Abstract
RtcB enzymes are novel RNA ligases that join 2′,3′-cyclic phosphate and 5′-OH ends. The phylogenetic distribution of RtcB points to its candidacy as a tRNA splicing/repair enzyme. Here we show that Escherichia coli RtcB is competent and sufficient for tRNA splicing in vivo by virtue of its ability to complement growth of yeast cells that lack the endogenous “healing/sealing-type” tRNA ligase Trl1. RtcB also protects yeast trl1Δ cells against a fungal ribotoxin that incises the anticodon loop of cellular tRNAs. Moreover, RtcB can replace Trl1 as the catalyst of HAC1 mRNA splicing during the unfolded protein response. Thus, RtcB is a bona fide RNA repair enzyme with broad physiological actions. Biochemical analysis of RtcB highlights the uniqueness of its active site and catalytic mechanism. Our findings draw attention to tRNA ligase as a promising drug target.
Keywords: Enzyme Catalysis, Enzyme Mutation, Protein Structure, RNA Splicing, Transfer RNA (tRNA)
Introduction
Escherichia coli RtcB exemplifies a new family of RNA ligases that directly seal 2′,3′-cyclic phosphate and 5′-OH ends (1–3). Direct ligation is thought to be the main pathway of tRNA splicing in animals and archaea (4–6). By contrast, yeast and plants rely on a different mechanism of tRNA splicing in which the broken 3′ and 5′ ends are healed (converted to a 3′-OH/2′-PO4 and 5′-PO4, respectively) and then sealed by a classical ATP-dependent RNA ligase (7) (see Fig. 1). RNA ligases of the RtcB family are present in metazoa and protozoa, but not in fungi and plants. RtcB homologs purified from archaeal and mammalian cells can seal broken tRNA halves and are thereby imputed to be the catalysts of archaeal and mammalian tRNA splicing (2, 3). However, this scenario is complicated by the existence of a yeast/plant-like tRNA splicing pathway in mammalian cells (8–12) and of analogous yeast-like RNA repair enzymes in many archaeal taxa (13–16). Definitive genetic evidence that RtcB is the sole essential agent of the repair phase of mammalian or archaeal tRNA splicing is lacking, and the available genetic evidence concerning the healing-sealing pathway in animals is equivocal. Genetic ablation of a murine homolog of a yeast-like pathway component Tpt1 (the enzyme that removes the 2′-phosphate at the splice junction; see Fig. 1) has no discernible phenotype (17), suggesting that the mammalian yeast-like pathway either is functionally redundant with direct ligation or is non-contributory to mammalian tRNA splicing. By contrast, siRNA-directed depletion of the mammalian RNA 5′-kinase (an ortholog of the kinase domain of yeast/plant tRNA ligase) elicited a defect in tRNA splicing in vitro (10). However, siRNA-directed depletion of mammalian RtcB was also reported to inhibit ligation of tRNA halves in cell extracts and to delay tRNA splicing in living cells (3). These findings leave unresolved the following key issues: (i) whether RtcB can suffice for tRNA splicing as the only source of tRNA ligase activity in a eukaryal cell and (ii) whether RtcB can perform other RNA repair functions in vivo. Here we address these questions by using budding yeast as a surrogate genetic model for tRNA splicing and RNA repair by heterologous enzymes (7).
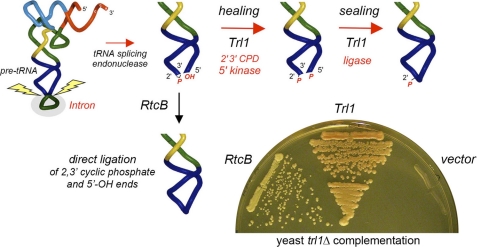
FIGURE 1.
RtcB can perform eukaryal tRNA splicing in vivo. The two pathways of tRNA splicing, via end-healing and end-sealing in yeast and plants (catalyzed in S. cerevisiae by the tRNA ligase Trl1) or via direct ligation in animals and archaea (where RtcB is a putative catalyst), are shown. The capacity of E. coli RtcB to complement an S. cerevisiae trl1Δ mutant was assayed by plasmid shuffle. Yeast trl1Δ p(CEN URA3 TRL1) cells transformed with a 2μ HIS3 plasmid (vector control) or 2μ HIS3 plasmids encoding Trl1 or RtcB under the control of a yeast TPI1 promoter were selected for histidine prototrophy and then streaked on agar medium containing 0.75 mg/ml 5-fluoroorotic acid (a drug that selects against the URA3 TRL1 plasmid). The plate was photographed after 3 days at 30 °C.
EXPERIMENTAL PROCEDURES
Expression Plasmids
The bacterial plasmid pET28b-His10Smt3-RtcB encodes wild-type E. coli RtcB fused to an N-terminal His10Smt3 tag (1). Missense mutations were introduced into the RtcB ORF by two-stage overlap extension PCR. The RtcB inserts were sequenced to confirm the desired mutations and exclude the acquisition of unwanted coding changes. The wild-type and mutated RtcB ORFs were excised from their respective pET plasmids and inserted into the yeast vector pRS423 (2μ HIS3) wherein RtcB expression is under the control of the yeast TPI1 promoter. The yeast p(CEN LEU2 GAL1PaT) plasmid for galactose-inducible expression of intracellular Pichia acaciae toxin (PaT)2 lacking the N-terminal 12-amino acid signal peptide was constructed by excising an NdeI/SalI fragment containing the PaT ORF from plasmid pPACBX (18) (a gift of Roland Klassen and Friedhelm Meinhardt) and inserting it into pRS415 (CEN LEU2) between GAL1 promoter and terminator elements.
RtcB Purification
The wild-type and mutant pET28b-His10Smt3-RtcB plasmids were transformed into E. coli BL21-CodonPlus(DE3). Induction of RtcB expression, preparation of soluble lysates, recovery of the His10Smt3-RtcB proteins by nickel-agarose chromatography, excision of the tags by treatment with the Smt3-specifc protease Ulp1, and separation of the tag-free RtcB proteins from His10Smt3 by a second round of nickel-agarose chromatography were performed as described (1). Protein concentrations were determined by using the Bio-Rad dye reagent with BSA as the standard. The polypeptide compositions of the RtcB preparations were analyzed by SDS-PAGE (supplemental Figs. S1 and S2).
RNA Repair Substrates
A synthetic RNA oligonucleotide R30 containing the anticodon stem-loop of yeast tRNAGlu(UUC) was 5′ 32P-labeled by reaction with T4 Pnkp and [γ-32P]ATP. R30 was then cleaved 3′ of the wobble uridine by reaction with Kluyveromyces lactis γ-toxin (1). The 32P-labeled R19>p strand with a 2′,3′ cyclic phosphate end (5′-pUGGCUCCGAUAUCACGCUU>p) was purified by preparative PAGE. Oligonucleotide HOR20 (5′-HOUCACCGUGGUAUCGGAGCGC) was employed to form a broken tRNA-like stem-loop (see Fig. 4) by mixture with the 5′ 32P-labeled R19 single strand at an R20:R19 ratio of 10:1. To test the RNA requirements for the RtcB ligation reaction, the 32P-labeled R19>p strand was also annealed to a synonymous deoxyuridine-containing HOD20 oligonucleotide to form a broken RNA-DNA hybrid stem-loop (see Fig. 5).
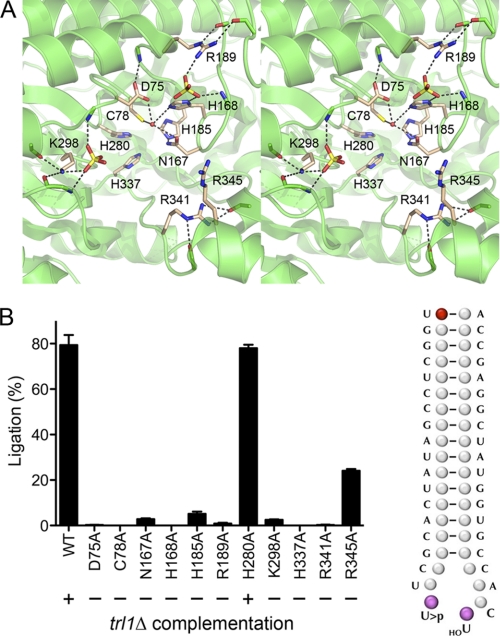
FIGURE 4.
Structure-guided mapping of the RtcB active site. Top, stereo view of the putative active site pocket of P. horikoshii RtcB (from Protein Data Bank (PDB) 1UC2). The conserved amino acid residues that were subjected to alanine scanning in E. coli RtcB are shown as stick models with beige carbons and are labeled according to their E. coli RtcB equivalents. Two sulfate anions bound to RtcB are rendered as stick models. A water molecule that could plausibly mimic the metal cofactor is depicted as a red sphere. Ionic and hydrogen-bonding interactions are denoted by dashed lines. Bottom, RNA ligase reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 2 mm MnCl2, 100 μm GTP, 100 nm of 5′-32P-labeled broken RNA stem-loop substrate (depicted at right), and 2 μm wild-type RtcB or RtcB-Ala mutants (see supplemental Fig. S1) were incubated for 30 min at 37 °C. The reactions were quenched by adding 10 μl of 40 mm EDTA/95% formamide, and the products were analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 m urea in 45 mm Tris borate, 1.25 mm EDTA. The extents of conversion of the radiolabeled 19-mer substrate strand into sealed 39-mer product (% ligation) were quantified by scanning the gel with a Fuji Film BAS-2500 imager. Each datum in the bar graph is the average of three separate ligation experiments ±S.E. The RtcB-Ala ORFs were cloned into yeast 2μ plasmids under the control of the TPI1 promoter and tested for trl1Δ complementation as described in the legend for Fig. 1. The results are summarized below the graph.
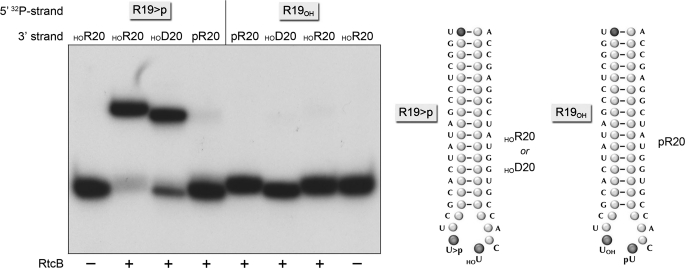
FIGURE 5.
RtcB substrate specificity. Reaction mixtures (10 μl) containing 50 mm Tris-HCl (pH 8.0), 2 mm MnCl2, 100 μm GTP, 100 nm of 5′-32P-labeled broken stem-loop substrates (illustrated at right) with 2′,3′ cyclic or 3′-OH ends on the proximal R19 strand and 5′-OH or 5′-PO4 ends on the distal 20-mer strand (as specified above the lanes), and 1 μm RtcB (where indicated by +) were incubated for 30 min at 37 °C. The products were analyzed by denaturing PAGE and visualized by autoradiography.
Alternative stem-loop RNA repair substrates with a 3′-OH end at the break site (see Fig. 5) were prepared using a synthetic R19OH oligonucleotide that had been 5′ 32P-labeled and then purified by PAGE. The labeled R19OH oligonucleotide was annealed to a 10-fold excess of unlabeled D20 and R20 strands.
RNA repair substrates with a 5′-PO4 at the break site (see Fig. 5) were formed by annealing radiolabeled R19>p or R19OH strands to a 10-fold excess of cold pR20 strand. The pRNA strand was generated by enzymatic phosphorylation of HOR20 using T4 Pnkp and cold ATP and then purified by PAGE.
RESULTS AND DISCUSSION
Here we interrogated the tRNA repair function of E. coli RtcB in vivo by complementation of a lethal deletion of the TRL1 gene encoding the tRNA ligase of Saccharomyces cerevisiae (7). Yeast Trl1 is a trifunctional tRNA repair enzyme (with 5′-OH kinase, 2′,3′ cyclic phosphodiesterase, and ATP-dependent RNA ligase activities) that heals and seals the broken tRNA halves with 2′,3′ cyclic phosphate and 5′-OH ends generated by incision of pre-tRNAs at the exon-intron junctions (Fig. 1). E. coli RtcB is a monofunctional ligase that directly joins 2′,3′ cyclic phosphate and 5′-OH ends (1) (Fig. 1). We expressed the 408-amino acid E. coli RtcB protein in S. cerevisiae under the control of a constitutive promoter on a 2μ plasmid and demonstrated by plasmid shuffle that RtcB could indeed sustain growth of a trl1Δ strain (Fig. 1). These results prove that RtcB is able and sufficient to perform the essential repair steps of eukaryal tRNA splicing in vivo.
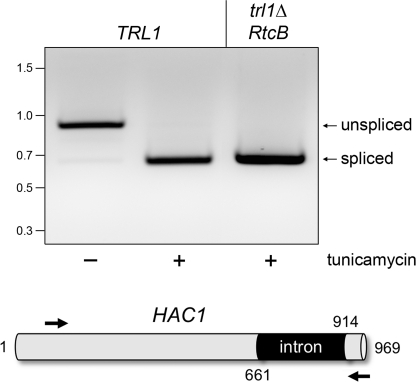
The discovery that the healing and sealing activities of yeast Trl1 are also responsible for unconventional splicing of the HAC1 mRNA in the yeast unfolded protein response (UPR) pathway (19, 20) extended the RNA repair paradigm to mRNA metabolism. Endoplasmic reticulum stress induces the Ire1 endonuclease to cleave HAC1 mRNA at two sites, which liberates a 252-nucleotide intron (Fig. 2) and leaves 2′,3′ cyclic phosphate and 5′-OH termini on the proximal and distal exons, respectively. Healing and sealing of the exons by Trl1 creates a new open reading frame encoding an active Hac1 transcription factor. The mammalian UPR entails stress-induced Ire1 cleavage and unconventional splicing of the XBP1 mRNA (21), but the enzyme responsible for sealing the broken XBP1 transcript is not known. Here we tested the capacity of RtcB to execute unconventional mRNA splicing during the yeast UPR. Yeast TRL1 cells exposed to tunicamycin shifted their HAC1 mRNA profile, as assayed by RT-PCR with primers flanking the intron, whereby the intron-containing long form was replaced by a spliced short form (Fig. 2). Sequencing of the isolated RT-PCR products confirmed their identity as unspliced and spliced HAC1 cDNAs, respectively. Yeast trl1Δ cells reliant on RtcB as their source of tRNA ligase were proficient in generating the spliced HAC1 RNA in the presence of tunicamycin (Fig. 2), signifying that RtcB is competent for mRNA splicing in the eukaryal UPR. Sequencing of the RT-PCR product verified that the Ire1 endonuclease cleavage sites were ligated faithfully.
FIGURE 2.
RtcB can perform HAC1 mRNA splicing in the unfolded protein response. The unprocessed yeast HAC1 open reading frame, containing a 252-nucleotide intron, is depicted at the bottom. To gauge HAC1 mRNA splicing, cultures (10 ml) of S. cerevisiae trl1Δ p(2μ HIS3 TRL1) and trl1Δ p(2μ HIS3 RtcB) cells were grown at 30 °C in YPD media until the A600 reached 0.6. The cultures were split into 5-ml aliquots, one of which was adjusted to 1.5 μg/ml tunicamycin to induce endoplasmic reticulum stress. One hour later, the cells were harvested by centrifugation. Total RNA was isolated by using a yeast RNA purification kit (Epicentre Biotechnologies). HAC1 transcripts were detected by RT-PCR using primers flanking the intron. The cDNAs were synthesized by using the SuperScript III system (Invitrogen) with 1.0 μg of total RNA as template and 0.1 μm of HAC1-specific antisense primer (5′-dCATGAAGTGATGAAGAAATCATTCAATTC; complementary to nucleotides 940–968). The cDNAs were amplified by 22 cycles of PCR with Herculase (Stratagene) using the antisense primer and a sense-strand primer (5′-dCCAAGGAAAAGAGCCAAGACAAAAGAGG; corresponding to nucleotides 82–109). The RT-PCR products were analyzed by 1.4% agarose gel electrophoresis. A negative image of the ethidium bromide-stained gel is shown. The positions and sizes (in bp) of marker dsDNAs are indicated at left. The predicted sizes of the unspliced and spliced HAC1 RT-PCR products are 886 and 634 bp, respectively.
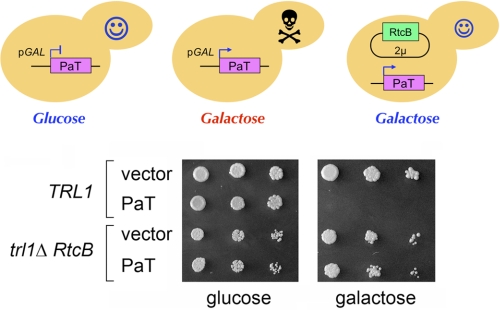
Another manifestation of tRNA repair is the capacity to protect cells from the cytotoxicity inflicted by tRNA-specific ribotoxins that incise the anticodon loop by a transesterification mechanism that leaves 2′,3′ cyclic phosphate and 5′-OH termini at the broken tRNA ends (22, 23). PaT is a secreted fungal defense molecule that penetrates S. cerevisiae cells and, upon accessing the cytoplasm, breaks the anticodon loop of tRNAGln(UUG). Consequent depletion of the tRNAGln(UUG) pool arrests yeast growth (18). Galactose-induced expression in S. cerevisiae of an intracellular form of PaT recapitulates its toxicity (18). The salient finding here was that replacing yeast Trl1 with RtcB as the source of tRNA ligase protected S. cerevisiae from PaT-mediated growth inhibition (Fig. 3). Taken together, these experiments establish that RtcB is a tRNA/mRNA repair enzyme with broad physiological actions.
FIGURE 3.
RtcB protects cells from cytotoxic tRNA damage. Top, schematic depiction of yeast cells harboring GAL1PaT in the presence of glucose or galactose as the carbon source. The right-most cell expresses RtcB in lieu of Trl1. Bottom, S. cerevisiae TRL1 and trl1Δ RtcB cells were transformed either with a CEN LEU2 GAL1PaT plasmid or with an empty CEN LEU2 vector. Cultures derived from single Leu+ colonies were grown in SD−Leu medium at 37 °C. After adjustment to A600 of 0.1, aliquots (3 μl) of serial 5-fold dilutions of the cells were spotted on −Leu agar plates containing either 2% glucose or 2% galactose. The plates were photographed after incubation for 3 days (glucose) or 4 days (galactose) at 37 °C.
The RtcB sealing reaction is posited to entail nucleophilic attack by the O5′ nucleophile on the cyclic phosphate, with expulsion of the ribose O2′. Our initial findings that RtcB is manganese-dependent (1) indicated that sealing is not merely reversal of the metal-independent cleavage transesterification mechanism used by tRNA splicing endoribonuclease and tRNA-damaging anticodon nucleases. Manganese might promote RtcB catalysis by coordinating the O5′ nucleophile to lower its pKa and/or engaging the cyclic phosphate oxygens to stabilize the transition state. The mechanistic novelty of RtcB is underscored by the crystal structure of the RtcB homolog from Pyrococcus horikoshii (24) (Fig. 4). RtcB has a distinctive tertiary structure with no similarity to any known ligases or phosphotransferases. RtcB has a deep and wide hydrophilic pocket lined by conserved histidines and a cysteine, suggestive of a metal-binding site that, being dominated by “soft” metal contacts to histidine nitrogens and cysteine sulfur, could account for the fact that E. coli RtcB requires manganese and is virtually inactive with magnesium (1). A water coordinated by the equivalents of E. coli RtcB side chains Asp-75, Cys-78, Asn-167, and His-168 is a potential mimic of the enzyme-bound metal (Fig. 4). Flanking the putative metal site in RtcB are two sulfate anions (potential mimetics of RNA phosphates) coordinated by basic amino acid side chains equivalent to E. coli RtcB residues Lys-298, His-168, and Arg-189 (Fig. 4).
Here we used the Pyrococcus RtcB structure to guide a mutational analysis of 11 conserved residues in E. coli RtcB that we thought might comprise an active site (Fig. 4). Recombinant RtcB and RtcB-Ala proteins were produced in bacteria purified from soluble lysates (supplemental Fig. S1). The proteins were tested for ligase activity with a broken tRNA-like substrate (1) modeled on the anticodon stem-loop of yeast tRNAGlu(UUC) (Fig. 4). Although RtcB and mutant H280A sealed ∼80% of the stem-loop substrate in vitro, mutants D75A, C78A, H168A, R189A, H337A, and R341A were virtually inert, with <1% ligation (Fig. 4). Mutants N167A, H185A, R189A and K298A were severely impaired (sealing 3, 5, and 3% of the substrate, respectively), and R345A was modestly impaired (24% sealing). Each of the RtcB-Ala mutants was tested for trl1Δ complementation in yeast; only H280A was functional (Fig. 4).
Structure-activity relations at seven of the RtcB amino acids defined as essential by the alanine scan were probed by introducing conservative substitutions (supplemental Fig. S2). Replacing Asp-75 by either asparagine or glutamate inactivated RtcB, attesting to the requirement for a carboxylate at this position and the steric constraints on the main chain to carboxylate distance (supplemental Fig. S2). Changing Cys-78 to serine abolished ligase activity, signifying that the Sγ atom is critical. His-168 and His-337 were replaced by glutamine and asparagine; the H168N, H168Q, H337N, and H337Q mutants were catalytically inactive (supplemental Fig. S2). The mutational effects are consistent with roles for Asp-75, Cys-78, His-168, Asn-167, His-185, and His-337 in metal binding and/or RNA transesterification. Although replacing the sulfate-binding Lys-298 with glutamine phenocopied K298A, the arginine substitution restored ligation to one-fourth of the wild-type RtcB (supplemental Fig. S2), highlighting positive charge as the key property of this residue. By contrast, neither Arg-189 nor Arg-341 could be functionally substituted by lysine or glutamine, implying that the multivalent ionic and hydrogen-bonding contacts of these arginines seen in the RtcB crystal structure (Fig. 4) are indeed pertinent to enzyme activity, either via binding to the RNA phosphates (a putative function of Arg-189) or in stabilizing the active site conformation (a likely role of Arg-341). None of the conservative mutants complemented trl1Δ (not shown).
Further insights to the substrate specificity of RtcB were gained by varying the 3′ and 5′ termini of the broken stem-loop substrate and testing the capacity of a 5′-OH DNA strand to serve as the nucleophile for the sealing reaction. A 5′ 32P-labeled 19-mer RNA with either a 2′,3′ cyclic phosphate end (R19>p) or a 3′-OH end (R19OH) was annealed to an unlabeled 20-mer strand composed of all ribonucleotides (R20) or all deoxynucleotides (D20) to form the broken stem-loops depicted in Fig. 5. The unlabeled 20-mer strands had either a 5′-OH terminus (HOR20 or HOD20) or a 5′-PO4 terminus (pR20). The results of the ligation assays established that: (i) 2′,3′ cyclic phosphate and 5′-OH ends are the only suitable reactants for E. coli RtcB among the combinations tested and (ii) RtcB is adept at joining the R19>p “donor” strand to either RNA or DNA “acceptor” strands so long as they have a 5′-OH end. The versatility of RtcB with respect to the 5′-OH acceptor suggests practical applications for this enzyme in tagging and/or cloning mature RNAs or RNA processing intermediates that have 2′,3′ cyclic phosphate ends.
In summary, we have provided convincing genetic and biochemical evidence that RtcB can serve as a genuine tRNA splicing/repair enzyme in a eukaryal cell. Our findings here and previously (7) attest to the portability of viral, fungal, plant, and bacterial tRNA repair systems. The salient theme is that sealing by any of several distinct enzymatic/chemical pathways suffices for tRNA splicing and unconventional mRNA splicing in vivo. The candidacy of mammalian RtcB homolog as an agent of tRNA splicing by direct ligation is supported by our genetic results. However, the jury is still out as to whether RtcB is functionally redundant with a mammalian tRNA repair pathway via sequential end-healing and end-sealing steps (8, 10).
Our findings also fortify the case for the Trl1-associated ligase as a promising antifungal drug target for the following reasons: (i) Trl1 is present in all fungal proteomes; (ii) ligase-inactivating mutations in the active site of Trl1 are lethal in vivo (25, 26); and (iii) mammalian taxa encode no homolog of the Trl1 ligase domain. Thus, any ATP-dependent ligase involved in the putative mammalian healing-sealing pathway (8) must either belong to a different enzyme family or have diverged so far from a fungal-type ancestor as to be unrecognizable. In either event, one predicts that a mechanism-based inhibitor of the Trl1 ligase should selectively block fungal growth without affecting mammalian cells. By the same token, an antagonist of RtcB might be useful in transiently impeding the UPR for therapeutic benefit in human diseases involving endoplasmic reticulum stress (21). The availability of isogenic yeast strains with orthogonal tRNA splicing systems should enable differential cell-based screening for bioactive molecules that selectively inhibit growth by targeting Trl1 versus RtcB and vice versa.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM46330 (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PaT
- P. acaciae toxin
- UPR
- unfolded protein response.
REFERENCES
- 1. Tanaka N., Shuman S. (2011) J. Biol. Chem. 286, 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Englert M., Sheppard K., Aslanian A., Yates J. R., 3rd, Söll D. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C. L., Lührmann R., Söll D., Martinez J. (2011) Science 331, 760–764 [DOI] [PubMed] [Google Scholar]
- 4. Filipowicz W., Shatkin A. J. (1983) Cell 32, 547–557 [DOI] [PubMed] [Google Scholar]
- 5. Laski F. A., Fire A. Z., RajBhandary U. L., Sharp P. A. (1983) J. Biol. Chem. 258, 11974–11980 [PubMed] [Google Scholar]
- 6. Zofallova L., Guo Y., Gupta R. (2000) RNA 6, 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwer B., Sawaya R., Ho C. K., Shuman S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zillmann M., Gorovsky M. A., Phizicky E. M. (1991) Mol. Cell. Biol. 11, 5410–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spinelli S. L., Malik H. S., Consaul S. A., Phizicky E. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14136–14141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weitzer S., Martinez J. (2007) Nature 447, 222–226 [DOI] [PubMed] [Google Scholar]
- 11. Ramirez A., Shuman S., Schwer B. (2008) RNA 14, 1737–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwer B., Aronova A., Ramirez A., Braun P., Shuman S. (2008) RNA 14, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato-Murayama M., Bessho Y., Shirouzu M., Yokoyama S. (2005) J. Mol. Biol. 348, 295–305 [DOI] [PubMed] [Google Scholar]
- 14. Jain R., Shuman S. (2009) RNA 15, 923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brooks M. A., Meslet-Cladiére L., Graille M., Kuhn J., Blondeau K., Myllykallio H., van Tilbeurgh H. (2008) Protein Sci. 17, 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Torchia C., Takagi Y., Ho C. K. (2008) Nucleic Acids Res. 36, 6218–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harding H. P., Lackey J. G., Hsu H. C., Zhang Y., Deng J., Xu R. M., Damha M. J., Ron D. (2008) RNA 14, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klassen R., Paluszynski J. P., Wemhoff S., Pfeiffer A., Fricke J., Meinhardt F. (2008) Mol. Microbiol. 69, 681–697 [DOI] [PubMed] [Google Scholar]
- 19. Sidrauski C., Cox J. S., Walter P. (1996) Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 20. Gonzalez T. N., Sidrauski C., Dörfler S., Walter P. (1999) EMBO J. 18, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hotamisligil G. S. (2010) Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amitsur M., Levitz R., Kaufmann G. (1987) EMBO J. 6, 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nandakumar J., Schwer B., Schaffrath R., Shuman S. (2008) Mol. Cell 31, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okada C., Maegawa Y., Yao M., Tanaka I. (2006) Proteins 63, 1119–1122 [DOI] [PubMed] [Google Scholar]
- 25. Sawaya R., Schwer B., Shuman S. (2003) J. Biol. Chem. 278, 43928–43938 [DOI] [PubMed] [Google Scholar]
- 26. Wang L. K., Shuman S. (2005) RNA 11, 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.