel assay. Here, we have measured the ability of umbilical cord blood endothelial colony-forming cell-derived cells to form tubules in growth factor reduced Matrigel in the presence or absence of two angiogenic inhibitors, suramin and SU6668, to compare the benefits and limitations of two quantification methods—Angiosys and Wimasis. These comparative analyses revealed that both Angiosys and Wimasis are easy to use, accurately quantify angiogenesis, and will suit the needs of different types of users.
Introduction
Neovascularization, which occurs by angiogenesis, vasculogenesis, or artheriogenesis, is important for physiologic tissue growth, in wound healing and tissue repair, and for pathological conditions such as the growth of some cancers and the development of diabetic eye disease (reviewed in Ref.1,2). Vasculogenesis, the de novo development of new blood vessels from endothelial colony-forming cells (ECFC) or their precursors, differs from angiogenesis, which is characterized by the sprouting of new vessels from existing vessels. In the latter case, three specialized endothelial cell types have been identified as tip, stalk, and phalanx cells (reviewed in Ref.3). The stalk cells proliferate and elongate the sprouting vessels, produce extracellular matrix, and form a vessel lumen; the tip cells that are located at the forefront of the sprouting vessels sense vessel guidance cues and allow migration and invasion toward proangiogenic stimuli, and the phalanx cells are quiescent endothelial cells that associate with perivascular cells to form vessels that regulate blood flow and hence the oxygenation of tissues.
Angiogenesis can be modeled in vitro by a number of assays, one being the short-term culture of endothelial cells in Matrigel, a gelatinous protein mixture obtained from Engelbreth-Holm-Swarm mouse sarcoma cells.4 This Matrigel assay is quick and easy to perform and also allows in vitro modeling of endothelial cell behavior, including survival, apoptosis, and the steps leading to capillary formation and invasion. It is also important for investigating the effects of drugs or small molecules on angiogenesis in vitro before these are developed into clinical therapies. Despite this, there is currently no common method used by researchers to quantitate vessels formed in this Matrigel assay. This may cause some variations in the analysis of such vessels among research groups and may make comparisons difficult.
In this study, we used umbilical cord blood (CB) ECFC-derived cells and examined their ability to form tubules in the Matrigel assay in the presence or absence of small molecule inhibitors, SU6668 and suramin. We then compared two novel methods for quantifying the resultant tubules, Angiosys and Wimasis, and provide data on these analyses and the benefits of each approach. These aim to assist researchers in tailoring their requirements for the quantification of vessel formation in the Matrigel assay under different experimental settings.
Materials and Methods
Cell lines and cell culture
Human umbilical CB units were sourced from the John Radcliffe Hospital in Oxford with ethical approval and informed written consent and were collected, processed, and stored under a Human Tissue Authority (HTA) license. Human ECFCs were generated in-house from these units by culturing CB mononuclear cells (2×107 cells/4 mL in each well of six-well plate) in complete EGM-2 medium5 supplemented with the EGM-2 bullet kit (Lonza Biologics, Cambridge, England) and 10% (v/v) Hyclone heat-inactivated fetal bovine serum (ThermoScientific, Waltham, MA) at 37°C in a humidified atmosphere with 10% CO2 air.6 Endothelial colonies that formed by 14 days of culture were selected using cloning rings and those from individual CB units were pooled and then passaged in the above medium. Passage 0 was the time of the appearance of the first ECFC-derived colonies in agreement with the E.U. Cascade program classification. Cells were subsequently used at passage four to six for FACS analysis and in the Matrigel assay.
Matrigel assay
CB-ECFC-derived cells (from three different CB batches) were trypsinized and resuspended in four different media with 2% (v/v) Hyclone fetal bovine serum as follows: (1) EBM-2 medium; (2) 10 μM suramin (Calbiochem, Darmstadt, Germany) in EBM-2 medium; (3) EBM-2 medium with 0.15% (v/v) DMSO; (4) 10 μM SU6668 (Calbiochem) in EBM-2 medium and 0.15% (v/v) DMSO. For each condition, CB-ECFC-derived cells were plated at a density of 1.5×104 cells/well in triplicate in 96-well plates coated with 50 μL of growth factor-reduced Matrigel (BD Biosciences, Oxford, England).7 Parameters used in this study are summarized in Appendix Table 1. Plates were incubated for 20 h at 37°C before photomicroscopy. After incubation, media were removed, plates washed with distilled water (PAA Laboratories, Yeovil, England) twice, and cells fixed using 100% (v/v) ice-cold methanol (Sigma-Aldrich Ltd., St. Louis, MO). Cells were washed with distilled water twice and next covered with fresh PBS. Each image from each well was taken at ×4 magnification using a Nikon Eclipse TE2000-U microscope (Nikon Ltd., London, England).
Umbilical CB ECFC-derived cell phenotyping
Three batches of CB-ECFC-derived cells at approximately 80% confluency were detached from tissue culture flasks using 100% (v/v) accutase (PAA Laboratories) for 5 min and washed and resuspended in magnetic activated cell sorting (MACS) buffer containing 1.0% (w/v) bovine serum albumin and Fc receptor blocking agent (Miltenyi Biotech, Bergisch-Gladbag, Germany) at 4°C before labeling with relevant conjugated monoclonal antibodies (Mabs) or isotype-matched negative-controls as described.5 The following mouse Mabs were used: PE-CD31 (mIgG1; clone WM59), PE-CD73 (clone AD2), PE-CD144 (clone 16B1), PE-CD166 (clone 105902), PE-CD133 (mIgG2b, clone 293C3), PE-mIgG1 isotype controls, FITC-CD105 (mIgG1; clone FAB10971F), FITC-CD146 (clone MAB16985F), FITC-CD90 (clone 5E10), FITC-mIgG1 isotype control, APC-CD34 (mIgG2a, clone AC136), APC-mIgG2a isotype control, PE-Cy7- CD14 (mIgG2a, clone M5E2), PE-Cy7-CD45 (mIgG1, clone 2D1), and PE-Cy7 mIgG1 and mIgG2a isotype controls (Appendix Fig. 1). Cells were analyzed on a BD LSR II flow cytometer using FACSDiva software (BD Biosciences) as previously described.5 Cells were stained using single conjugated antibodies and then incubated with 1:1000 Topro-3-dye in DMSO (Invitrogen Ltd., Paisley, Scotland) to select viable cells before CD antigen analysis.
Image processing and analysis methods
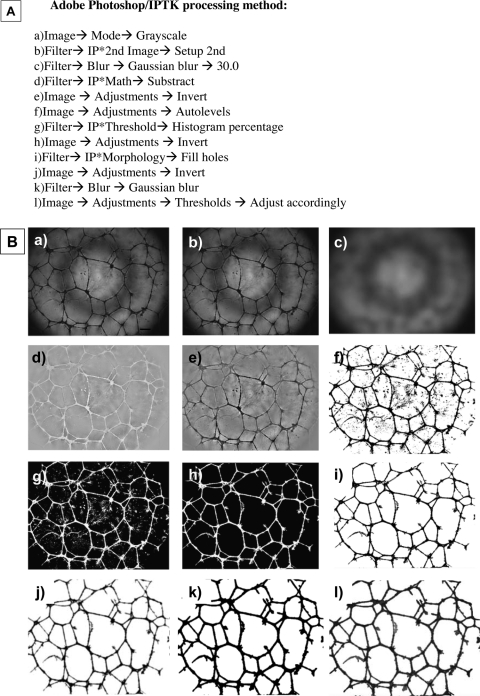
Phase-contrast images of CB-ECFC-derived cells in Matrigel were captured using an inverted-phase-contrast light microscope (TE2000-U; Nikon Ltd.) at×4 magnification, and saved as TIFF files. The acquisition software (Simple PCI version 6.6.0.0; Hamamatsu Corporation, Sewickley, PA) allowed the control of the bright field illumination of the microscope to acquire images of comparable mean brightness. Images were then processed using Adobe Photoshop CS2 9.0.2 (Adobe Systems, San Jose, CA) with the Image Processing Tool Kit plug-ins (Reindeer Graphics, Asheville, NC) and analyzed using Angiosys 1.0 (TCS Cellworks, Buckingham, England). The main processing steps applied to the images are shown in Appendix Figure 2a and b.
The second method used to quantitate tubules involved using a service provided by Wimasis, which permits users to upload their images online at anytime and from anywhere and allows their images to be analyzed and the results uploaded back to the researcher's server. Briefly, the image analysis process carried out by Wimasis was automated and involved filtering, segmenting, object recognition, and data processing. To date, Wimasis has used images from more than 100 institutes to stream line their processes and have generated a data readout system based on tubule characteristics (number of tubules, number and mean number of junctions, tubule area (%), total, mean and standard deviation of tubule length, number of independent tubules) and net characteristics (number of loops, mean perimeter loop and number of nets). In this study, the results were analyzed using the beta version of Wimasis WimTube. Wimasis tubule information video demonstrates this and can be found on the Wimasis Image Analysis Web site: www.wimasis.com/ and www.youtube.com/user/wimasis#p/u/3/d80tHnGkDA8
Statistics
Each treatment was carried out in triplicate and the experiments for each cell batch were repeated three times. Images of the whole Matrigel assay well were photographed at ×4 magnification as above and the average value of three images taken per cell batch per condition was considered as representative of that sample. Data were expressed as mean±standard error of the mean. Statistical significance was determined using Student's t-test where p≤0.05 was considered statistically significant.
Results
Phenotypic analysis of CB-ECFC-derived cells
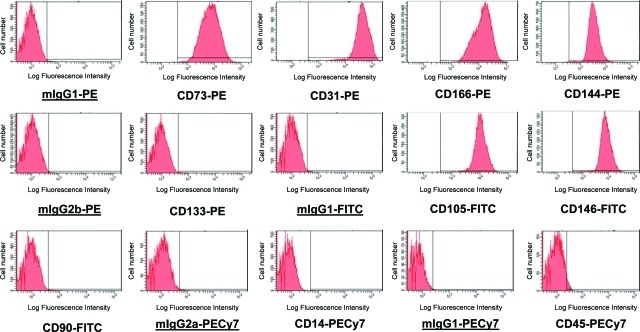
The CB-ECFC-derived cells formed typical endothelial colonies with cobblestone morphology (data not shown) as described previously.5 Three different batches of CB-ECFC-derived cells at passages four to six were phenotyped for cell surface markers using monoclonal antibodies and flow cytometry. These cells were negative for CD133, CD14, CD90, and CD45 but were strongly positive for CD31, CD166, and CD144 at the different passages indicating their endothelial lineage phenotype (Appendix Fig. 1).
Image processing and quantification using Angiosys
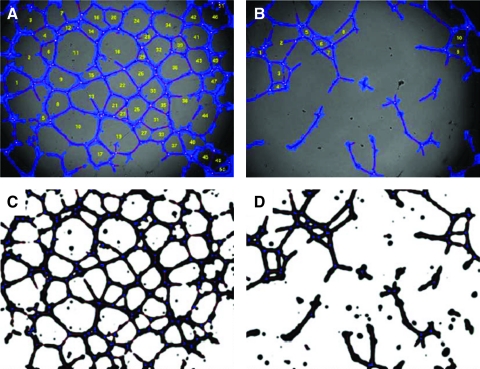
In Matrigel, the CB-ECFC-derived cells formed typical networks of endothelial tubules (Fig. 1). Phase-contrast images from same experiment were processed using an action script (Adobe Photoshop) (Appendix Fig. 2a). This script contained steps that include background subtraction, thresholding, and filling of holes in the inverted image, to produce a binary segmented image. These are detailed in Appendix Figure 2b and are summarized in Figure 1.
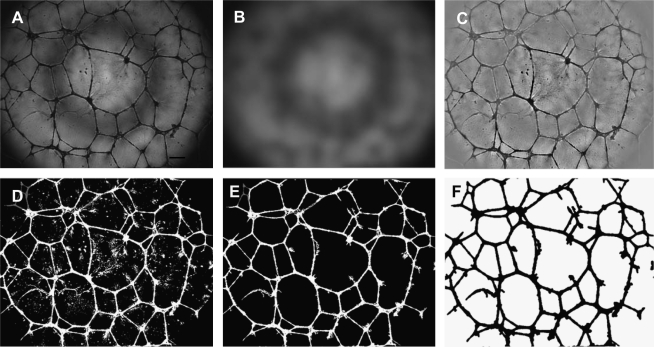
FIG. 1.
Image-processing steps of endothelial tubules formed in Matrigel. Single wells of the Matrigel assay were captured with an inverted microscope as detailed in Materials and Methods. (A) The original phase-contrast image. (B) Background image generated by Gaussian blur (30 pixels). (C) Background subtracted after conversion to the second image (B); the image underwent Gaussian blur. (D) Inverted image after thresholding on 15% darkest pixels. (E) Inverted image after hole filling. (F) Image used for analysis.
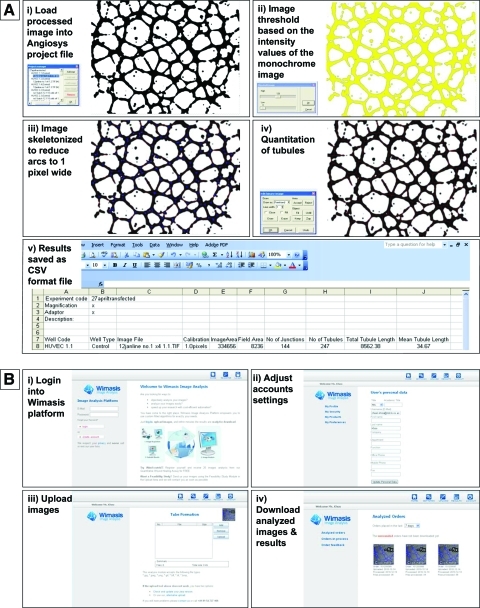
A script created in Angiosys (Fig. 2A) was used on the same batch of the images to minimize background variations and converted to a binary image by selecting a fixed proportion of the darkest pixels. This image was skeletonized (Fig. 2A iii) and the number of tubules, total tubule, and tubule length were measured. Results were then saved into a CSV file (Fig. 2A v), which could be analyzed using Microsoft Excel.
FIG. 2.
Tubule quantification using Angiosys and Wimasis. (A) Tubule quantification using Angiosys. (i) Load processed image into Angiosys project file. (ii) Image threshold based on the intensity values of the monochrome image. (iii) Image skeletonized to reduce arcs to 1 pixel wide. (iv) Quantitation of tubules. (v) Results saved as a CSV format file. (B) Tubule quantification using Wimasis. (i) Login into Wimasis platform. (ii) Adjust account settings. (iii) Upload images. (iv) Download analyzed images & results. Color images available online at www.liebertonline.com/tec
Image processing and quantification using Wimasis
The analysis of tubule formation images using Wimasis WimTube involved no batch scripting at all. The TIFF data were uploaded via the Web platform (https://mywim.wimasis.com) to the automated analysis tool WimTube and the resulting data were computed without any additional tweaking or coding required (Fig. 2B). The software tool conducted the conversion of data, noise filtering, particle detection, and tubule recognition automatically. This agile solution enabled the analysis of image data with highly variable data quality and made the inclusion of much of the image data in the analysis possible. The readout of the data was provided via e-mail and as a download from the Web platform. It included tubule length, branching points, cell-covered area, and nets in a CSV data file. Besides that, detailed overlay images were provided in which all branching points, tubules, and cells were viewable in an overlay image.
Effect of angiogenic inhibitors, suramin, and SU6668 on CB endothelial network formation
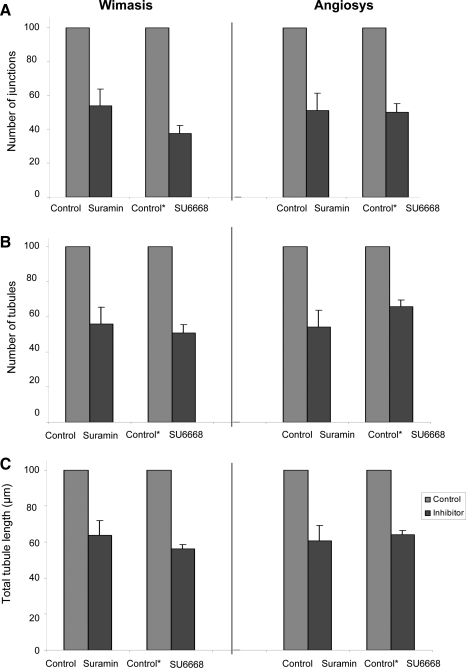
To compare the quantification methods, we analyzed the Matrigel tubules formed in the presence or absence of suramin and SU6668 (Fig. 3). Suramin is a specific and competitive inhibitor of G-protein-coupled receptor (GPCR) activity, whereas SU6668 is a tyrosine kinase receptor antagonist that binds to receptors such as platelet-derived growth factor receptor (PDGFR), an important receptor in cell migration and development of the microvasculature.8 Our results show that both suramin and SU6668 had a significant impact on tubule formation, causing a statistically significant reduction in the number of junctions, number of tubules, and the total tubule length (p<0.05) compared to cells in control EGM-2 media (Fig. 4). Images were analyzed and confirmed using Angiosys (Fig. 5A, B) and Wimasis (Fig. 5C, D) software.
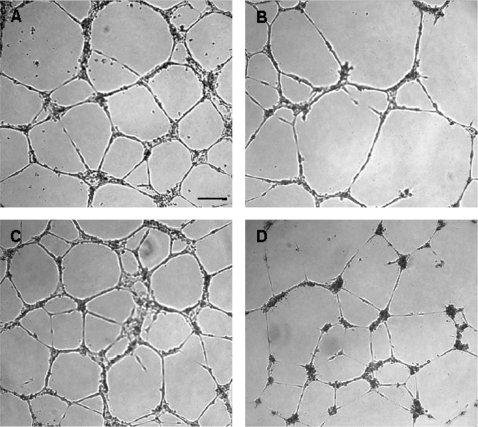
FIG. 3.
Representative images of umbilical CB endothelial tubules in growth factor-reduced Matrigel after incubation with control media or inhibitors CB-ECFC-derived cells were trypsinized and resuspended in respective test mediums and aliquoted in triplicate wells of a 96-well plate precoated with Matrigel and containing (A) EGM-2 medium; (B) 10 μM suramin in EGM-2 media; (C) EGM-2 medium and 0.15% DMSO; (D) 10 μM SU6668 in EGM-2 media (images were taken at ×4 magnification). CB, cord blood; ECFC, endothelial colony-forming cell.
FIG. 4.
Effects of suramin and SU6668 on CB-ECFC-derived cell angiogenesis measured by Angiosys and Wimasis. CB-ECFC-derived cells incubated for 20 h at 37°C were washed and fixed in 100% methanol. Images of tubules forming in each well were taken at ×4 magnification using Nikon Eclipse TE2000-U microscope and saved as TIFF format for processing using Angiosys or Wimasis service. Values demonstrated in the graphs are normalized values of means±standard error of the mean. (n=3 independent batches of cells), where absolute values of CB-ECFC-derived cells incubated in suramin or SU6668 were normalized against values of CB-ECFC-derived cells in basal media (control media). (A) Number of junctions. (B) Number of tubules. (C) Total tubule length (pixels). There was a significant difference observed in all experiments between the untreated and suramin- or SU6668-treated cells whether analyzed by either Wimasis or Angiosys (p-values <0.05; Student's t-test). Absolute values for tubule formation are shown in Figure 6.
FIG. 5.
Representative image of CB-ECFC-derived cells incubated with control media and test media (10 μm suramin) processed by Angiosys and Wimasis. (A) Processed image by Wimasis, of CB-ECFC-derived cells incubated with control media. (B) Processed image by Wimasis, of CB-ECFC-derived cells incubated with 10 μM suramin EGM-2 media. (C) Processed image by Angiosys, of CB-ECFC-derived cells incubated with control media. (D) Processed image by Angiosys, of CB-ECFC-derived cells incubated with 10 μM suramin EGM-2 media. Images were taken at ×4 magnification.
Comparison and benefits of quantification of angiogenesis using Angiosys and Wimasis methods
Images taken in our study were analyzed and confirmed using Angiosys and Wimasis software and the parameters and benefits of these methods are summarized in Table 1. The same parameters of the number of tubules, total tubule length and number of junctions could be measured using both Angiosys and Wimasis. In addition, Wimasis also allows a number of other parameters to be measured, including number of loops, mean perimeter of loop, number of independent tubules, and number of nets. Analysis with Wimasis incurs a cost per image, whereas Angiosys and preprocessing requires a one-off software payment (Table 1). However, in the latter case, the use of Angiosys requires increased processing time by the researchers themselves.
Table 1.
Comparison of Quantification of Angiogenesis Using Angiosys and Wimasis Methods
| Criteria | Angiosys | Wimasis |
|---|---|---|
| Image capture and processing | ||
| Easy to use | ✓ | ✓ |
| Uses phase-contrast image | ✓ | ✓ |
| Image acquisition | ✓ | ✓ |
| Simple PCI version 6.6.0.0 | Simple PCI version 6.6.0.0 | |
| Image processing before analysis | ✓ | Built in algorithm |
| Adobe photoshop processing | ||
| Semiautomated image batch processing | ✓ | Built in algorithm |
| Adobe photoshop processing | ||
| Parameters measured | ||
| Tubule length | ✓ | ✓ |
| Number of tubules | ✓ | ✓ |
| Number of junctions | ✓ | ✓ |
| Tubule area | ✓ | ✓ |
| Number of independent tubules | - | ✓ |
| Number of loops | - | ✓ |
| Mean perimeter loop | - | ✓ |
| Number of nets | - | ✓ |
| Cost of image analysis | €620 (Adobe Photoshop CS5 & plugins, Angiosys software) | €2 per image (1-200 images) |
| Turnaround time of image processing (100 images) | 48–72 h | 16 min |
| Turnaround time of tubule quantification (100 images) | 10 min | |
| Data analysis | Similar | Similar |
| Objectivity | Similar | Similar |
| Limitation | Turnaround time for image processing | Cost per image |
Comparison between both methods was based on factors such as image capture and processing, parameter measurements, cost, turnaround time for image processing and tubule quantification, data analysis, method limitations, and objectivity.
Angiosys and Wimasis can analyze phase-contrast images at low (∼4×), medium (∼10×) and high (∼20×) magnification. In this study, each of these methods relied on phase-contrast images at ×4 magnification. Wimasis accepts any images including unprocessed phase-contrast images, whereas Angiosys requires processed images. Therefore, to quantitate tubules using Angiosys, images first need to be processed using Adobe Photoshop.
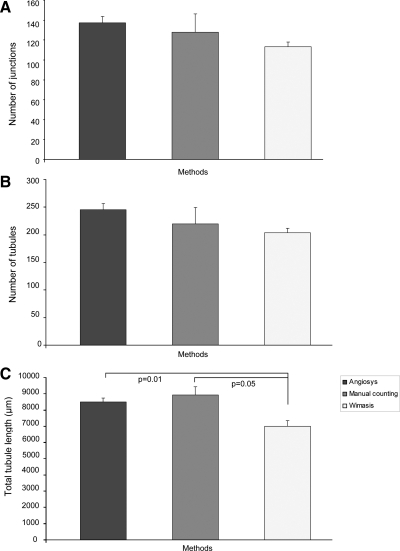
To determine if the results were similar for Angiosys, Wimasis, or manual counting systems, we compared the results by calculating the number of junctions, number of tubules, and tubule length and demonstrated that there were no significant differences in number of junctions and number of tubules generated by the CB-ECFC-derived cells incubated in basal media when analyzed using Wimasis and Angiosys compared to manual counting (Fig. 6A, B). However, based on manual counting, there is a significant difference in total tubule length compared to analysis using Wimasis (p=0.05) (Fig. 6C), but not using Angiosys (p=0.48).
FIG. 6.
Comparison of tubule quantification methods, Angiosys and Wimasis, with manual counting. All images used for comparison of methods were the same (i.e., images of CB-ECFC-derived cells incubated for 20 h at 37°C on Matrigel without any addition of inhibitors). Values are mean±standard error of the mean of absolute values for n=3 independent batches of cells analyzed for (A) number of junctions, (B) number of tubules, and (C) total tubule length (pixels). There was no significant difference in tubule or junction number among the methods of quantitation (p-values >0.05) but a significant difference in tubule length when measured by Wimasis as compared to Angiosys or manual counting as determined using Student's t-test (p-values ≤0.05).
Discussion
Popular assays such as the Matrigel assay have been used to rapidly screen for angiogenic and antiangiogenic factors and define genes functionally important for angiogenesis.9 The Matrigel method is a commonly used two-dimensional assay, which is easy to perform. It can also provide information on molecules affecting cell behavior in terms of cell adhesion, chemotaxis, and cytoskeleton rearrangements.10 CB-ECFC-derived cells plated in Matrigel and assessed in vitro follow similar processes seen for vascular tubule formation in vivo, where they first adhere onto Matrigel extracellular matrix 1 h after plating, then migrate toward each other over 2–4 h, and then form capillary-like tubules, which mature into tubular networks over 6–20 h.
Although accurate quantification of angiogenesis in Matrigel in vitro is important, there is still no consensus on the best method to use. Some researchers use commercial software, such as Optomax Image Analysis and QWin, whereas others prefer to use public domain Java-based image-processing programs such as NIH image/Scion Image and ImageJ.11–14 Based on the quantitative analyses of other researchers using different cell types and software, we analyzed two novel methods for Matrigel tubule quantification, Angiosys and Wimasis, and compared their benefits.
Tubular networks formed on Matrigel often vary in different physiological and pathophysiological environments. We studied the effect of suramin and the SU6668 inhibitor on tubule formation of CB-ECFC-derived cells using the Matrigel assay and compared the two quantification methods. Suramin was used because it inhibits angiogenesis by targeting fibroblast growth factor and vascular endothelial growth factor (VEGF) receptor signaling in cultured endothelial cells, whereas SU6668 targets such tyrosine kinase receptors as VEGFR, PDGFR, and, to a lesser extent, fibroblast growth factor-R2, inhibiting the phosphorylation of these receptors in endothelial cells, and thereby preventing tubule formation.15 Such analyses have been important for the initial rapid assessment of such drugs in the development of antiangiogenic cancer therapies.8
Various quantification parameters have been used to measure angiogenesis such as counting branching points, tubule area, tubule numbers, average tubule length, or various combinations of these measurements.16 The number of interconnections between tubules provides information on the way the endothelial cells organize themselves and grow in the presence of stimulators or inhibitors of angiogenesis.13 Tubule length and the number of tubules are also important parameters for measuring the effects of exogenous factors on angiogenesis, as demonstrated by Kumar et al.17 For example, human umbilical vein endothelial cells stimulated with VEGFA form a larger number of tubules and longer tubules than nonstimulated cells. Tubule length can also be affected by factors such as cell adhesion or chemoattractant gradients.10 Therefore, in our studies, we chose the combination of total tubule length, number of tubules, and number of junctions to measure angiogenesis, and to compare the two quantitation methods.
Two semi-automated image analysis tools were used here. The first was a two-step process that involves image-processing using Adobe Photoshop and quantification of tubules using Angiosys 1.0, a commercially available software package from TCS Cellworks. Tubule quantification can be carried out simply using Adobe Photoshop after the image has been processed, but is limited to just measuring branching points and tubule length. Angiosys software is easy to use, and allows the user to group image files and include automated batch processing using the same settings and enabling rapid and standardized analysis. In contrast, the second method, Wimasis, operates via an online platform, where users can load their images and have their images analyzed by Wimasis.com. Wimasis creates image analysis algorithms specific to the needs of the researcher, to help automate and standardize image analysis, thus giving a high degree of objectivity and data comparability between two test conditions. The development of the Wimasis platform is based on real-time images from over 100 participating institutes. In this study, images uploaded on to the Wimasis platform were identical photos (×4 magnification) to those processed using Adobe Photoshop and Angiosys.
Our data indicate that both Angiosys and Wimasis can calculate accurately features related to angiogenesis, such as the number of tubules and branching points. Wimasis is the method of choice when the user wishes to compare angiogenesis in many samples at the same time or wishes to measure specific angiogenic parameters that are not commonly quantified by other software. Tubule quantification using Angiosys, with initial image processing using Adobe Photoshop, is useful for researchers who have smaller sets of samples and wish to optimize image-processing and quantification steps. Our analysis using Angiosys and Wimasis showed that CB-ECFC-derived cells incubated in suramin or SU6668 have significantly reduced number of junctions, number of tubules, and total tubule length compared to noninhibited CB-ECFC-derived cells. When Angiosys and Wimasis are compared with manual counting, there are no significant differences in the number of tubules and number of junctions in noninhibited CB-ECFC-derived cells. However, there was a significant difference in total tubule length of the same image analyzed with Wimasis compared to manual counting. The difference in the total tubule length may be due to the general application in Wimasis being slightly less efficient in recognizing tubules. The preprocessing before Angiosys analysis is specific to this application and so can be expected to be more efficient. Manual counting has a disadvantage over these two methods in that it generally lacks objectivity and is generally more time consuming.
To date, Angiosys and Wimasis have mainly been used for the quantitation of tubules on Matrigel assay, as described here. Other potential imaging applications may include the automated measurement of microvessel densities, which are often quantitated manually in 5 μm cryosections of tissues or scaffold after staining consecutive sections with for endothelial biomarkers.18 This would require a clear definition of the vessels to be counted, as different investigators may score these as single or clusters of endothelial cells with or without lumina, or as complete circular structures possessing distinct lumina. A second potential application may lend itself to the scoring of vessels formed in three-dimensional scaffolds, either in vitro or after implantation in vivo and their removal for imaging in vitro. For this latter application, scaffolds containing GFP-labeled vessel networks could be fixed with paraformaldehyde in vitro, examined by confocal laser scanning microscopy and high-resolution confocal image stacks of the GFP-labeled vessels could then be reconstructed using such software packages as those supplied by Imaris to obtain a precise three-dimensional restoration of the vascular network and vessel morphology.19 The numbers of vascular tubules, tubule length, and branching points could then potentially be quantitated as described in this article using the Angiosys or Wimasis systems, although the accuracy of such an application and the difficulties in quantitating vessels that penetrate into different planes of the scaffold would need to be assessed in detail.
Revascularization of most damaged tissues and organs by endothelial cells and their precursors, proangiogenic cells, and the products of these cells represents what we have referred to as “the holy grail” for therapeutic intervention in tissue repair.1 Some examples include the revascularization required to treat the delayed wound healing that can occur in peripheral vascular disease and diabetes, for patients with major burns and for those with ischemic heart disease, the treatment of which constitutes a major burden on health providers.1 As highlighted by Arnaoutova et al., many of the cells and factors identified as being active in the Matrigel assay exhibit similar effects in vivo.11 Thus, researchers involved in tissue engineering and drug discovery applications for tissue repair will realize the benefit of the more defined quantitative analysis of the Matrigel assay described here, in further reducing the time required for and the accuracy of their first rapid high-throughput screens for sourcing and selecting appropriate human endothelial cell populations from different tissues and stages of development before their incorporation into GMP-grade scaffolds, for testing endothelial cells obtained from patients with genetic diseases or with genetic variability or following genetic modification, or in assaying the effects of novel pro- and antiangiogenic compounds before embarking on more costly and more complex testing in longer term in vitro and in vivo models.
In summary, both methods described can accurately quantify tubules formed by CB-ECFC-derived cells on Matrigel. An essential part of both methods is the semi-automated batch processing of images and quantification, which decreases the amount of time and concurrently increases accuracy compared to manual counting. Thus, the quantification methods investigated are easy to perform, reproducible with reliable readouts, and would cater for different end users. They also allow researchers to choose a method that suits their experimental needs.
Appendix
APPENDIX FIG. 1.
Surface phenotype of CB-ECFC-derived cells. Representative FACS histograms of CB-ECFC-derived cells at passage 4–6. Cells were 99.8%±0.1% CD73+, 99.3%±0.7% CD31+, 99.9%±0.04% CD166+, 95.9%±2.3% CD144+, 98.7%±0.8% CD105+, and 99.0%±0.9% CD146+, but did not express CD133, CD90, CD14, or CD45 (2.0%±0.5% CD133+, 0.9%±0.2% CD90+, 0.6%±0.2% CD14+, and 5.4%±0.4% CD45+). Isotype-negative control values were 1.53%±0.4% mIgG1-PE+, 1.12%±0.1% mIgG2b-PE+, 2.18%±0.6% mIgG2a-APC+, 0.83%±0.1% mIgG1-FITC+, 1.87%±0.2% mIgG2a-PeCy7+, and 0.62%±0.1% mIgG1-PeCy7+. Values were expressed as mean±standard error of the mean for n=3 independent batches of cells. CB, cord blood; ECFC, endothelial colony-forming cell. Color images available online at www.liebertonline.com/tec
APPENDIX FIG. 2.
Adobe Photoshop processing method. (A) Image-processing steps using that forms “script” on Adobe Photoshop; (B) representative photos of each image-processing step. (a) The original phase-contrast image; (b) image converted to grayscale; (c) after conversion to the second image (B), the image underwent Gaussian blur; (d) image subtracted; (e) image inverted and brightness and contrast adjusted by applying autolevels; (f) histogram percentage; (g) image inverted; (h) holes filled, that is, cleaning the image; (i) image inverted; (j) image underwent Gaussian blur; (k) threshold adjusted accordingly; (l) final image used for analysis. Images were taken at ×4 magnification.
Appendix Table1.
Parameters Used for Optimization of Cord Blood-Derived Endothelial Colony-Forming Cells on Matrigel Assay
| Factors | Parameters |
|---|---|
| Cells | Human CB-derived ECFCs |
| No. of cells | 1.5 × 104 cells |
| Reagents | GFR matrigel (BD) |
| Parameters | 20 hours of incubation |
| Media | EGM-2 media (2% FBS) |
| Matrigel amount | 50 μl per well of 96 well plate |
| Concentration of suramin | 10 μM |
| Concentration of SU6668 | 10 μM |
Factors potentially affecting assay variability include type of cells, number of cells, reagents, parameters, media, matrigel amount and concentration of inhibitors (suramin and SU6668)
CB, cord blood; ECFC, endothelial colony-forming cells.
Acknowledgments
The authors wish to acknowledge the support from NHS Blood and Transplant, National Institute of Health Research UK, Oxford National Institute for Health Research (NIHR) Biomedical Research Centre, Restore Burns and Wound Healing Trust, and EU Cascade project. This article presents independent research commissioned by the National Institute for Health Research (NIHR) under its Program Grants scheme (RP-PG-0310-1003). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. The authors wish to thank Mr. Kilian Schramm, Chief Executive Officer of Wimasis GmbH, who analyzed our data during the development of the trial version of Wimasis WimTube, and Ms. Janice Gotobed and Mrs. Sandra Britt, who collected the UCB units.
Disclosure Statement
All authors declare that no competing financial interests exist.
References
- 1.Watt S.M. Athanassopoulos A. Harris A.L. Tsaknakis G. Human endothelial stem/progenitor cells, angiogenic factors and vascular repair. J R Soc Interface. 2010;7(Suppl 6):S731. doi: 10.1098/rsif.2010.0377.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Critser P.J. Yoder M.C. Endothelial colony-forming cell role in neoangiogenesis and tissue repair. Curr Opin Organ Transplant. 2010;15:68. doi: 10.1097/MOT.0b013e32833454b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Smet F. Segura I. De Bock K. Hohensinner P.J. Carmeliet P. Mechanisms of vessel branching: filopodia on endothelial tip cells lead the way. Arterioscler Thromb Vasc Biol. 2009;29:639. doi: 10.1161/ATVBAHA.109.185165. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree B. Subramanian V. Behavior of endothelial cells on Matrigel and development of a method for a rapid and reproducible in vitro angiogenesis assay. In Vitro Cell Dev Biol Anim. 2007;43:87. doi: 10.1007/s11626-007-9012-x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. Fisher N. Newey S.E. Smythe J. Tatton L. Tsaknakis G. Forde S.P. Carpenter L. Athanassopoulos T. Hale S.J. Ferguson D.J. Tyler M.P. Watt S.M. The impact of proliferative potential of umbilical cord-derived endothelial progenitor cells and hypoxia on vascular tubule formation in vitro. Stem Cells Dev. 2009;18:359. doi: 10.1089/scd.2008.0071. [DOI] [PubMed] [Google Scholar]
- 6.Ingram D.A. Mead L.E. Tanaka H. Meade V. Fenoglio A. Mortell K. Pollok K. Ferkowicz M.J. Gilley D. Yoder M.C. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood. 2004;104:2752. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 7.Arnaoutova I. Kleinman H.K. In vitro angiogenesis: endothelial cell tube formation on gelled basement membrane extract. Nat Protoc. 2010;5:628. doi: 10.1038/nprot.2010.6. [DOI] [PubMed] [Google Scholar]
- 8.Liekens S. De Clercq E. Neyts J. Angiogenesis: regulators and clinical applications. Biochem Pharmacol. 2001;61:253. doi: 10.1016/s0006-2952(00)00529-3. [DOI] [PubMed] [Google Scholar]
- 9.Lin Y. Weisdorf D.J. Solovey A. Hebbel R.P. Origins of circulating endothelial cells and endothelial outgrowth from blood. J Clin Invest. 2000;105:71. doi: 10.1172/JCI8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merks R.M. Brodsky S.V. Goligorksy M.S. Newman S.A. Glazier J.A. Cell elongation is key to in silico replication of in vitro vasculogenesis and subsequent remodeling. Dev Biol. 2006;289:44. doi: 10.1016/j.ydbio.2005.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaoutova I. George J. Kleinman H.K. Benton G. The endothelial cell tube formation assay on basement membrane turns 20: state of the science and the art. Angiogenesis. 2009;12:267. doi: 10.1007/s10456-009-9146-4. [DOI] [PubMed] [Google Scholar]
- 12.Donovan D. Brown N.J. Bishop E.T. Lewis C.E. Comparison of three in vitro human “angiogenesis” assays with capillaries formed in vivo. Angiogenesis. 2001;4:113. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 13.Guidolin D. Vacca A. Nussdorfer G.G. Ribatti D. A new image analysis method based on topological and fractal parameters to evaluate the angiostatic activity of docetaxel by using the Matrigel assay in vitro. Microvasc Res. 2004;67:117. doi: 10.1016/j.mvr.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Tong E.Y. Collins G.C. Greene-Colozzi A.E. Chen J.L. Manos P.D. Judkins K.M. Lee J.A. Ophir M.J. Laliberte F.M. Levesque T.J. Motion-based angiogenesis analysis: a simple method to quantify blood vessel growth. Zebrafish. 2009;6:239. doi: 10.1089/zeb.2008.0554. [DOI] [PubMed] [Google Scholar]
- 15.Laird A.D. Vajkoczy P. Shawver L.K. Thurnher A. Liang C. Mohammadi M. Schlessinger J. Ullrich A. Hubbard S.R. Blake R.A. Fong T.A. Strawn L.M. Sun L. Tang C. Hawtin R. Tang F. Shenoy N. Hirth K.P. McMahon G. Cherrington SU6668 is a potent antiangiogenic and antitumor agent that induces regression of established tumors. Cancer Res. 2000;60:4152. [PubMed] [Google Scholar]
- 16.Staton C.A. Reed M.W. Brown N.J. A critical analysis of current in vitro and in vivo angiogenesis assays. Int J Exp Pathol. 2009;90:195. doi: 10.1111/j.1365-2613.2008.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R. Yoneda J. Bucana C.D. Fidler I.J. Regulation of distinct steps of angiogenesis by different angiogenic molecules. Int J Oncol. 1998;12:749. doi: 10.3892/ijo.12.4.749. [DOI] [PubMed] [Google Scholar]
- 18.Traktuev D.O. Prater D.N. Merfeld-Clauss S. Sanjeevaiah A.R. Saadatzadeh M.R. Murphy M. Johnstone B.H. Ingram D.A. March K.L. Robust functional vascular network formation in vivo by cooperation of adipose progenitor and endothelial cells. Circ Res. 2009;104:1410. doi: 10.1161/CIRCRESAHA.108.190926. [DOI] [PubMed] [Google Scholar]
- 19.Adighibe O. Micklem K. Campo L. Ferguson M. Harris A. Pozos R. Gatter K. Pezzella F. Is nonangiogenesis a novel pathway for cancer progression? A study using 3-dimensional tumour reconstructions. Br J Cancer. 2006;94:1176. doi: 10.1038/sj.bjc.6603039. [DOI] [PMC free article] [PubMed] [Google Scholar]