Abstract
The class III receptor-tyrosine kinase Flt3 regulates normal hematopoiesis. An internal tandem duplication (ITD) in the juxtamembrane domain of Flt3 (Flt3-ITD) contributes to transformation and is associated with poor prognosis in acute myeloid leukemia. Here, we demonstrate that, as compared with wild-type Flt3 (Flt3-WT), Flt3-ITD more rapidly undergoes degradation through the proteasomal and lysosomal pathways in model hematopoietic 32D cells and in human leukemic MV4-11 cells. The Hsp90 inhibitor 17-allylaminodemethoxygeldanamycin (17-AAG) preferentially induced the polyubiquitination and proteasomal degradation of Flt3-ITD autophosphorylated on Tyr-591 in these cells. The E3 ubiquitin ligases c-Cbl and to a lesser extent Cbl-b facilitated at least partly Lys-48-linked polyubiquitination of autophosphorylated Flt3-ITD when coexpressed in 293T cells. Moreover, c-Cbl and Cbl-b facilitated degradation of Flt3-ITD in 293T cells and significantly enhanced the 17-AAG-induced decline in autophosphorylated Flt3-ITD. The enhancement of Flt3-ITD degradation was also observed in 32D cells inducibly overexpressing c-Cbl or Cbl-b. Furthermore, overexpression of loss-of-function mutants of both c-Cbl (c-Cbl-R420Q) and Cbl-b (Cbl-b-C373A) together in 32D cells retarded the degradation of autophosphorylated Flt3-ITD and significantly inhibited the 17-AAG-induced degradation of Flt3-ITD to confer the resistance to cytotoxicity of 17-AAG on these cells. These results suggest that c-Cbl as well as Cbl-b may play important roles in Hsp90 inhibitor-induced degradation of Flt3-ITD through the ubiquitin proteasome system and in regulation of the basal expression level of Flt3-ITD in leukemic cells.
Keywords: Cancer Therapy, Leukemia, Receptor-tyrosine kinase, Ubiquitin Ligase, Ubiquitination, 17-AAG, Cbl-b, Flt3-ITD, HSP90, c-Cbl
Introduction
Fms-like tyrosine kinase 3 (Flt3)2 also known as fetal liver kinase-2 (Flk-2) is a receptor-tyrosine kinase expressed on hematopoietic progenitors and regulates early steps of hematopoietic progenitor cell proliferation, survival, and differentiation (1, 2). Wild-type Flt3 (Flt3-WT) has been shown to be highly expressed in several hematopoietic malignancies including 70–100% cases of acute myeloid leukemia (AML). Oncogenic internal tandem duplication (ITD) mutations in the juxtamembrane domain of Flt3 (Flt3-ITD) and point mutations within the tyrosine kinase domain, such as the most predominant D835Y mutation, are the most frequent kinase mutations in AML, occurring in 25–30 and 5–10% of cases, respectively, and are associated with poor prognosis (1–3). Flt3-ITD results in ligand-independent autophosphorylation and activation of the receptor with subsequent activation of multiple downstream targets, including the signal transducer and activator of transcription 5 (STAT5), mitogen-activated protein kinase, and Akt pathways. Flt3-ITD has been shown to induce a myeloproliferative disorder in murine models. Flt3-ITD exists partially in an immature, underglycosylated, constitutively phosphorylated form (4). Aberrant intracellular localization and activity of Flt3-ITD generate oncogenic phosphorylation patterns and aberrantly activate signaling cascades (5). Because Flt3 or Flt3-ligand (FL) knock-out mice have only a subtle hematopoietic stem/progenitor cell deficit and show no significant disadvantage in viability, interference with deregulated Flt3 functions appears as a promising treatment option for AML (1–3). Thus, several Flt3 inhibitors are currently under evaluation for their efficacy in AML patients with Flt3-ITD.
Recent studies showed that both activated Flt3 and mutant Flt3 are degraded through the proteasome (6–11). The proteasomal degradation of targeted proteins, including protein kinases, requires polyubiquitination catalyzed by a series of enzymes containing ubiquitin activating enzymes (E1), ubiquitin conjugases (E2), and ubiquitin ligases (E3) (12, 13). E3 ubiquitin ligases confer substrate specificity and are responsible for mediating the transfer of ubiquitin from E2s to the substrates. The Cbl proteins are a highly conserved family of RING finger E3 ubiquitin ligases that regulate signaling by receptor and non-receptor-tyrosine kinases, including EGF receptor, PDGF receptor α, and FMS (14). There are three mammalian Cbl proteins encoded by separate genes: c-Cbl, Cbl-b, and Cbl-c. These proteins contain an N-terminal phosphotyrosine binding domain that allows direct interaction with activated receptor-tyrosine kinases and a RING finger domain that classifies Cbl proteins as E3 ubiquitin ligases. Recently, mutations of c-Cbl as well as Cbl-b have been found in various hematological neoplasms, including myelodysplastic/myeloproliferative neoplasms, myeloproliferative neoplasms, and secondary AML (15). Activated Flt3 recruits c-Cbl via the autophosphorylation sites Tyr-589 and Tyr-599 to induce tyrosine phosphorylation of c-Cbl, which is required for the E3 activity (1, 14–16). It was also shown that a dominant negative form of c-Cbl, Cbl-70Z, inhibited FL-induced ubiquitination of Flt3 when coexpressed in COS7 cells (10). Furthermore, overexpression of loss-of-function c-Cbl mutants identified in AML cases, such as c-Cbl-R420Q, with Flt3 in IL-3-dependent murine model hematopoietic cell lines 32D and BaF3-induced autophosphorylation of Flt3, activation of its downstream signaling events, and IL-3-independent proliferation (10, 16). More recently, mice with a loss-of-function mutation of c-Cbl (c-Cbl-C379A) or Cbl-b (Cbl-b-C373A) have been shown to develop a myeloproliferative disease that progressed to leukemia, which exhibited augmented Flt3 signaling and was prevented through mating with FL knock-out mice (17). Thus, c-Cbl as well as Cbl-b is strongly implicated in regulation of Flt3 signaling and in leukemogenesis through its disturbance. However, little is known about the involvement of Cbls in regulation of Flt3-ITD expression and about its molecular mechanisms.
Oncogenic kinases have been shown to adopt various mechanisms that spare them from negative regulatory processes. One mechanism that attenuates the conformational instability of oncogenic proteins is their association with the Hsp90 chaperone, which controls the folding and intracellular trafficking of diverse cellular proteins involved in signal transduction and cell cycle regulation (18, 19). Hsp90 has been shown to form complexes with many cellular proteins that are important for cancer or leukemic cells, such as BCR/ABL and Flt3-ITD. Thus, more than a dozen Hsp90 inhibitors are currently undergoing clinical evaluation in cancer and leukemic patients (19, 20). One of the promising Hsp90 inhibitors is 17-allylaminodemethoxygeldanamycin (17-AAG), which interacts with the ATP binding pocket of Hsp90, thereby inhibiting ATP binding and chaperone function of Hsp90. Previously, 17-AAG was shown to disrupt the chaperone association of Hsp90 with Flt3-ITD (8). Inhibition of the Hsp90 by 17-AAG or geldanamycin has also been demonstrated to result in degradation of Flt3-ITD and apoptosis in myeloid cell lines transfected with Flt3-ITD as well as in primary AML cells expressing Flt3-ITD (21–23). But molecular mechanisms involved in degradation of Flt3-ITD induced by Hsp90 inhibitors and in regulation of its sensitivity still remain to be examined.
In this study we examine the mechanisms regulating stability and 17-AAG-induced degradation of Flt3-ITD in the IL-3-dependent hematopoietic progenitor model cell line 32Dcl3, human leukemic MV4-11 cells, and in the transient expression system using 293T cells. This study reveals important roles for c-Cbl and Cbl-b in constitutive and 17-AAG-induced degradation of autophosphorylated Flt3-ITD through the ubiquitin proteasome system, which should shed light on the pathogenesis of refractory AML with Flt3-ITD as well as with mutations in c-Cbl or Cbl-b and may contribute to the development of its effective therapeutic strategies.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Murine IL-3-dependent 32Dcl3 cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and 10% WEHI conditioned medium as the source of IL-3 as described previously (24). 293T cells were cultured in Dulbecco modified Eagle's medium (DMEM) containing 10% FCS. MV4-11 cells (expressing Flt3-ITD) (25) were purchased from ATCC and cultured in Iscove's modified Dulbecco medium containing 10% FCS. PLAT-A (26), an amphotropic virus packaging cell line, was kindly provided by Dr. Toshio Kitamura and maintained in DMEM supplemented with 10% FCS.
Doxycycline (DOX), cycloheximide (CHX), and chloroquine were purchased from Sigma. G418 and hygromycin B were purchased from Wako (Osaka, Japan). MG132 and t-butoxycarbonyl-D-fmk were purchased from Calbiochem and BioVision (Mountain View, CA), respectively. Benzyloxycarbonyl-VAD-fmk was purchased from R&D System (Minneapolis, MN). FL was purchased from Pepro Tech (Rocky Hill, NJ). Sorafenib was purchased from LTK Laboratories (St. Paul, MN). Antibodies against Flt3, Cbl, Cbl-b, Nedd4, Hsp90, and α-tubulin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antiphosphotyrosine monoclonal antibody (4G10) and anti-ubiquitin (FK2) were purchased from Millipore (Billerica, MA). Phosphospecific antibodies against Flt3-Tyr-591 and STAT5-Tyr-694 were from Cell Signaling (Beverly, MA). Anti-β-actin was purchased from Sigma. Anti-HisG was purchased from Invitrogen. Anti-polyubiquitin (FK1) and anti-Ubi-Lys-63 were purchased from Enzolifesciences (Farmingdale, NY).
Immunoprecipitation and Immunoblotting
For immunoprecipitation experiments, cells were lysed in a lysis buffer containing 1% Triton X-100, 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each of aprotinin and leupeptin. Cell lysates were subjected to immunoprecipitation and immunoblotting as described previously (24). For immunoblot analysis of total cell lysates, samples were prepared by mixing an aliquot of cell lysates with an equal volume of 2× Laemmli sample buffer and heating at 100 °C for 5 min. The results shown are representative of experiments repeated at least three times.
Expression Plasmids
Retrovirus vectors, pRevTRE and pMIG (Addgene plasmid 9044), were obtained from Clontech (Palo Alto, CA) and Addgene (Cambridge, MA), respectively. pMXs-IG (27) was kindly provided by Dr. Toshio Kitamura. pRK5-Ubi-WT, -KO, -Lys-48, -K48R (Addgene plasmids 17608, 17603, 17605, 17604, respectively) (28), and pcDNA-HA-Nedd4 (Addgene plasmid 11426) (29) were obtained from Addgene. pRK5-Ubi-K63R (30) was kindly provided by Dr. J. Held. pcDNA3-Flt3-WT and pcDNA3-Flt3-ITD (4) were kindly provided by Dr. F. Böhmer. Expression plasmids for c-Cbl, Cbl-b, and their loss-of-function mutants (pCEFL-c-Cbl, -Cbl-b, -c-Cbl-C381A, and -Cbl-b-C373A) (31) were kindly provided by Dr. S. Lipkowitz. An expression plasmid for a loss-of-function mutant of c-Cbl, pMIG-c-Cbl-R420Q (16), was kindly provided by Dr. K. Spiekermann. pDG-Ubi-GFP (32, 33) was kindly provided by Dr. D. Gray.
Expression plasmids, pTRE-tight-Flt3-WT and pTRE-tight-Flt3-ITD, were constructed by subcloning the NotI/HindIII fragments coding for human Flt3-WT and Flt3-ITD from pcDNA3-Flt3-WT and pcDNA3-Flt3-ITD, respectively, into pTRE-tight. Retroviral expression plasmids, pMXs-IG-Flt3-ITD and pRevTRE-Flt3-ITD, were constructed by subcloning the NotI/HindIII fragment from pcDNA3-Flt3-ITD into pMXs-IG and pRevTRE, respectively. A retroviral expression plasmid, pRevTRE-Flt3-D835Y, was constructed by exchanging a Flt3 cDNA fragment (nucleotides 1673–3049 of GenBankTM accession number BC144040 coding for Pro-536 to the C terminus) obtained from primary AML cells expressing Flt3-D835Y with the corresponding region of pRevTRE-Flt3-ITD. The plasmid DNA coding for the intracellular portion of Flt3 was sequenced to confirm that it contained the D835Y mutation but not the ITD mutation. Retroviral expression plasmids, pRevTRE-c-Cbl and pRevTRE-c-Cbl-C381A, were constructed by subcloning the BamHI fragment coding for c-Cbl and c-Cbl-C381A from pCEFL-c-Cbl and pCEFL-c-Cbl-C381A, respectively, into pRevTRE. Retroviral expression plasmids, pRevTRE-Cbl-b and pRevTRE-Cbl-b-C373A, were constructed by subcloning the HindIII/EcoRV fragments coding for Cbl-b and Cbl-b-C373A from pCEFL-Cbl-b and pCEFL-Cbl-b-C373A, respectively, into pRevTRE.
Transfection and Infection
For inducible expression of Flt3-WT or Flt3-ITD in 32Dcl3 cells, 32Dcl3 cells were first transfected with pTet-On (Clontech) by electroporation using Gene Pulser (Bio-Rad) at 330 V and 960 microfarads. Transfected cells were selected in medium containing G418, and clones were selected by limiting dilution. A selected clone, Ton.32D, was used for control experiments and for cotransfection with pTRE-tight-Flt3-WT or pTRE-tight-Flt3-ITD and pMAM2-BSD (Funakoshi, Tokyo, Japan) by electroporation. Transfected cells were selected in medium containing G418 and blasticidin. Several clones were isolated by limiting dilution and examined for the induction of Flt3-WT or Flt3-ITD expression by the addition of DOX. A selected clone, Ton.32D/Flt3-WT or Ton.32D/Flt3-ITD, was used for subsequent experiments.
To obtain 32Dcl3 cells that inducibly express Flt3-D835Y, PLAT-A cells were first transfected with pRevTRE-Flt3-D835Y using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instruction. The recombinant retrovirus was harvested 60 h after transfection and used to infect Ton.32D cells. Infected cells were selected in DOX-containing medium without IL-3 to give Ton.32D/Flt3-D835Y cells. Pools of these infected cells were used for subsequent experiments. To obtain Flt3-ITD-expressing cells that inducibly overexpress c-Cbl, c-Cbl-C381A, Cbl-b, or Cbl-b-C373A and control cells, Ton.32D cells were infected with the recombinant retrovirus obtained from PLAT-A transfected with pRevTRE-c-Cbl, -c-Cbl-C381A, -Cbl-b, -Cbl-b-C373A, or pRevTRE. Infected cells were then selected in medium containing hygromycin to give Ton.32D/pRevTRE-c-Cbl, -c-Cbl-C381A, -Cbl-b, -Cbl-b-C373A, and Ton.32D/pRevTRE cells. These cells were subsequently infected again with the recombinant retrovirus obtained from PLAT-A transfected with pMXs-IG-Flt3-ITD. Infected cells were selected in medium without IL-3 to give Ton.32D/Flt3-ITD/pRevTRE-c-Cbl, -c-Cbl-C381A, -Cbl-b, -Cbl-b-C373A, and Ton.32D/Flt3-ITD/pRevTRE cells. Pools of these infected cells were used for subsequent experiments.
To obtain Ton.32D/Flt3-ITD or Ton.32D/Flt3-WT cells that overexpress both c-Cbl-R420Q and Cbl-b-C373A, these cells were first infected with the recombinant pMIG-c-Cbl-R420Q retrovirus. Cells expressing green fluorescent protein (GFP) were sorted using a BD Biosciences FACSscan flow cytometer (Mountain View, CA). GFP-positive cells were infected again with the recombinant pRevTRE-Cbl-b-CA retrovirus and selected in medium containing hygromycin to give Ton.32D/Flt3-ITD/c-Cbl-RQ/Cbl-b-CA and Ton.32D/Flt3-WT/c-Cbl-RQ/Cbl-b-CA cells. Pools of infected cells were used for subsequent experiments.
For transient expression in 293T cells, cells were transfected with the indicated plasmids using the Lipofectamine reagent according to the manufacture's instruction. Cells were harvested 48 h after transfection for immunoprecipitation and immunoblotting.
Cell Viability Assay
Cell viability was measured by the sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate (XTT) colorimetric assay using the Cell Proliferation Kit II (Roche Molecular Biochemicals) according to the manufacturer's instructions. In brief, cells were cultured in a 96-well plate at 2 × 105 cells/ml in the indicated concentrations of 17-AAG and cultured for 36 h. The XTT labeling mixture was added for 4 h before the optical density at 490 nm was measured with a microplate reader.
RESULTS
Constitutively Activated and Autophosphorylated Flt3-ITD Rapidly Undergoes Both Proteasomal and Lysosomal Degradation
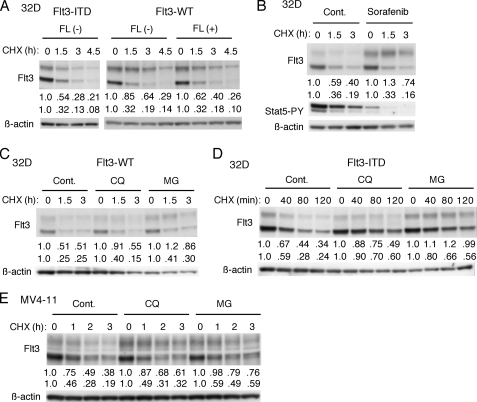
To examine the kinetics of Flt3-WT and Flt3-ITD degradation in hematopoietic cells, we first analyzed Ton.32D/Flt3-WT and Ton.32D/Flt3-ITD cells, which express Flt3-WT or Flt3-ITD when cultured with DOX. Previously, it has been shown that Flt3 migrates in SDS-PAGE as two bands that correspond to the immature intracellular receptor of 130 kDa and the mature glycosylated receptor of 155 kDa expressed on the cell surface (34, 35). As shown in Fig. 1A, treatment of cells with the protein synthesis inhibitor CHX revealed that FL facilitated degradation of the mature form of Flt3-WT, which however, was not as unstable as that of Flt3-ITD. On the other hand, the turnover of the intracellular form was more rapid, with no significant difference observed between Flt3-WT and Flt3-ITD. To examine the possible effect of activation and autophosphorylation of Flt3 on its stability, we examined the effect of sorafenib, which very effectively inhibits the kinase activity of Flt3-ITD (36). As shown in Fig. 1B, sorafenib increased the expression level of mature Flt3-ITD and drastically inhibited its degradation in CHX-treated cells, whereas it significantly inhibited tyrosine phosphorylation of STAT5, thus indicating that Flt3-ITD was effectively inhibited. These data suggest that the degradation of Flt3 expressed on the cell surface may be facilitated by its activation and autophosphorylation.
FIGURE 1.
Constitutively activated and autophosphorylated Flt3-ITD rapidly undergoes both proteasomal and lysosomal degradation. A, Ton.32D/Flt3-ITD (Flt3-ITD) or Ton.32D/Flt3-WT (Flt3-WT) cells were cultured with 1 μg/ml DOX for 24 h to induce the Flt3-ITD or -WT expression, respectively. Ton.32D/Flt3-WT cells were then pretreated for 15 min with 20 ng/ml FL or left untreated as indicated. Cells were then treated with 50 μg/ml CHX and lysed after the indicated times. Cell lysates were subjected to immunoblot analysis with anti-Flt3 followed by reprobing with anti-β-actin as indicated. Relative expression levels of the mature (upper rows) and immature (lower rows) forms of Flt3, determined by densitometric analysis, are shown. B, after culture with DOX, Ton.32D/Flt3-ITD cells were pretreated for 30 min with 200 nm sorafenib or left untreated as the control as indicated and subjected to immunoblot analysis using antibodies against indicated proteins. C and D, after culture with DOX, Ton.32D/Flt3-ITD (Flt3-ITD) or Ton.32D/Flt3-WT (Flt3-WT) cells were pretreated for 2 h with 100 μm chloroquine (CQ) where indicated and with 50 ng/ml FL for Ton.32D/Flt3-WT. Cells were subsequently cultured for 30 min with or without 5 μm MG132 (MG) where indicated in the presence of 100 μm t-butoxycarbonyl-D-fmk. Cells were then treated with 10 μg/ml CHX for the indicated times and analyzed. E, MV4-11 cells were treated for 1 h with 25 μm chloroquine or 5 μm MG132 or left untreated as control (Cont.) as indicated in the presence of 50 μm benzyloxycarbonyl-VAD-fmk. Cells were then treated with 2 μg/ml CHX and analyzed.
We then explored the cellular degradation pathways of Flt3-WT and Flt3-ITD by using the proteasome inhibitor MG132 and the lysosome inhibitor chloroquine. In addition, we used the pan-caspase inhibitor t-butoxycarbonyl-D-fmk or benzyloxycarbonyl-VAD-fmk to exclude putative apoptotic degradation of Flt3-ITD (11). As shown in Fig. 1, C and D, MG132 and, to a lesser degree, chloroquine inhibited degradation of the mature forms of Flt3-WT and Flt3-ITD. These two inhibitors similarly inhibited degradation of the immature form of Flt3-ITD without significantly affecting that of Flt3-WT. Consistent with these data, MG132 and chloroquine retarded the degradation of both mature and immature forms of Flt3-ITD in human leukemic MV4-11 cells as well (Fig. 1E). These data indicate that Flt3-ITD constitutively undergoes both proteasomal and lysosomal degradation.
Hsp90 Inhibitor 17-AAG Preferentially Facilitates the Proteasomal Degradation of Flt3-ITD
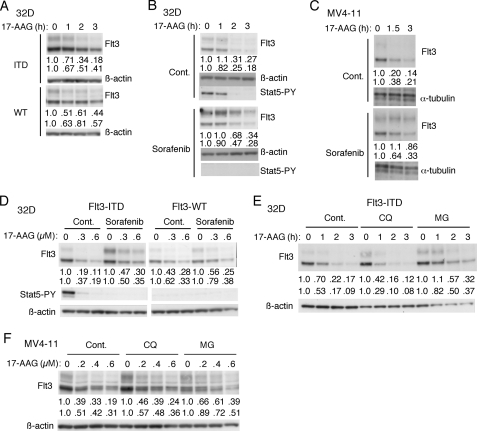
Previously, it has been reported that Hsp90 inhibitors including 17-AAG induced apoptosis selectively in Flt3-ITD-positive leukemia cells (22, 23). Furthermore, Hsp90 inhibitors have been shown to degrade Flt3-ITD in a dose- and time-dependent manner in both Flt3-ITD-positive leukemia cell lines and in primary AML cells (7, 21, 23, 37, 38). As shown in Fig. 2A, 17-AAG induced the decline in expression of Flt3-ITD in both mature and immature forms more rapidly than that of Flt3-WT. Next, we examined the effect of tyrosine phosphorylation on degradation induced by 17-AAG. Pretreatment with sorafenib retarded the 17- AAG-induced decline in expression of Flt3-ITD in both Ton.32D/Flt3-ITD (Fig. 2B) and MV4-11 cells (Fig. 2C). As shown in Fig. 2D, 17-AAG dose dependence studies confirmed that the 17-AAG sensitivity of Flt3-ITD was higher than that of Flt3-WT and was decreased by pretreatment with sorafenib. These data suggest that, as compared with Flt3-WT, constitutively activated and autophosphorylated Flt3-ITD may more rapidly undergo degradation when Hsp90 is inhibited. We next examined the degradation mechanisms of Flt3-ITD in cells treated with 17-AAG. Pretreatment of cells with MG132 but not with chloroquine significantly inhibited the 17-AAG-induced decline in expression level of Flt3-ITD in both Ton.32D/Flt3-ITD (Fig. 2E) and MV4-11 (Fig. 2F), which indicates that Flt3-ITD may be mostly degraded through the proteasomal pathway when Hsp90 is inhibited.
FIGURE 2.
Hsp90 inhibitor 17-AAG preferentially induces proteasomal degradation of autophosphorylated Flt3-ITD. A, after treatment with DOX for 24 h, Ton.32D/Flt3-ITD (ITD) or Ton.32D/Flt3-WT (WT) cells were treated with 0.5 μm 17-AAG for the indicated times and subjected to immunoblot analysis with anti-Flt3 followed by reprobing with anti-β-actin as indicated. Relative expression levels of the mature (upper rows) and immature (lower rows) forms of Flt3 determined by densitometric analysis are shown. B and C, Ton.32D/Flt3-ITD cells pretreated with 1 μg/ml DOX for 24 h (B) or MV4-11 cells (C) were pretreated with 200 nm sorafenib for 1 h or left untreated as the control as indicated. Cells were then treated with 0.5 μm 17-AAG for the indicated times and subjected to immunoblot analysis using antibodies against indicated proteins. D, after culture with DOX, Ton.32D/Flt3-ITD (Flt3-ITD) or Ton.32D/Flt3-WT (Flt3-WT) cells were either pretreated for 30 min with 200 nm sorafenib or left untreated as the control as indicated and with 50 ng/ml FL for Ton.32D/Flt3-WT. Cells were then treated for 2 h with indicated concentrations (μm) of 17-AAG and analyzed. E and F, Ton.32D/Flt3-ITD cells precultured with DOX (E) or MV4-11 cells (F) were pretreated for 1 h with 25 μm chloroquine (CQ) or 5 μm MG132 (MG) or left untreated as the control as indicated. Cells were then treated for 2 h with indicated concentrations of 17-AAG and analyzed.
Hsp90 Preferentially Binds Autophosphorylated Flt3-ITD to Protect Its Polyubiquitination and Degradation
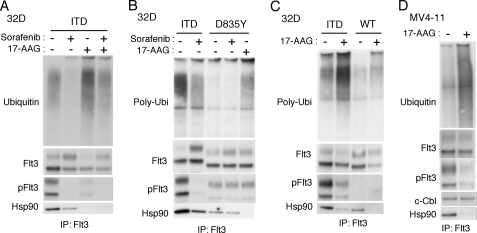
Because ubiquitination plays a critical role in regulation of proteasomal degradation, we next examined ubiquitination of Flt3-ITD. As shown in Fig. 3A, immunoblot analysis of Flt3 immunoprecipitates with anti-ubiquitin antibody revealed that Flt3-ITD was ubiquitinated in Ton.32D/Flt3-ITD pretreated with MG132. However, treatment of cells with sorafenib remarkably inhibited ubiquitination of Flt3-ITD as well as its autophosphorylation on Tyr-591. On the other hand, 17-AAG significantly enhanced ubiquitination of Flt3-ITD while predominantly reducing the expression level of Flt3-ITD phosphorylated on Tyr-591. Sorafenib reduced ubiquitination of Flt3-ITD in 17-AAG-treated cells as well. It was also noted that 17-AAG and, to a lesser degree, sorafenib inhibited the physical association of Flt3-ITD with Hsp90. To rule out the possibility that the inhibitory effect of sorafenib on Flt3-ITD binding to Hsp90 is indirectly mediated through inhibition of other kinases by this multi-kinase inhibitor or through other effects, we examined Ton.32D cells expressing the Flt3-tyrosine kinase domain mutant Flt3-D835Y, which is constitutively activated like Flt3-ITD but is significantly resistant to sorafenib as compared with Flt3-ITD (36, 39). As shown in Fig. 3B, Flt3-D835Y was autophosphorylated to a lesser degree than Flt3-ITD and showed lesser degrees of Hsp90 binding and ubiquitination than Flt3-ITD. Notably, sorafenib did not significantly affect either autophosphorylation of Flt3-D835Y or its binding with Hsp90, whereas17-AAG abrogated the binding of the mutant with Hsp90. On the other hand, sorafenib unambiguously inhibited the association of Flt3-ITD with Hsp90 under the same conditions. These results strongly suggest that sorafenib inhibits the binding of Flt3-ITD to Hsp90 specifically through inhibition of its kinase activity and autophosphorylation.
FIGURE 3.
Hsp90 preferentially binds autophosphorylated Flt3-ITD to protect its polyubiquitination and degradation. A, after culture with DOX, Ton.32D/Flt3-ITD cells were pretreated for 1 h with 200 nm sorafenib or left untreated as indicated and further treated with 5 μm MG132 for 1 h with 0.5 μm 17-AAG added or not 30 min before harvest as indicated. Flt3-ITD was immunoprecipitated (IP) and subjected to immunoblot analysis with anti-ubiquitin followed by reprobing with anti-Flt3, anti-phospho-Tyr-591-Flt3 (pFlt3) and anti-Hsp90 as indicated. B, Ton.32D/Flt3-ITD (ITD) or Ton.32D/Flt3-D835Y (D835Y) cells cultured with DOX were pretreated for 1 h with 50 μm sorafenib or left untreated as indicated and further treated with 5 μm MG132 for 1 h with 0.5 μm 17-AAG added or not 30 min before harvest as indicated. Flt3 was immunoprecipitated and subjected to immunoblot analysis. C, after culture with DOX, Ton.32D/Flt3-ITD (ITD) or Ton.32D/Flt3-WT (WT) cells were treated with 5 μm MG132 for 1 h with 0.5 μm 17-AAG added or not 30 min before harvest as indicated. Flt3 was immunoprecipitated and subjected to immunoblot analysis. D, MV4-11 were treated with 5 μm MG132 for 90 min with 1 μm 17-AAG added or not 60 min before harvest as indicated. Flt3 was immunoprecipitated and subjected to immunoblot analysis.
Using a monoclonal antibody specific for polyubiquitin, it was confirmed that 17-AAG significantly enhanced polyubiquitination of Flt3-ITD (Fig. 3C). Flt3-WT associated with Hsp90 to a lesser degree as compared with Flt3-ITD, which was also inhibited by 17-AAG. However, polyubiquitination of Flt3-WT was much less obvious as compared with that of Flt3-ITD even after treatment with 17-AAG. As shown in Fig. 3D, it was confirmed that 17-AAG significantly enhanced ubiquitination of Flt3-ITD expressed in MV4-11. 17-AAG also remarkably inhibited binding of Flt3-ITD with Hsp90. Intriguingly, the amount of c-Cbl that co-immunoprecipitated with Flt3-ITD was not decreased even though that of autophosphorylated Flt3-ITD was preferentially and significantly reduced after treatment of MV4-11 cells with 17-AAG. Together, these results suggest that the activated and autophosphorylated form of Flt3-ITD preferentially binds to Hsp90 and, upon inhibition of Hsp90 by 17-AAG, it dissociates from Hsp90 and undergoes polyubiquitination and proteasomal degradation. It should be noted, however, that the interaction of Flt3 with Hsp90 should depend on its autophosphorylation only partially, because Flt3-WT also evidently associates with Hsp90 even though its autophosphorylation was hardly detectable (Fig. 3D).
c-Cbl Facilitates Lys-48-linked Polyubiquitination of Autophosphorylated Flt3-ITD
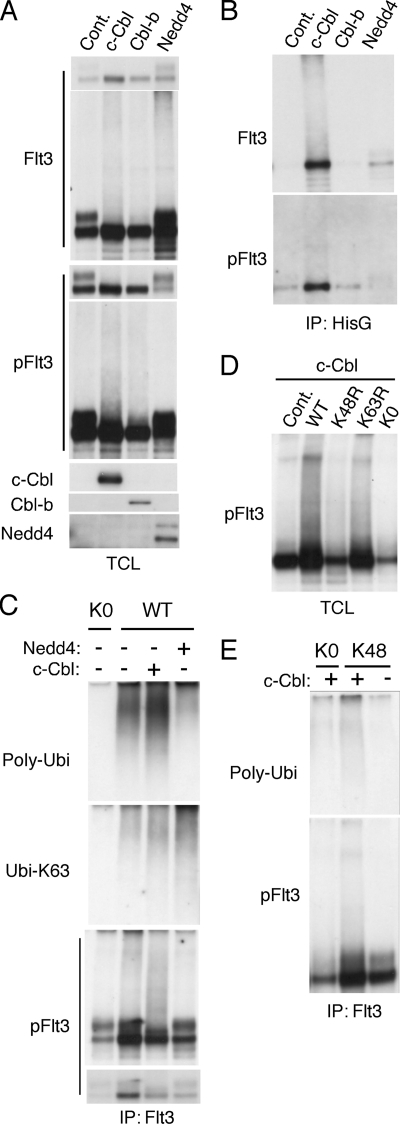
To gain insights into possible E3 ubiquitin ligases capable of ubiquitinating Flt3-ITD, we coexpressed several E3 ubiquitin ligases, including c-Cbl, Cbl-b, and Nedd4, with Flt3-ITD and HisG-tagged ubiquitin in 293T cells. As shown in Fig. 4A, coexpression of c-Cbl or Cbl-b remarkably decreased the expression of mature Flt3-ITD, whereas it gave a smeary pattern suggestive of ubiquitination by immunoblot analysis with anti-Flt3 as well as anti-phospho-Tyr-591-Flt3. On the other hand, Nedd4 reduced immature but not mature Flt3-ITD and gave a smeary pattern only in anti-Flt3 but not in anti-phospho-Tyr-591-Flt3 immunoblotting. To confirm the effects of these ubiquitin ligases on Flt3-ITD ubiquitination, HisG-tagged ubiquitin was immunoprecipitated from the cell lysates and analyzed by immunoblotting. As shown in Fig. 4B, c-Cbl and, to a much lesser extent, Cbl-b increased the amount of Flt3-ITD immunoprecipitated with anti-HisG, which also gave a smeary pattern and was reactive with anti-phospho-Tyr-591-Flt3. On the other hand, Nedd4 increased the amount of Flt3-ITD immunoprecipitated, which however, did not react with anti-phospho-Tyr-591-Flt3. These data suggest that c-Cbl and, to a lesser extent, Cbl-b may preferentially facilitate ubiquitination of autophosphorylated mature form of Flt3-ITD, whereas Nedd4 may facilitate ubiquitination of the immature form not phosphorylated on Tyr-591.
FIGURE 4.
c-Cbl facilitates polyubiquitination of autophosphorylated Flt3-ITD at least partly in the Lys-48-linked manner. A and B, 293T cells were transfected on 6-well plate with 0.4 μg of pCEFL-c-Cbl (c-Cbl), pCEFL-Cbl-b (Cbl-b), pcDNA-HA-Nedd4 (Nedd4), or pcDNA3 (Cont.), as indicated along with 0.4 μg each of pcDNA3-Flt3-ITD and pDG-Ubi-GFP. Cells were harvested 48 h after transfection, and HisG-tagged ubiquitin was immunoprecipitated using anti-HisG antibody. Total cell lysates (TCL) and immunoprecipitates (IP) were analyzed by immunoblot analysis using anti-Flt3 followed by reprobing with anti-phospho-Tyr-591-Flt3 (pFlt3), anti-c-Cbl, anti-Cbl-b, and anti-Nedd4 as indicated. For anti-Flt3 and anti-anti-phospho-Tyr-591-Flt3, shorter and longer exposure films are shown as upper and lower panels, respectively. C, 293T cells were transfected on a 12-well plate with 0.2 μg of pCEFL-c-Cbl (c-Cbl) or pcDNA-HA-Nedd4 (Nedd4) as indicated or 0.2 μg of pcDNA3 and 0.2 μg of pRK5-Ubi-KO (KO) or pRK5-Ubi-WT (WT) as indicated along with 0.2 μg of pcDNA3-Flt3-ITD. Flt3-ITD was immunoprecipitated and analyzed by immunoblotting. D, 293T cells were transfected with 0.4 μg each of pcDNA3-Flt3-ITD and pCEFL-c-Cbl along with 0.2 μg of pRK5 (Cont.), pRK5-Ubi-WT (WT), pRK5-Ubi-K48R (K48R), pRK5-Ubi-K63R (K63R), or pRK5-Ubi-KO (KO) as indicate on 6-well plates. Cell lysates were harvested and analyzed. E, 293T cells were transfected with 0.4 μg of pCEFL-c-Cbl (c-Cbl) or pCEFL as indicated along with 0.2 μg of pRK5-Ubi-KO (KO) or pRK5-Ubi-Lys-48 (K48) as indicated and 0.4 μg of pcDNA3-Flt3-ITD on 6-well plates. Flt3-ITD was immunoprecipitated and analyzed.
To examine the ubiquitination pattern more precisely, we then coexpressed mutant forms of ubiquitin with Flt3-ITD and the ubiquitin ligases. As shown in Fig. 4C, polyubiquitination of Flt3-ITD was observed when coexpressed with wild-type ubiquitin but not with the ubiquitin mutant lacking all the lysine residues and was increased or decreased by coexpression of c-Cbl or Nedd4, respectively. On the other hand, Nedd4 specifically increased the Lys-63-linked polyubiquitination of Flt3-ITD. Moreover, it was confirmed that c-Cbl but not Nedd4 facilitated polyubiquitination of Flt3-ITD autophosphorylated on Tyr-591. As shown in Fig. 4D, the ubiquitin mutant lacking Lys-48, but not that lacking Lys-63, significantly inhibited polyubiquitination of autophosphorylated Flt3-ITD observed when coexpressed with c-Cbl. Finally, it was demonstrated that c-Cbl facilitated polyubiquitination of Flt3-ITD autophosphorylated on Tyr-591 when coexpressed with the ubiquitin mutant lacking all the lysine residues except for Lys-48 (Fig. 4E). Together, these data suggest that Lys-48-linked polyubiquitination of autophosphorylated Flt3-ITD may be specifically enhanced by c-Cbl, which strongly implicates c-Cbl in regulation of Flt3-ITD through the ubiquitin proteasome system, because the Lys-48-linked polyubiquitination plays a critical role in targeting proteins for proteasomal degradation (12, 13).
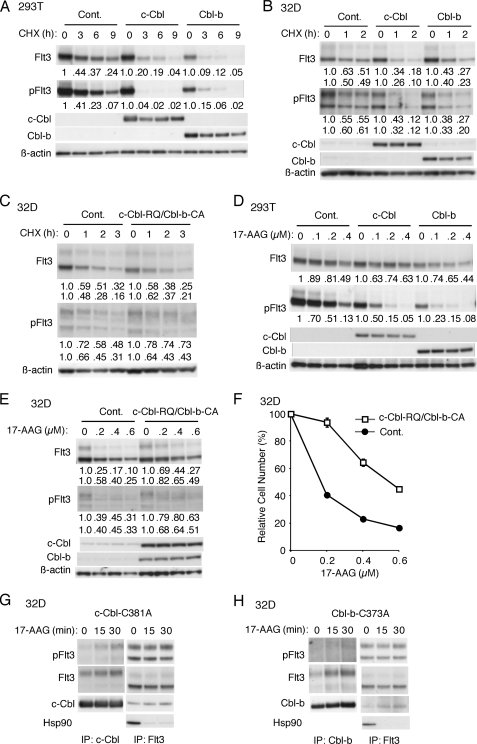
c-Cbl and Cbl-b Play Important Roles in Constitutive and 17-AAG-induced Degradation of Autophosphorylated Flt3- ITD
Because c-Cbl and Cbl-b were implicated in polyubiquitination of Flt3-ITD, we examined their effects on degradation of Flt3. First, c-Cbl or Cbl-b was overexpressed with ubiquitin and Flt3-WT or Flt3-ITD in 293T cells, and cells were treated with CHX for varying times. As shown in Fig. 5A, c-Cbl as well as Cbl-b remarkably enhanced the degradation of Flt3-ITD, with the most drastic effect observed for c-Cbl on Flt3-ITD autophosphorylated on Tyr-591. In contrast, neither c-Cbl nor Cbl-b significantly affected the stability of Flt3-WT (negative data not shown). Next, we examined Flt3-ITD-expressing 32D cells that inducibly overexpress c-Cbl or Cbl-b when treated with DOX. As shown in Fig. 5B, the stability of both mature and immature Flt3-ITD was reduced in 32D cells overexpressing c-Cbl or Cbl-b, although the effects were not as remarkable as those observed in 293T cells. Overexpression of c-Cbl or Cbl-b failed to affect the stability of Flt3-WT in 32D cells as well (negative data not shown). When a loss-of-function mutant of c-Cbl or Cbl-b (c-Cbl-C381A or Cbl-b-C373A, respectively) was overexpressed inducibly in Flt3-ITD-expressing 32D cells, we failed to observe any significant effects on stability of Flt3-ITD (negative data not shown). Thus, we overexpressed loss-of-function mutants of both c-Cbl and Cbl-b (c-Cbl-R420Q and Cbl-b-C373A, respectively) in 32D cells expressing Flt3-ITD. As shown in Fig. 5C, overexpression of both mutants together significantly stabilized the mature form of Flt3-ITD autophosphorylated on Tyr-591. These data suggest that c-Cbl and Cbl-b preferentially facilitate degradation of autophosphorylated Flt3-ITD. It was also implied that endogenous c-Cbl or Cbl-b alone may be sufficient for an efficient degradation of Flt3-ITD, because inhibition of both was required to inhibit the degradation significantly.
FIGURE 5.
c-Cbl and Cbl-b play important roles in constitutive and 17-AAG-induced degradation of autophosphorylated Flt3-ITD. A, 293T cells were transfected on 6-well plates with 0.5 μg each of pCEFL (Cont.), pCEFL-c-Cbl (c-Cbl), or pCEFL-Cbl-b (Cbl-b) as indicated and 0.5 μg of pcDNA3-Flt3-ITD. Two days after transfection cells were treated with 100 μg/ml CHX for the indicated times and harvested. Total cell lysates were analyzed by immunoblot analysis using anti-Flt3 followed by reprobing with anti-phospho-Tyr-591-Flt3 (pFlt3), anti-c-Cbl, anti-Cbl-b, and anti-β-actin as indicated. Relative expression levels of Flt3-ITD and Flt3-ITD phosphorylated on Tyr-591 were determined by densitometric analysis of the anti-Flt3 and anti-pFlt3 data, respectively, and are shown below each panel. B, after culture with 1 μg/ml DOX for 24 h, Ton.32D/Flt3-ITD/pRevTRE (Cont.), Ton.32D/Flt3-ITD/pRevTRE-c-Cbl (c-Cbl), and Ton.32D/Flt3-ITD/pRevTRE-Cbl-b (Cbl-b) cells were treated with 25 μg/ml CHX for the indicated times and lysed. Cells lysates were subjected to Western blot analysis. Relative expression levels of the mature (upper rows) and immature (lower rows) forms of Flt3-ITD and Flt3-ITD phosphorylated on Tyr-591 were determined by densitometric analysis of the anti-Flt3 and anti-pFlt3 data, respectively, and are shown below each panel. C, after culture with DOX, Ton.32D/Flt3-ITD (Cont.) and Ton.32D/Flt3-ITD/c-Cbl-R420Q/Cbl-b-C373A (c-Cbl-RQ/Cbl-b-CA) cells were treated with 10 μg/ml CHX for the indicated times, and cell lysates were analyzed. D, 293T cells were transfected on 6-well plates with 0.4 μg each of pCEFL (Cont.), pCEFL-c-Cbl (c-Cbl), or pCEFL-Cbl-b (Cbl-b) as indicated and 0.4 μg of pcDNA3-Flt3-ITD. Two days after transfection cells were treated for 5 h with the indicated concentrations of 17-AAG and harvested. Total cell lysates were analyzed by immunoblot analysis. E, after culture with DOX, Ton.32D/Flt3-ITD (Cont.) and Ton.32D/Flt3-ITD/c-Cbl-R420Q/Cbl-b-C373A (c-Cbl-RQ/Cbl-b-CA) cells were treated for 2 h with indicated concentrations of 17-AAG, and cell lysates were analyzed. F, after culture with DOX, Ton.32D/Flt3-ITD (Cont.) and Ton.32D/Flt3-ITD/c-Cbl-R420Q/Cbl-b-C373A (c-Cbl-RQ/Cbl-b-CA) cells were further cultured for 36 h with the indicated concentrations of 17-AAG and 1 μg/ml DOX in the absence of IL-3. Numbers of viable cells were measured by the XTT assay and are expressed as percentages of the cell number without 17-AAG. Each data point represents the mean of triplicate determinations, with error bars indicating S.E. G and H, Ton.32D/Flt3-ITD/pRevTRE-c-Cbl-C381A (G) or -Cbl-b-C373A (H) cells cultured with DOX were pretreated with 5 μm MG132 for 30 min and then treated with 0.5 μm 17-AAG for the indicated times. Cells were lysed and subjected to immunoprecipitation with anti-c-Cbl, anti-Cbl-b, or anti-Flt3, as indicated and analyzed with immunoblot analysis using indicated antibodies.
To examine whether c-Cbl or Cbl-b may be involved in 17-AAG-mediated degradation of Flt3-ITD, c-Cbl or Cbl-b was overexpressed with ubiquitin and Flt3-WT or Flt3-ITD in 293T cells, and cells were treated with varying concentrations of 17-AAG. As shown in Fig. 5D, 17-AAG dose-dependently decreased the expression level of Flt3-ITD, whereas the effects were more prominently observed on that of Flt3-ITD autophosphorylated on Tyr-591. Although the 17-AAG sensitivity of Flt3-ITD as a whole was not significantly enhanced by overexpression of c-Cbl or Cbl-b, that of Flt3-ITD phosphorylated on Tyr-591 was prominently enhanced. On the other hand, overexpression of c-Cbl-R420Q and Cbl-b-C373A together in Flt3-ITD-expressing 32D cells significantly decreased the 17-AAG sensitivity of both mature and immature forms of Flt3-ITD as well as their autophosphorylated forms (Fig. 5E). In accordance with this, when cultured with varying concentrations of 17-AAG, overexpression of both mutants together significantly decreased the suppression of proliferation and viability of Flt3-ITD-dependent 32D cells by 17-AAG (Fig. 5F). Together, these data suggest that c-Cbl and Cbl-b may play important roles in the 17-AAG-induced degradation of activated and autophosphorylated Flt3-ITD though the ubiquitin proteasome system.
To gain insights into the mechanisms by which c-Cbl and Cbl-b play roles in 17-AAG-induced degradation of Flt3-ITD, we examined the physical interaction of these E3 ligases with Flt3-ITD. In MV4-11 cells treated with 17-AAG and MG132, the amount of c-Cbl associated with Flt3-ITD was not decreased, although the expression level of Flt3-ITD, particularly that of autophosphorylated Flt3-ITD, was decreased by 17-AAG (Fig. 3D), thus suggesting that the relative amount of c-Cbl associated with autophosphorylated Flt3-ITD may be increased after the treatment. To pursuit this possibility, we examined in 32D cells the effects of 17-AAG on the interaction of Flt3-ITD with c-Cbl-C381A and Cbl-b-C373A, loss-of-function mutants expected not to induce degradation of Flt3-ITD through the ubiquitin/proteasome pathway. As shown in Fig. 5G, c-Cbl-C381A was associated preferentially with the mature form of Flt3-ITD. 17-AAG rapidly dissociated Flt3-ITD from Hsp90 but moderately increased the association between c-Cbl-C381A and Flt3-ITD. Similar results were obtained with Cbl-b-C373A (Fig. 5H). Thus, these results support the idea that c-Cbl and Cbl-b preferentially interact with the mature form of Flt3-ITD that is dissociated from Hsp90 by 17-AAG to facilitate its degradation through the ubiquitin/proteasome pathway.
DISCUSSION
In this study we have demonstrated that the mature form of Flt3-ITD was more rapidly degraded than that of Flt3-WT in 32D cells, which was significantly retarded by inhibition of its kinase activity, thus suggesting that autophosphorylation may play a role in triggering the degradation (Fig. 1). Although the constitutive degradation of Flt3 was dependent on both proteasome and lysosome, 17-AAG enhanced only the proteasome-dependent degradation, which also affected Flt3-ITD more remarkably than Flt3-WT in a kinase activity-dependent manner and was associated with profound polyubiquitination of Flt3-ITD (Figs. 2 and 3). Flt3-ITD bound Hsp90 more significantly than Flt3-WT did in a kinase activity-dependent manner. In 293T cells, c-Cbl significantly enhanced polyubiquitination of a Flt3-ITD preferentially of the autophosphorylated form at least partly in the Lys-48-linked manner. Overexpression of c-Cbl or Cbl-b also enhanced constitutive and 17-AAG-induced degradation of Flt3-ITD, whereas that of their loss-of-function mutants together in 32D cells retarded the degradation and reduced the cytotoxicity of 17-AAG (Fig. 5). These results suggest that c-Cbl and Cbl-b play significant and complementary roles in constitutive and 17-AAG-mediated degradation of autophosphorylated Flt3-ITD through the ubiquitin proteasome system.
Our findings showed that the overexpression of c-Cbl or Cbl-b promoted the degradation of Flt3-ITD in both 293T and 32D cells (Fig. 5, A and B), whereas the degradation of Flt3-WT was not influenced by the overexpression of Cbl proteins (data not shown). On the contrary, several previous reports showed that c-Cbl could not catalyze ubiquitination or degradation of mutant forms of receptor-tyrosine kinases, such as mutant EGF receptors and hybrid receptors of PDGF receptor α/β (40, 41). These reports proposed that receptor mislocalization, aberrant phosphorylation, and lack of c-Cbl recruitment sites might account for the reduced efficiency of c-Cbl-mediated degradation of these aberrant proteins. Although Flt3-ITD is also mislocalized (4, 5), it was previously reported that the interaction of Flt3 with c-Cbl depends on the phosphorylation of Tyr-589 or Tyr-599, autophosphorylation sites in the juxtamembrane domain of Flt3 (16). Most of ITD mutants, including the mutant we used in this study, carry a duplication containing at least one of the juxtamembrane tyrosine residues Tyr-589 or Tyr-599, which should facilitate binding of c-Cbl with Flt3-ITD. In accordance with this, we confirmed the association of c-Cbl with Flt3-ITD in both 32D (data not shown) and MV4-11 (Fig. 3D), which is also in accordance with a previous report (10) and should explain the ability of c-Cbl to ubiquitinate Flt3-ITD for proteasomal degradation.
This study has shown that c-Cbl and Cbl-b could degrade both mature and immature forms of Flt3-ITD. However, a recent study suggested that c-Cbl could degrade mainly Flt3-WT and only the mature form of Flt3-ITD (11). This suggestion was based on an observation that the mature form of Flt3-ITD was diminished when Flt3-ITD was coexpressed with c-Cbl in HEK293T cells. The authors speculated that the immature intracellular form of Flt3-ITD was resistant to c-Cbl-mediated degradation due to its mislocalization. We also observed that the mature form of Flt3-ITD was diminished when coexpressed with c-Cbl or Cbl-b in 293T cells (Fig. 4A). However, time course studies using CHX unequivocally showed that the immature form of Flt3-ITD undergoes a rapid degradation when coexpressed in these ubiquitin ligases in both 293T and hematopoietic 32D cells (Fig. 5, A and B). This is important because it has been reported that the intracellularly retained immature form of Flt3-ITD can aberrantly activate STAT5 and is associated with transforming potential of Flt3-ITD (5, 42). It is also important to note that the degradation of Flt3-ITD phosphorylated on Tyr-591, which is most likely phosphorylated also on other autophosphorylation sites, was particularly fast and dependent on c-Cbl and Cbl-b (Fig. 5). This should account for the fact that Flt3-ITD is degraded more rapidly than Flt3-WT in a kinase activity-dependent manner (Fig. 1) and implicates c-Cbl and Cbl-b in regulation of leukemogenic signaling from Flt3-ITD, because this phosphorylation is required for the activation of STAT5 signaling by Flt3-ITD (43, 44).
In this study we indicated for the first time that c-Cbl facilitates the Lys-48-linked polyubiquitination of Flt3-ITD, which has been reported to serve as the signal to target proteins for degradation by proteasome (Fig. 4, D and E) (12, 13). A recent report showed that E3 ubiquitin-ligase SIAH1 and E2 ubiquitin-conjugase UBCH8 physically interacted with and targeted Flt3-ITD for proteasomal degradation (11). Additionally, in this study we showed that Nedd4, a homologous to E6-AP (E6-associated protein) C terminus (HECT) family ubiquitin ligase involved in regulation of other receptor-tyrosine kinases (13, 45), could also facilitate ubiquitination of Flt3-ITD mostly via Lys-63-linked polyubiquitin chains. The Lys-63-linked polyubiquitination has been shown to be adopted by a number of HECT family ubiquitin ligases and implicated in various cellular processes, such as endocytosis and vesicular sorting (46). Thus, it is likely that several types of ubiquitin ligases may be involved in degradation of Flt3-ITD through the lysosome and proteasome system, although further studies are needed to evaluate the significance of Nedd4 in regulation of Flt3-ITD in hematopoietic cells.
Previous reports showed that 17-AAG inhibited chaperone association of Hsp90 with Flt3-ITD and enhanced its ubiquitination and proteasomal degradation (7, 8). We have extended these observations by formally showing that Flt3-ITD undergoes polyubiquitination by using a specific antibody (Fig. 3C). More importantly, we have revealed by using the Flt3 kinase inhibitor sorafenib that the association of Flt3-ITD with Hsp90- and 17-AAG-induced ubiquitination and degradation significantly depends on the aberrant kinase activity and autophosphorylation (Figs. 2, B and C, and 3, A and B). This would explain the observed differences in 17-AAG sensitivity of Flt3-ITD and Flt3-WT for degradation and ubiquitination (Figs. 2A and 3C). This would also shed light on the controversy that exists in the literature for the chaperoning of Flt3-WT by Hsp90. One study indicated that Hsp90 could form the molecular complex with both Flt3-ITD and Flt3-WT, whereas the other reports indicated that Flt3-WT was not the client of Hsp90 in both myeloid cell line and primary AML cells (21–23). This study equivocally showed that Hsp90 could physically associate with both Flt3-ITD and Flt3-WT in 32D cells (Fig. 3C). Furthermore, we found that Flt3-D835Y also associated with Hsp90 (Fig. 3B). However, the binding was most significantly observed for Flt3-ITD, most likely because of its high kinase activity and autophosphorylation status. Together, these observations indicate that autophosphorylated Flt3-ITD depends more significantly on Hsp90 than Flt3-WT or Flt3-ITD that is inhibited by its inhibitor for its stability. This should be taken into consideration in developing new combination therapies using Flt3 and Hsp90 inhibitors.
Most importantly, this study revealed that c-Cbl and Cbl-b are involved in 17-AAG-induced proteasomal degradation of Flt3-ITD. In this regard, it is notable that c-Cbl was coimmunoprecipitated with Flt3-ITD from 17-AAG-treated MV4-11 cell lysates in Fig. 3D. Even though the amount of autophosphorylated Flt3-ITD was drastically reduced after treatment with 17-AAG, that of c-Cbl coimmunoprecipitated was not at all reduced. Furthermore, in 32Dcl3 cells overexpressing c-Cbl-C381A or Cbl-b-C373A, 17-AAG was shown to enhance the physical association of Flt3-ITD with these loss-of-function mutants (Fig. 5, G and H). Thus, it is tempting to hypothesize that 17-AAG may alter the conformation of Flt3-ITD to enhance the recruitment of c-Cbl and Cbl-b. This hypothesis as well as possible involvement of other E3 ligases needs to be addressed in future study to shed more light on the molecular basis for Hsp90 inhibitor therapy against AML.
Acknowledgments
We thank Drs. F. Böhmer, K. Spiekermann, T. Kitamura, J. Held, S. Lipkowitz, D. Gray, T. Dawson, and A. Weissman for the generous gifts of experimental materials.
This work was supported in part by grants from Ministry of Education, Culture, Sports, Science, and Technology of Japan.
- Flt3
- Fms-like tyrosine kinase 3
- AML
- acute myeloid leukemia
- ITD
- internal tandem duplication
- FL
- Flt3-ligand
- 17-AAG
- 17-allylaminodemethoxy geldanamycin
- XTT
- sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate
- DOX
- doxycycline
- CHX
- cycloheximide
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Meshinchi S., Appelbaum F. R. (2009) Clin. Cancer Res. 15, 4263–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Masson K., Rönnstrand L. (2009) Cell. Signal. 21, 1717–1726 [DOI] [PubMed] [Google Scholar]
- 3. Kindler T., Lipka D. B., Fischer T. (2010) Blood 116, 5089–5102 [DOI] [PubMed] [Google Scholar]
- 4. Schmidt-Arras D. E., Böhmer A., Markova B., Choudhary C., Serve H., Böhmer F. D. (2005) Mol. Cell. Biol. 25, 3690–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choudhary C., Olsen J. V., Brandts C., Cox J., Reddy P. N., Böhmer F. D., Gerke V., Schmidt-Arras D. E., Berdel W. E., Müller-Tidow C., Mann M., Serve H. (2009) Mol. Cell 36, 326–339 [DOI] [PubMed] [Google Scholar]
- 6. Bali P., George P., Cohen P., Tao J., Guo F., Sigua C., Vishvanath A., Scuto A., Annavarapu S., Fiskus W., Moscinski L., Atadja P., Bhalla K. (2004) Clin. Cancer Res. 10, 4991–4997 [DOI] [PubMed] [Google Scholar]
- 7. George P., Bali P., Cohen P., Tao J., Guo F., Sigua C., Vishvanath A., Fiskus W., Scuto A., Annavarapu S., Moscinski L., Bhalla K. (2004) Cancer Res. 64, 3645–3652 [DOI] [PubMed] [Google Scholar]
- 8. George P., Bali P., Annavarapu S., Scuto A., Fiskus W., Guo F., Sigua C., Sondarva G., Moscinski L., Atadja P., Bhalla K. (2005) Blood 105, 1768–1776 [DOI] [PubMed] [Google Scholar]
- 9. Robinson L. J., Xue J., Corey S. J. (2005) Exp. Hematol. 33, 469–479 [DOI] [PubMed] [Google Scholar]
- 10. Sargin B., Choudhary C., Crosetto N., Schmidt M. H., Grundler R., Rensinghoff M., Thiessen C., Tickenbrock L., Schwäble J., Brandts C., August B., Koschmieder S., Bandi S. R., Duyster J., Berdel W. E., Müller-Tidow C., Dikic I., Serve H. (2007) Blood 110, 1004–1012 [DOI] [PubMed] [Google Scholar]
- 11. Buchwald M., Pietschmann K., Müller J. P., Böhmer F. D., Heinzel T., Krämer O. H. (2010) Leukemia 24, 1412–1421 [DOI] [PubMed] [Google Scholar]
- 12. Jung T., Catalgol B., Grune T. (2009) Mol. Aspects Med. 30, 191–296 [DOI] [PubMed] [Google Scholar]
- 13. Lu Z., Hunter T. (2009) Annu. Rev. Biochem. 78, 435–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swaminathan G., Tsygankov A. Y. (2006) J. Cell Physiol. 209, 21–43 [DOI] [PubMed] [Google Scholar]
- 15. Kales S. C., Ryan P. E., Nau M. M., Lipkowitz S. (2010) Cancer Res. 70, 4789–4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reindl C., Quentmeier H., Petropoulos K., Greif P. A., Benthaus T., Argiropoulos B., Mellert G., Vempati S., Duyster J., Buske C., Bohlander S. K., Humphries K. R., Hiddemann W., Spiekermann K. (2009) Clin. Cancer Res. 15, 2238–2247 [DOI] [PubMed] [Google Scholar]
- 17. Rathinam C., Thien C. B., Flavell R. A., Langdon W. Y. (2010) Cancer Cell 18, 341–352 [DOI] [PubMed] [Google Scholar]
- 18. Pearl L. H., Prodromou C., Workman P. (2008) Biochem. J. 410, 439–453 [DOI] [PubMed] [Google Scholar]
- 19. Trepel J., Mollapour M., Giaccone G., Neckers L. (2010) Nat. Rev. Cancer 10, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerji U. (2009) Clin. Cancer Res. 15, 9–14 [DOI] [PubMed] [Google Scholar]
- 21. Al Shaer L., Walsby E., Gilkes A., Tonks A., Walsh V., Mills K., Burnett A., Rowntree C. (2008) Br. J. Haematol. 141, 483–493 [DOI] [PubMed] [Google Scholar]
- 22. Minami Y., Kiyoi H., Yamamoto Y., Yamamoto K., Ueda R., Saito H., Naoe T. (2002) Leukemia 16, 1535–1540 [DOI] [PubMed] [Google Scholar]
- 23. Yao Q., Nishiuchi R., Li Q., Kumar A. R., Hudson W. A., Kersey J. H. (2003) Clin. Cancer Res. 9, 4483–4493 [PubMed] [Google Scholar]
- 24. Miura O., Cleveland J. L., Ihle J. N. (1993) Mol. Cell. Biol. 13, 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quentmeier H., Reinhardt J., Zaborski M., Drexler H. G. (2003) Leukemia 17, 120–124 [DOI] [PubMed] [Google Scholar]
- 26. Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 27. Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. (2003) Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 28. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Magnifico A., Ettenberg S., Yang C., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. (2003) J. Biol. Chem. 278, 43169–43177 [DOI] [PubMed] [Google Scholar]
- 30. Marx C., Held J. M., Gibson B. W., Benz C. C. (2010) Cancer Res. 70, 3709–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davies G. C., Ettenberg S. A., Coats A. O., Mussante M., Ravichandran S., Collins J., Nau M. M., Lipkowitz S. (2004) Oncogene 23, 7104–7115 [DOI] [PubMed] [Google Scholar]
- 32. Tsirigotis M., Thurig S., Dubé M., Vanderhyden B. C., Zhang M., Gray D. A. (2001) Biotechniques 31, 120–126 [DOI] [PubMed] [Google Scholar]
- 33. Tsirigotis M., Zhang M., Chiu R. K., Wouters B. G., Gray D. A. (2001) J. Biol. Chem. 276, 46073–46078 [DOI] [PubMed] [Google Scholar]
- 34. Mizuki M., Fenski R., Halfter H., Matsumura I., Schmidt R., Müller C., Grüning W., Kratz-Albers K., Serve S., Steur C., Büchner T., Kienast J., Kanakura Y., Berdel W. E., Serve H. (2000) Blood 96, 3907–3914 [PubMed] [Google Scholar]
- 35. Choudhary C., Schwäble J., Brandts C., Tickenbrock L., Sargin B., Kindler T., Fischer T., Berdel W. E., Müller-Tidow C., Serve H. (2005) Blood 106, 265–273 [DOI] [PubMed] [Google Scholar]
- 36. Zhang W., Konopleva M., Shi Y. X., McQueen T., Harris D., Ling X., Estrov Z., Quintás-Cardama A., Small D., Cortes J., Andreeff M. (2008) J. Natl. Cancer Inst. 100, 184–198 [DOI] [PubMed] [Google Scholar]
- 37. Yao Q., Nishiuchi R., Kitamura T., Kersey J. H. (2005) Leukemia 19, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 38. Yao Q., Weigel B., Kersey J. (2007) Clin. Cancer Res. 13, 1591–1600 [DOI] [PubMed] [Google Scholar]
- 39. Kancha R. K., Grundler R., Peschel C., Duyster J. (2007) Exp. Hematol. 35, 1522–1526 [DOI] [PubMed] [Google Scholar]
- 40. Padrón D., Sato M., Shay J. W., Gazdar A. F., Minna J. D., Roth M. G. (2007) Cancer Res. 67, 7695–7702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toffalini F., Kallin A., Vandenberghe P., Pierre P., Michaux L., Cools J., Demoulin J. B. (2009) Haematologica 94, 1085–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmidt-Arras D., Böhmer S. A., Koch S., Müller J. P., Blei L., Cornils H., Bauer R., Korasikha S., Thiede C., Böhmer F. D. (2009) Blood 113, 3568–3576 [DOI] [PubMed] [Google Scholar]
- 43. Rocnik J. L., Okabe R., Yu J. C., Lee B. H., Giese N., Schenkein D. P., Gilliland D. G. (2006) Blood 108, 1339–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vempati S., Reindl C., Wolf U., Kern R., Petropoulos K., Naidu V. M., Buske C., Hiddemann W., Kohl T. M., Spiekermann K. (2008) Clin. Cancer Res. 14, 4437–4445 [DOI] [PubMed] [Google Scholar]
- 45. Yang B., Kumar S. (2010) Cell Death Differ. 17, 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]