Abstract
The 26 S proteasome is the eukaryotic protease responsible for the degradation of most cellular proteins. As such it accommodates the ability to function under diverse conditions that the cell may encounter. This function is supported by various adaptors that modulate various aspects in protein degradation, these include regulation of substrate delivery, deubiquitination, unfolding, and 20 S gate dilation. Here we show a new functional complex between the P97 and the proteasome that is assembled in response to proteasomal impairment. This entails P97 binding to the 26 S proteasome via the 19 S particle thereby forming an additional hexameric ATPase ring to relieve repression. P97-bound proteasomes showed selective binding toward the Npl4-ufd1 P97 co-factors, indicating a unique cellular role for P97 binding to proteasomes. P97-bound proteasomes display enhanced activity, showing a relief in proteolysis impairment. Our findings place P97 directly in non-ERAD proteasomal functions and establish a new checkpoint in UPS impairment. The ability to modulate proteasome activity and properly respond to protein misfolding, is of great importance in cellular regulation.
Keywords: ATPases, Proteasome, Protein Degradation, Protein Folding, Ubiquitin, ERAD, P97, Protein Misfolding, Proteotoxicity
Introduction
Maintaining a stable environment is a fundamental requirement to retain the ability of proteins to function properly. The misfolding of proteins whether imposed by encoded mutations or inflicted by environmental changes, serves as a continuous challenge the cell must deal with. Whereas accumulation of a mutant, misfolded protein deprives the cell of that specific protein activity, a nonspecific effect referred to as “proteotoxicity” challenges cellular homeostasis. This occurs by promiscuous interaction the misfolded protein engages, thereby nonspecifically depriving proper proteome function in a dominant manner. The importance of properly folding proteins is exemplified in pathologic situations and diseases (1–3). In light of this, several cellular mechanisms have evolved to maintain homeostasis, thus enabling proper function even during severe conditions. Of particular importance in this context are the many molecular chaperones present in all types of cells and cellular compartments that bind and neutralize proteotoxins (4). A second mode of cellular response to rid cells of potential proteotoxins is the activation of the protein degradation apparatus. The ubiquitin proteasome system (UPS)2 is the cellular moiety in eukaryotic cells that is responsible for the degradation of most cellular proteins in an ATP-dependent manner (5). Protein degradation by the UPS occurs in two steps, a ubiquitin-tagging reaction generally composed of a series of ubiquitination events initiated by a E1-activating enzyme, followed by an E2-conjugating enzyme and a E3 ubiquitin ligase (6). It is the combination of the E2-E3 enzymes that renders the specificity of the poly-ubiquitin chain produced to accommodate a degradation signal and the substrate modified. While these initial steps of protein ubiquitination have received most of the attention as the regulatory step in protein degradation, it has become apparent in recent years that the 26 S proteasome is a dynamic particle that can be modified to handle a variety of substrates. Although the 20 S catalytic particle is the entity in which proteolysis occurs, most steps post-ubiquitination are intended to prepare the substrate to be properly suitable for degradation. Processes executed by the 19 S particle include de-ubiquitination, unfolding requiring ATP hydrolysis and increasing catalytic accessibility of the 20 S particle (7, 8). These steps can be modified by sets of adaptors that bind the 19 S particle including adaptors that are transiently associated with the 19 S particle and serve as shuttling factors composed of ubiquitin-like (UBL) and ubiquitin associated (UBA) domains, thus increasing efficiency of substrate delivery to the 19 S particle resident ubiquitin receptors (9). Another set of adaptors are those that bind the 19 S particle and aid in the turnover of specific substrates or during specific conditions (10, 11). Thus the combination of both particles enables efficient proteolysis of designated proteins.
P97 (VCP/Cdc48) is a barrel shaped homo-hexameric ATP-dependent chaperone that belongs to type II AAA ATPase superfamily, however it is not associated with a protease. P97 contains three distinct domains; an N-terminal N-domain followed by two adjacent ATPase domains, D1 and D2. P97, similar to other AAA proteins, is involved in many cellular processes such as membrane fusion, ER-associated degradation (ERAD), transcription activation, cell cycle control, apoptosis and mitosis (12–16). In most of these cellular activities P97 functions either as a segregase that binds poly-ubiquitinated proteins and dissociates them from their binding partners or extracts them from protein complexes. By translating ATP hydrolysis into a mechanical force P97 execute its segregase activity (12, 17, 18). Much like the 19 S particle, P97 specificity is obtained via numerous adaptors including a family of ubiquitin regulatory X (UBX)-UBA domain containing adaptors (18, 19). The relation between P97 and the 19 S ATPases is re-enforced by the notion that several UPS substrates require P97 to be properly processed by the 26 S proteasome (14, 20). In this study we have revealed the association of P97 with proteasomes under conditions that confer UPS impairment. Our data show direct interaction between P97 and the 26 S proteasome complex and establish a regulation step in UPS impairment.
EXPERIMENTAL PROCEDURES
Cell Transfection, Lysis, and Protein Purification
Plasmids expressing Flag P97 truncations were constructed by amplifying the relevant amino acids of rat P97 cDNA using pQE9-P97 plasmid as a template (21) and ligating into the mammalian expression plasmid PCDNA3.1 containing an N-terminal Flag tag. For hexamerizatiom evaluations, wildtype p97 or truncated forms were expressed in TNT reticulocyte expression system (Promega) and subjected to glycerol gradient centrifugation. The collected fractions were resolved on SDS-PAGE and exposed to autoradiograph. Mammalian expression plasmids of GST and YFP proteasomal subunits and Flag-tagged P97 cofactors have been previously described (10, 21). Knock-down of human P97 and Ufd1 expression was obtained using a predesigned RNAi expression vectors (Sigma SCHLNG-NM_007126 and NM_005659, respectively) to produce stable clones. Transient transfections were performed using 293T cells transfected using the calcium-phosphate precipitation method. Stable CHO cell clones expressing YFP proteasomal subunits were obtained by antibiotic selection with 450 μg/ml G418.
Cell lysis was performed in TNH buffer (20 mm HEPES-7.9, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1.5 mm MgCl2, 1 mm DTT, 5 mm ATP, and protease inhibitors), clarified at 20,000 × g for 10 min. Where indicated 400 μl of lysate were layered over a 10–40% continuous glycerol gradient and centrifuged in a TH660 rotor for 4 h at 374,400 × g. Gradients were divided into 14 fractions. For direct proteasome purifications, cell lysates were immunopurified against a PSMA1 Ab. for 2 h and beads were extensively washed with lysis buffer subsequently eluted with Laemmli buffer. Where indicated, cells were treated with arsenite for the indicated time (0.5 mm Sigma S7400) MG132 or Velcade as indicated (10 μm 60min). Conditions for all other protein misfolding agents are indicated in figure legends.
The diamide-mediated chemical crosslinking was conducted by metabolically labeling the transfected cells with 35S-TRANSlabel (ICN 15 μCi/ml overnight), followed by MG132 treatment, velocity gradient centrifugation and Psma1 immunoprecipitation from the proteasome-containing fractions. The purified complex was reacted with Diamide (100 mm Sigma D3648) in TNH buffer (−DTT) on ice for 45 min. The sample was denatured in 2% SDS at 95 °C, diluted to 0.1% SDS, and immunoprecipitated with a Flag antibody. Where indicated, the cross-linker was reversed with DTT, and the samples were resolved by SDS-PAGE.
20 S purifications were performed by immunoprecipitating Psma1 followed by washes with 1 m NaCl to dissociate 19 S particles. No 19 S subunits were detected upon evaluation of purified 20 S particles (data not shown).
Proteasome Assays
Peptide activation assays were performed as previously described (22). Briefly, 20 S particles were resuspended in a final volume of 50 μl (45 mm Tris-HCl, pH 8.0, 5 mm DTT, 50 μm Suc-LLVY-AMC (Sigma S6510)), and the indicated peptide. Peptides corresponding to the last ten C-terminal amino acids of human Rpt5 and P97 were obtained from Synthetic Biomolecules. P97 proteasome complex activity assays were obtained from the indicated cells and were performed as previously described (10). Briefly, enzymatic activity of proteasomes immobilized on a P97 purification matrix was assayed after their purification from 293T cells untreated or treated with arsenite (to induce P97-proteasome association). Psma1 IP served as a reference for the activity of conventional proteasomes and GST purifications as a background control. All catalytic activities were normalized to proteasome content evaluated using a quantitative Rpt1 immunoblot (Odyssey, Li-COR Biosciences).
Antibodies
P97 antisera for immunoblot was produced by immunizing rabbits against the rat P97 (Covance). P97 antisera for IP was a kind gift from Yihong Ye. Psmd14 antisera was produced by immunizing rats against the mouse Psmd14. Anti-Psma1 (ABR) or Mab. 2–17 (a kind gift from Keiji Tanaka), anti-ubiquitin (Zymed Laboratories Inc.), anti-Rpt1 (Biomol), anti-Flag M2 (Sigma), anti-GAPDH, and UFD1 (Santa Cruz Biotechnology). Anti-GST and -GFP antibodies were a kind gift from David Ron.
RESULTS
P97 Interacts with the Proteasome upon Proteasome Impairment
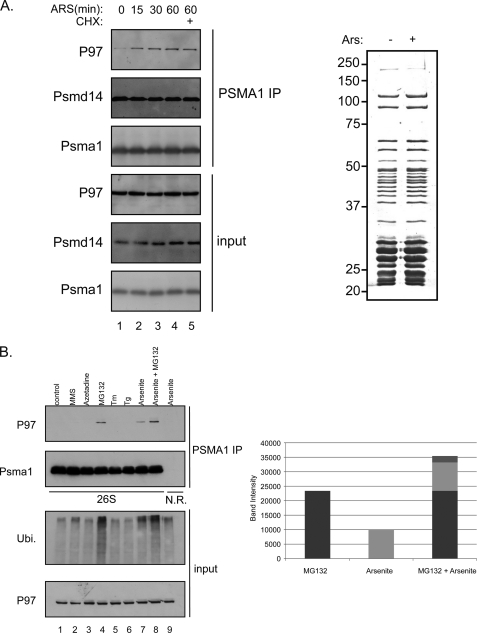
P97 acts as a molecular chaperone in many diverse cellular activities (12, 14–16). Interestingly, most of these activities are related to the UPS (18). The involvement of P97 in the UPS degradation pathway is best exemplified by the findings that P97 acts in conjunction with the proteasome to degrade a wide variety of ubiquitinated targets, probably acting upstream of the proteasome (23–26). However the notion of P97 involvement in proteasome substrate degradation comes mainly from genetic interactions showing the requirement of P97 in proteasome degradation of specific substrates. While P97 and the yeast homologue Cdc48 could be detected in proteasome purifications by highly sensitive methods such as mass-spectrometry (27, 28), their robustness and lack of quantitative interpretation did not allow correlating P97 recruitment to the UPS system upon proteasome impairment (29). These results imply that either the interaction is transient or only minute quantities from each population (P97 and proteasome) are engaged in this interaction, as this interaction could not be verified biochemically. As P97 is viewed as a segregase with an unfolding activity (20), P97 presumably interacts with proteasomes at initial steps to assist in substrate processing toward the 20 S catalytic chamber and as such may do so in a very transient manner. To enrich and/or stabilize the population of P97-associated proteasomes, we reasoned that high proteasome workload conditions may be a means to gain such enrichment, as P97 proteasome association under these conditions may be stabilized. To this end, we treated cells with arsenite an agent known to induce protein misfolding and poly-ubiquitin accumulation (10). Cells were treated with arsenite for the indicated time course, and the amount of P97 associated with proteasomes was evaluated. As seen in Fig. 1A, as short as 15 min after arsenite treatment, one can detect an increase in P97 content in purified 26 S proteasomes, an increase that is steadily increased with time and does not require de novo protein synthesis. In contrast to the observed change in P97, global proteasome profiling did not change upon arsenite treatment as evaluated from the Coomassie-stained purified proteasomes (Fig. 1A). Our previous quantitative analysis indicates the specificity in the observed increase of P97 content in arsenite-treated proteasomes (30).
FIGURE 1.
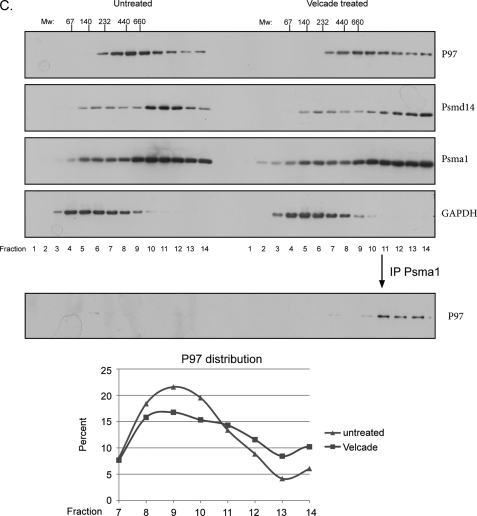
Proteasome-P97 interaction. A, left panel shows a time course induction of P97-proteasome interaction upon arsenite treatment (0.5 mm). Untreated or arsenite-treated lysates were subjected to PSMA1 (26 S) IP followed by a PSMD14 and P97 immunoblots. Cycloheximide (10 μg/ml) was added to prevent de novo protein synthesis. Right panel indicates the protein content (as evaluated by Coomassie-stained SDS-PAGE) of the 26 S proteasomes purified from control and arsenite-treated cells. Input represents 50 μg of total cell extract while the IP was performed using 2 mg of cell extract. B, HEK 293 cells were treated with several stress agents and subjected to PSMA1 (26 S) IP and P97 immunoblot. The stress agents indicated are: 2, the electrophile methylmethanesulfonate (MMS, 2.4 mm 1 h); 3, the amino acid analog Azetidine (5 mm, overnight); 4, proteasome inhibitor MG132 (10 μm, 1 h); 5, the ER stress-inducing agents tunicamycin (Tm, 20 μg/ml, 1 h); 6, thapsigargin (Tg, 2 μm, 1 h) 7, arsenite (0.5 mm, 1 h); and 8, MG132 (10 μm, 1 h)+arsenite (0.5 mm, 1 h). Lane 1 represents the untreated cells. As a negative control we used a non-relevant IP against cells treated with arsenite. Right panel shows the quantitation of the P97 signal indicating the accumulative effect of MG132 and arsenite. Input represents 50 μg of total cell extract while the IP was performed using 2 mg of cell extract. C, immunoblot of P97, the 19S lid component PSMD14, the 20 S subunit PSMA1 and GAPDH in fractions of glycerol gradients prepared from untreated and Velcade-treated cells (1 h 10 μm). The immunoblot of P97 in PSMA1 immunoprecipitations from fractions of glycerol gradients is shown in the bottom panel. The migration of complexes of known size is indicated above. The migration of P97 from the indicated fractions was quantified and presented as fractions from the total signal. Note the increase in P97 in the HMW fractions upon Velcade treatment. GAPDH IB served as a nonspecific migration control.
Next we wanted to examine the specificity of the interaction between P97 and the proteasome and to survey the effect of several protein misfolding agents and proteasome inhibition conditions. Cells were treated with the indicated agents, proteasome purified and P97 content evaluated. As seen in Fig. 1B only arsenite and proteasome inhibition induced an increase in P97 proteasome association (Fig. 1B, lanes 4, 7, and 8). This specificity was in agreement with the enhanced ability of these two conditions to increase the cellular content of poly-ubiquitinated proteins (Fig. 1B). This correlation implies that P97 recruitment to the proteasome is aimed toward such particles that are stalled in their ability to process substrates (see “Discussion”).
To evaluate what fraction of proteasomes and P97 are interacting with each other under impaired UPS conditions, we analyzed the shift in sedimentation properties of endogenous P97 from control and proteasome-inhibited cells. We noted an increase in P97 population in the high molecular weight (HMW) fractions upon proteasome inhibition (Fig. 1C). This shift is better visualized if proteasome purification is first performed from each fraction prior to a P97 immunoblot (Fig. 1C, lower panel). These results are in agreement with previous reports that noted the shift in P97 migration upon proteasome inhibition (31). Taken together our results show the rapid recruitment of P97 to proteasomes upon accumulation of poly-ubiquitinated proteins.
P97 Proteasome Complex Contains Both 19 S and 20 S Particles
Two possibilities come to mind when addressing the issue of P97-proteasome formation. One entails the binding of P97 to the 26 S particle similar to the interaction of other proteasomal adaptors with the 26 S proteasome (10, 32). The other possibility predicts the formation of a hybrid proteasome in which the P97 hexamer displaces at least one 19 S regulatory particle, thereby binding to the 20 S particle, in analogy to PA28 (33). To address the question whether the P97-proteasome complex includes both 19 S and 20 S particles, we stably expressed YFP (as control) or YFP fusion proteins of both 19 S and 20 S components in CHO cells. YFP proteasome incorporation was evaluated by the percentage of YFP that was incorporated into the HMW proteasomal fractions (Fig. 2A, right panel). Lysates prepared from control and proteasome inhibited cells were subjected to YFP immunopurification and the presence of P97, 19 S and 20 S components were monitored. As seen in Fig. 2A (left panel), P97 is present in the purification of both the 20 S and 19 S complexes after proteasome inhibition, implying that the P97-proteasome complex contains a 20 S particle and at least one 19 S cap. Evaluation of endogenous P97 was performed by using P97 immunopurifications and 19 S or 20 S immunoblots (Fig. 2B). As noted, both the 20 S (Psma1) and the 19 S (Rpt1) particles were purified with P97 and neither one purified in the non-relevant immunopurification, confirming once more the existence of at least one 19 S particle in the P97-proteasome complex. However based on these results, one cannot distinguish whether P97 binds or replaces 19 S particles.
FIGURE 2.
The P97-proteasome complex contains both the 19 S and the 20 S particles. A, immunoblots of P97, Psmd14, and YFP after YFP IP's from CHO clones expressing the indicated proteins obtained from untreated or Velcade-treated cells. The right panels reveal the glycerol gradient YFP immunoblots of the various clones. B, 293 cells untreated or MG132 treated were lysed and purified with P97 antibody, P97 IP followed by Rpt1 and Psma1 immunoblots. As a negative control, we used a non-relevant IP (NR IP).
P97 Binds Directly to the 19 S Particle
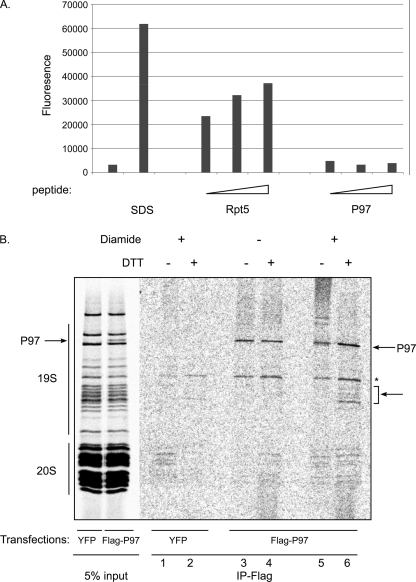
Recently, a HbYX motif was identified as regulating PAN and the 19 S ATPase association with the catalytic particle (22, 34). These studies showed that the C-terminal peptides of PAN, Rpt2, or Rpt5 bind to the catalytic particle and are sufficient to increase catalytic accessibility as evaluated by their ability to enhance hydrolysis of a short peptide substrate. As P97 (but not its close homologue Nsf) also contains a HbYX motif (amino acids LYG) in the C terminus (supplemental Table S1), the existence of this sequence in P97 C termini would support a model in which a hybrid proteasome is formed through P97's HbYX and the 20 S α subunits. To examine this possibility we evaluated the ability of P97's C terminus to induce gate opening thereby allowing the 20S particle to hydrolyze a small peptide substrate. Latent 20 S particles were purified and incubated with increasing amount of P97 or Rpt5 C termini peptides. Full activation was achieved by incubation of 20 S particles in the presence of 0.01% SDS as a control. As shown in Fig. 3A only Rpt5 but not P97 C termini peptides stimulated 20 S peptide hydrolysis activity. The inability of P97 C-terminal peptide to induce activation of 20 S hydrolysis indicates that although both ATPases interact with the proteasome, their mode of binding is different in nature. Similar results were obtained using the full-length P97 (supplemental Fig. S1).
FIGURE 3.
P97 binds the 19 S particle of the proteasome. A, 0.01% SDS and several concentrations of peptides corresponding to the C-terminal residues of P97 and Rpt5 (50, 200, 400 μm) were incubated with 20 S proteasomes and activity against the N-succinyl-LLVY-AMC (100 μm) fluoregenic peptide was measured. B, autoradiograph of a SDS-PAGE of metabolically labeled proteins purified by glycerol gradient centrifugation followed by PSMA1 immunoprecipitation from cells transfected with Flag P97 or YFP-treated with MG132. The purified complex was displayed as is (lanes 1 and 2) or exposed to diamide (100 μm), followed by disruption in 2% SDS. Samples were diluted and immunoprecipited with an anti-Flag antibody as indicated (lanes 3–6). Note the appearance of several bands with the approximate mass of 50 kDa in a P97 and diamide-dependent manner.
To identify the direct binding partners of P97 in the proteasome complex we performed a cross linking assay. Cells ectopically expressing Flag-P97 or Flag-YFP (as a control), were metabolically labeled with [35S]methionine/cysteine, subjected to a velocity gradient centrifugation and proteasomes were affinity purified from HMW fractions. The purified complex was displayed as is (Fig. 3B, input), or treated with diamide, forming disulfide bonds between adjacent cysteines. Non-covalent interactions were then disrupted by denaturing the complex, and P97 (and crosslinked partners) was immunoprecipitated under stringent conditions. Samples were either reduced by DTT to reveal the individual radiolabeled proteins, or directly resolved by SDS-PAGE. Autoradiography revealed that in addition to radiolabeled Flag-P97 (Fig. 3B, lanes 3–6), the immune complex contained several diamide-dependent bands that appeared as HMW proteins in the absence of DTT (Fig. 3B, lane 5) and as lower molecular weight proteins with mobility corresponding to 19 S ATPases Rpt1–6 subunits, upon cross-linking reversion (+DTT, Fig. 3B, lane 6). The appearance of the cross-linked band was detected in a cross-linker dose-dependent manner (supplemental Fig. S2). These findings indicate that P97 is in direct proximity to the 19 S regulatory particle. Additionally, we note the appearance of P97 already in the input of the purified proteasomes in cells overexpressing Flag-P97 (see “Discussion”).
Characterizing P97 Binding Domains and Adaptor Selectivity in Proteasome Association
P97 consists of an N-terminal domain and two AAA ATPase domains (D1 and D2) in tandem (18). To elucidate the domains required for the interaction of P97 with the proteasome, Flag-tagged P97 deletion mutants were ectopically expressed in cells. Immunopurified proteasomes were evaluated toward their P97 content with a Flag immunoblot (Fig. 4A). We observed that deleting both the N-terminal (1–185) and the C-terminal domain (761–806) of P97 did not perturb the ability of P97 to bind proteasomes. However, deleting either one of the AAA ATPase domains disrupted the binding of P97 to proteasomes. This result is in agreement with previous data showing the inability of the C-terminal HbYX motif of P97 to activate the catalytic 20 S particle (Fig. 3A). As P97 functions as a homohexamer, we evaluated the ability of each one of the above deletion mutants to form such hexameric structures. In vitro coupled transcription and translation of the various deletions were subjected to velocity gradient centrifugation and P97 distribution was evaluated. As seen in Fig. 4B, we noted the correlation between the ability of P97 to bind proteasomes and the ability to form hexamers. We conclude that the D1 and D2 ATPase domains of P97 are required both for hexamer formation and proteasome binding.
FIGURE 4.
Mapping of the proteasome binding region of P97. A, full-length p97 or truncated varients were expressed as FLAG-tagged fusion proteins in HEK 293T cells treated with MG132. Expression of FLAG-tagged p97 proteins was detected by immunoblotting before (input left panel) and after IP of PSMA1 (right panel). Note the importance of D1 and D2 domains for P97-proteasome complex formation. B, wild type p97 or truncated proteins were expressed in TNT reticulocyte expression system and subjected to glycerol gradient centrifugation. The collected fractions were resolved on SDS-PAGE and exposed to autoradiography. The hexameric form of P97 proteins is indicated. Flag P97 linker +D2 (461–806) and Flag P97 ΔD2 (1–480) fail to form hexamers.
Although many of the characterized cellular roles of P97 involve the UPS, several P97 adaptors involve P97 in non-proteolytic processes. One such example is P47 that recruits P97 toward membrane fusion processes and autophagosome biogenesis (35, 36). To evaluate the presence and selectivity of P97 co-factors in proteasome association, we ectopically expressed various Flag-tagged P97 co-factors (P47, Npl4, and Ufd1). Proteasome purifications were carried out from cell extracts and P97 content was evaluated. As seen in Fig. 5A, both Npl4 and Ufd1 were co-purified with Psma1 only upon treatment with a proteasome inhibitor, however P47 was absent from this purified complex. Furthermore unlike Npl4-ufd1, P47 expression did not elevate the amount of P97 purified with proteasomes upon UPS impairment. This result indicates that P97 complexes that are associated with the proteasome are selective, therefore P97 molecules that are committed to cellular processes involving P47 co-factors are not recruited to proteasomes. Ufd1-Npl4 are co-factors that are associated with numerous P97 adaptors in various UPS-related P97 cellular processes (31). Thus the P97 proteasome interaction described above involves a specific P97 subset of adaptors. To evaluate the requirement of these selective P97 adaptors for proteasomal binding, we performed proteasome purifications from control and Ufd1 knock-down cells treated with a proteasome inhibitor. As seen in Fig. 5B, upon Ufd1 knock-down we could detect a decrease in P97 co-purification with proteasomes. These results ascertain the requirement and specificity of a selective group of P97 adaptors required for proteasomal binding.
FIGURE 5.
P97 cofactors specificity and requirement in P97-proteasome co-purifications. A, P97 cofactors were expressed as FLAG-tagged fusion proteins in HEK 293T cells untreated or treated with Velcade (as indicated). Lysates were subjected to a direct immunoblot (input) or immunprecipitated with a PSMA1 antibody. P97 and FLAG expressed protein content co-purified with PSMA1 were evaluated on the immune-purified material (Psma1 IP). B, stable clones expressing a Ufd1 knock-down expression vector were produced to evaluate the requirement of Ufd1 toward P97-proteasome co-purification. Proteasomal-P97 co-purifications were performed as described in Fig. 1.
P97 Binding to Proteasomes Induces Proteasomal Activation
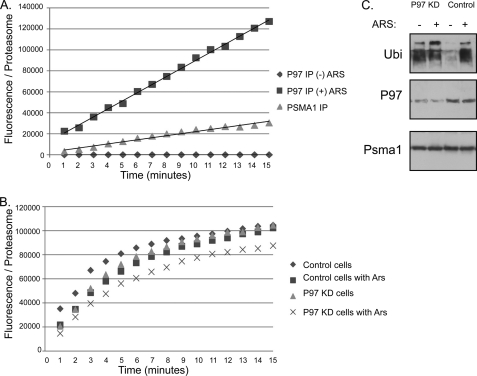
P97 and proteasome are involved in numerous cellular processes and are both essential entities. In addition, only a fraction of both abundant complexes interact upon protein misfolding (Fig. 1C). As proteasome adaptors have been previously shown to change various features in the proteasome (10, 32, 33), we decided to compare conventional proteasomes to P97 containing proteasomes. The hydrolysis of a small proteasome substrate peptide was monitored on P97 purifications performed in the absence or presence of arsenite. As noted in Fig. 6A, no hydrolysis was detected in P97 immunopurifications in the absence of arsenite, indicating the lack of background activity from P97 preparations. When proteasome activity was compared between conventional proteasomes and P97-proteasomes (Fig. 6A), we noted a 4-fold increase in activity of proteasomes containing P97. To address the role of P97 in proteasome protection during arsenite treatment, we have compared the activity of proteasomes purified from control and arsenite challenged cells, upon P97 knock-down. As seen in Fig. 6B, P97 is required to compensate the decrease in activity upon arsenite treatment as the activity decrease could only be observed in the arsenite-treated P97 knockdown cells. These results demonstrate that the proteasome-P97 complex significantly up-regulates the catalytic accessibility of proteasomes under protein misfolding conditions and that this increase is required in order to maintain proper proteasomal activity during the protein misfolding conditions. In line of these results we noted that the knock-down of P97 expression had a global effect on the accumulation of cellular poly-ubiquitin even in non-stressed cells (Fig. 6C) probably revealing the role of P97 in steady state conditions (for example in ERAD processes). It is the very high Mw poly-ubiquitin that increases further in the arsenite treated P97 knockdown cells that may be inhibiting proteasomal activity during protein misfolding conditions.
FIGURE 6.
Induced activity and role of the P97-proteasome complex. A, digestion of a small, unstructured peptide (N-succinyl-LLVY-AMC), monitored by fluorescence in samples of proteasomes purified by P97 immunoprecipitation from cells untreated or arsenite-treated. PSMA1 IP served as a reference for the activity of conventional proteasomes and GST purifications as a background control. All observed activity was inhibited upon proteasomal inhibitor treatment (data not shown). Arbitrary fluorescence units were normalized to proteasome content assessed by a quantitative immunoblot for Rpt1 (see supplemental Fig. S4). B, similarly, peptide hydrolysis assay were performed on proteasomes purified from untreated and arsenite-treated cells purified from control and P97 knock-down cells. C, cells with a knock-down expression of P97 (P97KD) were evaluated toward their basal and arsenite-induced poly-ubiquitination levels. PSMA1 and P97 immunoblots confirm the equal loading and P97-reduced expression, respectively.
DISCUSSION
Our data presented describe the association of an additional ATPase hexamer with the 26 S proteasome. While the role of P97 in numerous cellular processes is tightly linked to the UPS, no direct interaction with the proteasome was demonstrated. Recently a P97-dependent impairment in substrate proteolysis was reported to be linked to the folded status of the substrate (20). This result complements our observation in which P97 association with proteasomes is apparent only during conditions in which the population of challenging misfolded substrates rises (caused either by misfolding or proteasome inhibition). The nature of this association is that of a rapid response not requiring any de novo protein synthesis (as soon as 15 min after arsenite treatment; Fig. 1A). The association seems to be induced only after specific conditions, as a variety of different protein misfolding agents could not promote such association (Fig. 1B). As all the selected misfolding agents induce a strong UPR response, we conclude that increasing UPS substrates per se is not sufficient for stable complex formation between P97 and the proteasome and indicate the role of P97 binding to proteasomes in an ERAD independent manner. In contrast, stalling proteasomal processing (as evident by the accumulation of poly-ubiquitinated proteins) constitutes a cellular check point inducing the stable formation of the P97-proteasome complex. This hybrid configuration is presumably attempting to relieve this attenuation. As the proteasome is a highly abundant cellular moiety (37), it is capable of buffering fluctuations that occur in substrate availability. Hence only dramatic changes cause inefficient substrate turnover leading to poly-ubiquitin accumulation. It is only under such conditions (arsenite and proteasome inhibition tested in this report) were we able to detect P97 recruitment to proteasomes. This specificity may imply that P97 recognizes only proteasomes associated with substrates. While under normal conditions this configuration is highly transient, severe protein misfolding stabilizes this status. In this scenario, P97 is constantly associated with the proteasome in a highly transient fashion that does not withstand our normal purification methodologies. This interaction is only detected during conditions that impair substrate turnover.
The steady state population of P97 hexamers that are bound to proteasomes is minute (Fig. 1C). This finding is expected as P97 is in greater abundance over proteasomes (37, 38) and normally only small fractions of proteasomes are processing substrates. The P97-proteasome complex was further characterized as containing both 19 S and 20 S particles (Fig. 2) and cross-linking experiments confirmed the close proximity of P97 to 19 S subunits rather than 20 S subunits (Fig. 3B) thereby supporting the direct binding of P97 to the 26 S proteasome. In addition, the inability of the P97 to stimulate 20 S hydrolysis of a small peptide (Fig. 3A and supplemental Fig. S1), argues against a direct 20 S binding model of P97. Careful examination of radiolabeled proteasomes purified from proteasome inhibitor treated cells overexpressing P97, showed the appearance of a single additional band with a mass of 97 kDa. While this may imply the ability of P97 to bind directly to proteasomes without the necessity of adaptors, it is probably due to the inability to distinguish between the various adaptors and proteasome subunits, as the mass of P97 cofactors overlaps with proteasome subunits. We reason this as certain P97 cofactors could be detect by immunoblots in proteasome purifications (Fig. 5A).
In an attempt to map the domains required for proteasome binding of P97, we demonstrated the requirement for P97 hexamerization. Deletion of the N and C-terminal portions of P97 (186–761) did not perturb proteasome binding and hexamerization (Fig. 4). Hexamerization of AAA ATPase is an evolutionary conserved feature for protease binding. It is a required for ClpA binding to ClpP (39) and is observed during 26 S biogenesis (40, 41). Regarding the ATPase activity requirement for P97-proteasome binding, we were unable to evaluate this in vivo, as expression of a P97 ATP hydrolysis defective mutant (P97QQ), hetero-hexamerized with the endogenous P97 in vivo, thereby preventing us from determining this requirement (supplemental Fig. S3).
ATPase substitution by a proteolytic component is observed in the ClpP complex that can bind various ATPases (ClpX and ClpA) thereby enabling versatility in the unfolding capacity of the protease complex, affecting its ability to degrade loss versus tightly folded substrates (42). Another more extreme change is the replacement of the cap of the protease forming a hybrid protease. In this scenario the nature of the peptides generated by the hybrid protease are modified to become more suitable for immune presentation (43). In this respect the binding of P97 to the proteasome is unique as it is not replacing a 19 S regulatory particle but rather additive (Fig. 3). Why would the addition of an ATPase hexamer to an existing one (19 S Rpt's) benefit the protease during protein misfolding? We can consider two scenarios that would explain this addition. The first would claim that the addition of P97 increases the unfolding capacity of the 19 S thereby increasing its ability to process hard to unfold substrates that clog substrate processing by the proteasome (similar to the case of ClpP; (42)). This feature of P97 would be supported by P97's unfolding activity (44) and by the ability of ClpB to promote substrate degradation performed by ClpP (45). In this case P97 would provide the required excess unfolding capacity while the 19 S is required for dilating the catalytic particle, thus enabling translocation. In an alternative scenario, we envision P97 acting as a segregase to unclog proteasomes that are stalled with a hard to unfold substrate thereby relieving the impaired flow of accumulated UPS substrates. Our findings showing increase in hydrolysis of the fluorogenic peptide (Fig. 5A) may demonstrate the capability of P97 to enhance the capacity of the 19 S to accommodate translocation into the portal leading to the catalytic chamber, and argue that the P97-proteasome complex is highly processive, thus supporting the first scenario. This alteration in processivity enables the proteasome to handle the increase in burden imposed by increasing amounts of poly-ubiquitinated substrates, We could not observe major changes in activity of proteasomes purified from unstressed or arsenite-stressed cells (Fig. 6B) despite the fact that more poly-ubiquitinated substrates are co-purified during these conditions (10). These findings argue toward a role for P97 in proteasomal activity enhancement that is not mediated by the poly-ubiquitinated substrate (46). Recent reports have shown that the active sites status of the catalytic particle subunits, regulate association with the regulatory particle and complex assembly chaperones (47, 48) thus arguing toward a mechanism that enable allosteric changes across the catalytic chamber. We envision that P97 binding to the proteasome may transmit similarly changes across the 26 S complex that may affect several components along the particle. In our attempts to observe proteolytic changes using a poly-ubiquitinated substrate, we were unable to produce comparable amounts of P97-proteasomes as done in the kinetic peptide assay (Fig. 6). Revealing the identity of the misfolded challenging proteasome substrates would be required to show the differences in the P97-proteasome complex using a poly-ubiquitinated substrate assay. Despite the low abundance of proteasome-P97 complexes, the biological importance of P97 in maintaining UPS homeostasis is exemplified by our observations showing the high basal levels of poly-ubiquitin accumulation observed in P97 knock-down cells (Fig. 6C) and by the reduced peptide hydrolysis observed mainly in the P97 knock-down arsenite-treated cells (Fig. 6B). Notably, our finding extends P97 functions as a UPS cofactor, in ERAD-independent processes. How does the P97 complex recognize stalled proteasomes and what is the nature/identity of the misfolded substrates this complex deals with, are questions for further studies that are required to unravel the mechanistic role of P97 in supporting proteasome impairment.
Supplementary Material
Acknowledgments
We thank H. Mayer, D. Ron, K. Tanaka, and Y. Ye for reagents and A. Navon for fruitful discussions.
This work was supported by the Israeli Science Foundation (ISF 497/08), ERC-FP7-IRG (to A. S.), and the Brain Power for Israel Foundation (to E. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- UPS
- ubiquitin proteasome system
- ERAD
- ER-associated degradation.
REFERENCES
- 1. Dobson C. M. (2003) Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 2. Dobson C. M. (2003) Nat. Rev. Drug Discov. 2, 154–160 [DOI] [PubMed] [Google Scholar]
- 3. Vendruscolo M., Zurdo J., MacPhee C. E., Dobson C. M. (2003) Philos. Transact. A Math Phys. Eng. Sci. 361, 1205–1222 [DOI] [PubMed] [Google Scholar]
- 4. Macario A. J., Conway de Macario E. (2007) Front. Biosci. 12, 2588–2600 [DOI] [PubMed] [Google Scholar]
- 5. Baumeister W., Lupas A. (1997) Curr. Opin. Struct. Biol. 7, 273–278 [DOI] [PubMed] [Google Scholar]
- 6. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 7. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanaka K. (2009) Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartmann-Petersen R., Gordon C. (2004) Curr. Biol. 14, R754–R756 [DOI] [PubMed] [Google Scholar]
- 10. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 11. Wilk S., Chen W. E., Magnusson R. P. (2000) Arch. Biochem. Biophys. 383, 265–271 [DOI] [PubMed] [Google Scholar]
- 12. Jentsch S., Rumpf S. (2007) Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 13. Fu X., Ng C., Feng D., Liang C. (2003) J. Cell Biol. 163, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bar-Nun S. (2005) Curr. Top. Microbiol. Immunol. 300, 95–125 [DOI] [PubMed] [Google Scholar]
- 15. Cao K., Nakajima R., Meyer H. H., Zheng Y. (2003) Cell 115, 355–367 [DOI] [PubMed] [Google Scholar]
- 16. Meyer H., Drozdowska A., Dobrynin G. (2010) Biochem. Cell Biol. 88, 23–28 [DOI] [PubMed] [Google Scholar]
- 17. Tucker P. A., Sallai L. (2007) Curr. Opin. Struct. Biol. 17, 641–652 [DOI] [PubMed] [Google Scholar]
- 18. Wang Q., Song C., Li C. C. (2004) J. Struct. Biol. 146, 44–57 [DOI] [PubMed] [Google Scholar]
- 19. Schuberth C., Buchberger A. (2008) Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beskow A., Grimberg K. B., Bott L. C., Salomons F. A., Dantuma N. P., Young P. (2009) J. Mol. Biol. 394, 732–746 [DOI] [PubMed] [Google Scholar]
- 21. Ye Y., Meyer H. H., Rapoport T. A. (2003) J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai R. M., Chen E., Longo D. L., Gorbea C. M., Li C. C. (1998) J. Biol. Chem. 273, 3562–3573 [DOI] [PubMed] [Google Scholar]
- 25. Dai R. M., Li C. C. (2001) Nat. Cell Biol. 3, 740–744 [DOI] [PubMed] [Google Scholar]
- 26. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 27. Besche H. C., Haas W., Gygi S. P., Goldberg A. L. (2009) Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiseman R. L., Chin K. T., Haynes C. M., Stanhill A., Xu C. F., Roguev A., Krogan N. J., Neubert T. A., Ron D. (2009) J. Biol. Chem. 284, 15233–15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C. M., Xu C. F., Neubert T. A., Mor A., Philips M. R., Ron D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma C. P., Slaughter C. A., DeMartino G. N. (1992) J. Biol. Chem. 267, 10515–10523 [PubMed] [Google Scholar]
- 34. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kondo H., Rabouille C., Newman R., Levine T. P., Pappin D., Freemont P., Warren G. (1997) Nature 388, 75–78 [DOI] [PubMed] [Google Scholar]
- 36. Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., Eskelinen E. L., Thumm M. (2010) J. Cell Biol. 190, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hendil K. B. (1988) Biochem. Int. 17, 471–477 [PubMed] [Google Scholar]
- 38. Peters J. M., Walsh M. J., Franke W. W. (1990) EMBO J. 9, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kress W., Mutschler H., Weber-Ban E. (2007) Biochemistry 46, 6183–6193 [DOI] [PubMed] [Google Scholar]
- 40. Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 42. Martin A., Baker T. A., Sauer R. T. (2008) Nat. Struct. Mol. Biol. 15, 139–145 [DOI] [PubMed] [Google Scholar]
- 43. Cascio P., Call M., Petre B. M., Walz T., Goldberg A. L. (2002) EMBO J. 21, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thoms S. (2002) FEBS Lett. 520, 107–110 [DOI] [PubMed] [Google Scholar]
- 45. Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E. U., Dougan D. A., Tsai F. T., Mogk A., Bukau B. (2004) Cell 119, 653–665 [DOI] [PubMed] [Google Scholar]
- 46. Bech-Otschir D., Helfrich A., Enenkel C., Consiglieri G., Seeger M., Holzhütter H. G., Dahlmann B., Kloetzel P. M. (2009) Nat. Struct. Mol. Biol. 16, 219–225 [DOI] [PubMed] [Google Scholar]
- 47. Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 48. Kusmierczyk A. R., Kunjappu M. J., Kim R. Y., Hochstrasser M. (2011) Nat. Struct. Mol. Biol. 18, 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.