Abstract
Heparanase acts as a master regulator of the aggressive tumor phenotype in part by enhancing expression of proteins known to drive tumor progression (e.g. VEGF, MMP-9, hepatocyte growth factor (HGF), and RANKL). However, the mechanism whereby this enzyme regulates gene expression remains unknown. We previously reported that elevation of heparanase levels in myeloma cells causes a dramatic reduction in the amount of syndecan-1 in the nucleus. Because syndecan-1 has heparan sulfate chains and because exogenous heparan sulfate has been shown to inhibit the activity of histone acetyltransferase (HAT) enzymes in vitro, we hypothesized that the reduction in nuclear syndecan-1 in cells expressing high levels of heparanase would result in increased HAT activity leading to stimulation of protein transcription. We found that myeloma cells or tumors expressing high levels of heparanase and low levels of nuclear syndecan-1 had significantly higher levels of HAT activity when compared with cells or tumors expressing low levels of heparanase. High levels of HAT activity in heparanase-high cells were blocked by SST0001, an inhibitor of heparanase. Restoration of high syndecan-1 levels in heparanase-high cells diminished nuclear HAT activity, establishing syndecan-1 as a potent inhibitor of HAT. Exposure of heparanase-high cells to anacardic acid, an inhibitor of HAT activity, significantly suppressed their expression of VEGF and MMP-9, two genes known to be up-regulated following elevation of heparanase. These results reveal a novel mechanistic pathway driven by heparanase expression, which leads to decreased nuclear syndecan-1, increased HAT activity, and up-regulation of transcription of multiple genes that drive an aggressive tumor phenotype.
Keywords: Cancer Tumor Promoter, Cell Surface Enzymes, Histone Acetylase, Histone Modification, Membrane Enzymes, Proteoglycan Structure, Proteoglycan Synthesis, Heparanase, Syndecan-1
Introduction
Heparanase, an endoglycosidase that cleaves heparan sulfate, is up-regulated in many cancers where it promotes tumor growth, angiogenesis, and metastasis (1, 2). High levels of heparanase in cancer patients are associated with shorter postoperative survival time compared with patients with low levels of heparanase (1). Although some of the tumor promoting effects of heparanase can be attributed to its ability to remodel the extracellular matrix barrier by cleaving heparan sulfate, heparanase is also known to regulate cell signaling and gene transcription (1, 3–5). Elevation of heparanase levels in myeloma cells, either by transfection of cells or by addition of recombinant active heparanase enzyme to cells, up-regulates expression of MMP-9, VEGF, HGF,2 and RANKL, which together drive an aggressive tumor phenotype (6–9). Although the mechanism whereby heparanase drives gene expression remains unknown, the enzyme is present and active in the nucleus where it could act locally to regulate gene expression (10).
Acetylation of the N-terminal tails of histones by histone acetyltransferase enzymes has been known for many years to be a process correlating with transcriptional activation (11–14). This process is balanced by the activity of histone deacetylases (HDACs), which selectively remove acetyl groups from histone protein. A shift in the balance between HAT and HDAC activity can drive cells to undergo apoptosis, proliferation, and/or malignancy (14). Evidence is mounting that heparan sulfates or other glycosaminoglycans can negatively regulate HAT activity causing inhibition of gene expression. Exogenous heparan sulfate can block the activity of HATs in cell-free assays, and addition of heparin to pulmonary fibroblasts was shown to reduce histone H3 acetylation by 50% (15). It was recently demonstrated that anti-proliferative glycosaminoglycans are taken up selectively by tumor cells and cause a decrease in histone H3 acetylation (16). Although the mechanism of HAT inhibition by heparan sulfate is unknown, the inhibitory activity is dependent upon heparan sulfate chain length and sulfation pattern, indicating that there is some degree of specificity rather than just random inhibition (15, 16). It has been speculated that heparan sulfate blocks HAT activity either by binding directly to HAT or by binding to histone proteins in a way that blocks their acetylation by HAT.
There are numerous reports that heparan sulfate proteoglycans localize within the nucleus (17–20). We recently discovered that the syndecan-1 heparan sulfate-bearing proteoglycan is present in the nucleus of myeloma tumor cells and that the amount of nuclear syndecan-1 is dramatically reduced upon elevation of heparanase expression (21). In the present study, we demonstrate that the elevation of heparanase expression in myeloma cells coupled with the loss of syndecan-1 from the nucleus results in an increase in HAT activity leading to enhanced transcription of genes that contribute to the aggressive behavior of this cancer. High levels of nuclear HAT activity were inhibited in a dose-dependent fashion by syndecan-1 or by heparin, consistent with the observation that reduced nuclear syndecan-1 levels in heparanase-high cells leads to enhanced gene transcription. In addition, we demonstrate that heparan sulfate binds to the HAT enzyme p300 and thus may act to directly inhibit enzyme activity. These findings reveal a new function for heparanase in regulating the activity of HAT enzymes and provide further mechanistic insight into how heparanase acts as a multifunctional regulator of the aggressive tumor phenotype.
EXPERIMENTAL PROCEDURES
Cell Lines
U266, MM.1S, and CAG human myeloma cells were cultured in RPMI 1640 growth medium supplemented with 10% fetal bovine serum. CAG cells were transfected as described previously with empty vector or vector containing the cDNA for human heparanase to prepare heparanase low (HPSE-low) and heparanase high (HPSE-high) cells, respectively (6).
HAT Activity Assay
HAT activity was measured using a commercially available, nonradioactive HAT assay kit (Millipore, Temecula, CA). The kit utilizes biotin-histone H3 or H4 substrate peptides (peptides representing the HAT-modifying tails, amino acid residues 1–21), linked to streptavidin-coated plates. Following exposure of wells to nuclear extracts, the extent of reaction is measured using an anti-acetyl lysine antibody. Samples were assayed for 60 min in a final reaction volume of 50 μl/well according to manufactures recommended protocol. In some experiments the HPSE-high cells were treated with a heparanase inhibitor, SST0001 (125 μg/ml), and HAT activity was assessed.
For preparing nuclear extracts, 7 × 106 cells were resuspended with ice-cold TEMP buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 4 mm MgCl2, 0.5 mm PMSF) and left on ice for 10 min. Cells were then dounce-homogenized (6–8 strokes) and pelleted by centrifugation at 1500 × g for 10 min. The nuclear pellets were resuspended in a small volume of TEP buffer (same as TEMP but lacking MgCl2), and an equal volume of 1 m NaCl was added. After incubation on ice for 60 min, pellets were spun down at 15,000 × g for 15 min, and the supernatant (nuclear extract) was collected. Protein concentration of the nuclear extract was measured using a BCA protein assay kit (Pierce), and equal protein was used for HAT activity assays. To determine the role of syndecan-1 in regulating HAT activity, purified syndecan-1 (prepared as described (7)) was added to the nuclear extracts from heparanase-high cells, and HAT activity was measured. For some experiments, the nuclear extracts were desalted using Centricon YM-30 filters (Millipore, Bedford, MA), and the heparan sulfate chains were removed from the extracts by addition of 5 milliunits/ml heparinase III (Seikagaku) at 37 °C for 2 h. Control samples were treated with the same amount of heat-inactivated heparinase III.
HDAC Activity Assay
Nuclear lysates were assayed for HDAC activity using the Fluor-de-Lys HDAC fluorimetric activity assay kit (Enzo Life Sciences, Plymouth Meeting, PA) using a Hitachi F7000 fluorimeter with microplate accessory. Briefly, 25 μl of sample was incubated with 50 μl Fluor-de-Lys developer solution at 25 °C for 1 min. The assay was initiated by addition of 25 μl Fluor-de-Lys substrate (0.5 mm). A time scan was performed for 5 min at 25 °C with the following parameters: λex = 360 nm, λem = 460 nm, slit widths = 5.0 nm, and a PMT voltage of 700 V. The relative HDAC activity was determined from the slope. Significance was determined using the Mann-Whitney Rank Sum Test.
BIAcore Assay
Real-time analysis of the interactions of the full-length form of human p300 (Active Motif, Carlsbad, CA) with biotinylated heparan sulfate from porcine intestinal mucosa (Sigma), heparan sulfate from bovine kidney (Seikagaku Co, Tokyo, Japan), and heparin from porcine intestine (Nacalai tesque, Kyoto, Japan) were performed with a BIAcore 2000 biosensor (BIAcore AB, Uppsala, Sweden). The biotinylated heparan sulfate and heparin were prepared as described previously (22). A streptavidin-coated sensor chip was used to immobilize the biotinylated heparan sulfate and heparin. Comparable amounts of samples were used for immobilization. The injection of biotinylated heparan sulfate and heparin onto the surface of the sensor was controlled to obtain a response of 670–720 resonance units, corresponding to 0.8–0.9 ng of the immobilized polysaccharides. The conditions for interaction analysis are as follows; flow rate, 30 μl/min; association and dissociation times, each 2 min; running buffers, 10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% Tween 20 (22).
Western Blot
Antibodies against acetyl histone H3 and total histone H3 were purchased from Millipore (Temecula, CA). For some experiments, MM.1S and U266 cells were treated with recombinant human heparanase (200 ng/ml; provided by Dr Israel Vlodavsky) and incubated at 37 °C for 12 h. Western blot analysis was performed as reported previously (6).
Real-Time PCR
Approximately 1 × 106 cells were treated with either the HAT inhibitor anacardic acid (30 μm) (EMD4 Biosciences, Darmstadt, Germany) for 3 h or with the HDAC inhibitor trichostatin A (1 μm) (Enzo Life Sciences, Plymouth Meeting, PA) for 5 h as indicated. RNA was extracted (RNeasy Mini Kit, Qiagen), and cDNA was synthesized (Clontech, Mountain View, CA). Real-time PCR was conducted using the following primers: MMP-9 (F), TGACAGCGACAAGAAGTG; MMP-9 (R), CAGTGAAGCGGTACATAGG; VEGF (F), CTTGCCTTGCTGCTCTAC; VEGF (R), TGGCTTGAAGATGTACTCG); and SYBR Green Supermix (Bio-Rad). Expression was determined relative to 28 S rRNA.
Immunohistochemistry and Fluorescent Microscopy
Sections from tumors formed from HPSE-low or HPSE-high cells were stained for acetylated histones using anti-acetyl histone H3 antibody (Millipore, Temecula, CA), as described previously (6). Fluorescent staining for acetylated histone H3 in cells was performed as described (21).
Statistical Analyses
All results are representative of at least three independent experiments. Except where noted, comparisons between two groups were analyzed by Student's t test, and a p value < 0.05 was considered statistically significant. Data are mean ± S.D.
RESULTS
Heparanase Enhances Activity of Histone Acetyltransferase
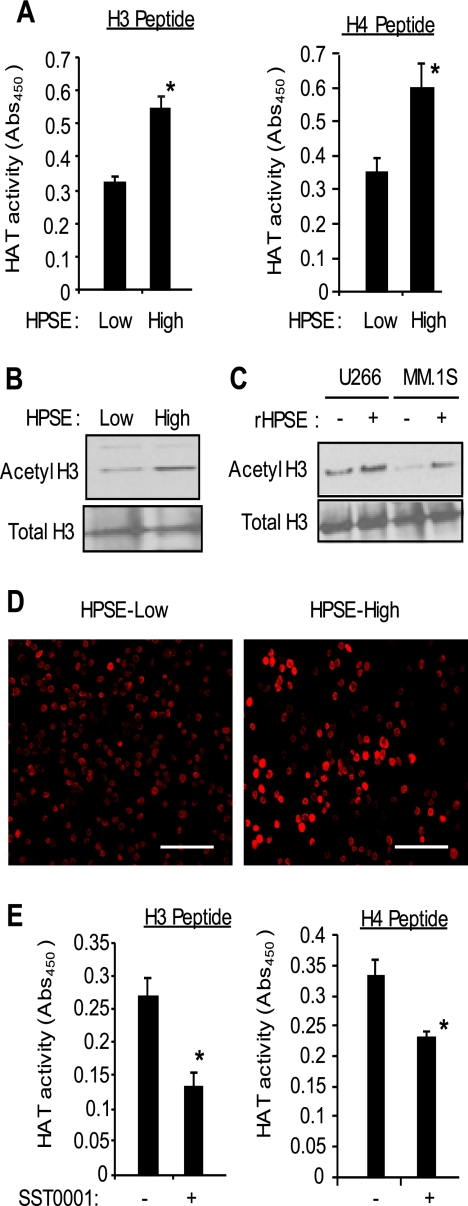
Using histone H3 and histone H4 peptides as acetylation substrates, HAT enzyme activity was assessed in CAG myeloma cells engineered to express low or high levels of heparanase. Results demonstrate that HPSE-high cells had significantly elevated levels of HAT activity in their nucleus compared with HPSE-low cells (Fig. 1A). Western blotting and immunocytochemistry confirmed that there was more acetylated histone H3 present in the HPSE-high cells than in the HPSE-low cells (Fig. 1, B and D). Western blots probed for HAT enzymes CBP, P300/CBP-associated factor, and p300 revealed that each enzyme was present in equivalent amounts in the HSPE-high and HPSE-low cells (supplemental Fig. S1A), indicating that the increased level of acetylated histone in the HPSE-high cells is due to enhanced HAT enzyme activity, not to an increase in the amount of HAT present. Also, it is important to note that the level of heparanase expression and activity in the HPSE-high CAG cells is similar to that found in some myeloma patient tumors (23, 24). Thus, the increase in HAT activity by these cells is not due to an enhancement of heparanase expression beyond levels that are likely to be found in the human cancer microenvironment.
FIGURE 1.
Heparanase up-regulates HAT activity and acetylation of histones. A, nuclear extracts isolated from CAG human myeloma cells expressing either low or high levels of HPSE were assayed for their level of HAT activity by ELISA using histone H3 and histone H4 peptide substrates (*, p < 0.05). Data are expressed as absorbance ± S.D. B, cell lysates from CAG HPSE-low or HPSE-high cells were assessed for their level of acetylated and total histone H3 by Western blotting. C, U266 and MM.1S human myeloma cells were incubated without or with recombinant heparanase (rHPSE; 200 ng/ml for 12 h), and cell lysates were assessed for their level of acetylated and total histone H3. D, immunofluorescence images of CAG HPSE-low and HPSE-high cells stained for acetylated histone H3. Bar, 100 microns. E, SST0001, a potent inhibitor of heparanase activity, inhibits HAT activity in myeloma cells. HPSE-high cells were treated with the heparanase inhibitor SST0001 (125 μg/ml) for 4 h, and HAT activity was assessed by ELISA using histone H3 and histone H4 peptides as substrates. Data are expressed as absorbance ± S.D. *, p < 0.05 versus untreated cells.
To further examine the effect of heparanase on HAT activity, we utilized two other myeloma cell lines, U266 and MM1.S, and exposed these cells to recombinant human heparanase. Previous work has shown that exogenous heparanase is taken up by cells and can induce cellular behaviors similar to those seen in cells transfected with a cDNA for heparanase (3, 6, 7, 25). Results reveal that 12 h after addition of recombinant human heparanase, levels of acetylated histones were elevated and significantly higher than in cells not receiving exogenous heparanase (Fig. 1C). This indicates that exogenous heparanase can enhance a relatively rapid up-regulation of HAT activity and that the effect on HAT activity seen in HPSE-high CAG myeloma cells is not simply an artifact related to their transfection.
We have demonstrated previously that tumors formed by HPSE-high cells grow aggressively in animals and display high levels of MMP-9, VEGF, and HGF compared with tumors formed by HPSE-low cells (6–8). Treatment of animals with SST0001, an inhibitor of heparanase, blocks the aggressive tumor growth of HPSE-high cells and dramatically lowers tumor expression of MMP-9, VEGF, and HGF, indicating that inhibition of heparanase in vivo down-regulates expression of these genes (26). Consistent with this in vivo finding, when SST0001 was added to HPSE-high cells in culture for 12 h, HAT activity present in the nucleus was diminished as measured by decreased acetylation of histones H3 and H4 (Fig. 1E). We then examined whether SST0001 was having a direct effect on HAT activity. This is an important question because SST0001 is composed of a chemically modified heparin (26), and studies have shown that when heparin is added to cells, it can be transported to the nucleus (17, 27). Because heparin can inhibit HAT activity (15), it is possible that when SST0001 is added to cells it is transported to the nucleus where it directly inhibits HAT activity by a mechanism unrelated to its ability to inhibit heparanase. To test this, SST0001 or heparin was added directly to nuclear extracts of HPSE-high CAG cells. Results demonstrated that SST0001 did not inhibit HAT activity in the nuclear extracts while heparin at the same concentration as SST0001 clearly inhibited HAT activity (supplemental Fig. S1B). Thus, SST0001 appears to inhibit HAT activity by inhibiting heparanase.
Acetylation of histones is tightly regulated during the cell cycle and is up-regulated during the S phase in mammalian cells (28). Thus, the high level of HAT activity in HPSE-high cells could be due to a higher number of cells in S phase than are present in the HPSE-low cells. However, this is not the case with the cells being studied here because cell cycle analysis revealed no significant difference in the number of cells in S phase in HPSE-high and HPSE-low cells (supplemental Fig. S1C).
Heparanase Enhances Acetylation of Histone in Myeloma Tumors Growing in Vivo
Because heparanase promotes tumor progression (24) and because regulation of histone modifications and HAT overexpression play a central role in cancer progression (14, 29), we investigated whether the heparanase mediated up-regulation of HAT activity that we see in vitro is also present within tumors growing in vivo. Immunohistochemistry of tumor xenografts revealed that tumors formed by HPSE-high cells have high levels of acetylated histone H3 compared with cells within tumors formed by HPSE-low cells (Fig. 2). This dramatic increase in the level of histone acetylation in tumors formed by HPSE-high cells correlates with increased tumor cell expression of MMP-9, VEGF, HGF, and RANKL, four proteins that promote the aggressive phenotype of myeloma tumors (9, 26). Importantly, the correlation between high heparanase expression and elevated levels of HGF and RANKL has also been demonstrated in bone marrow biopsies of myeloma patients (8, 9), indicating that our data from laboratory models of myeloma parallel clinical observations.
FIGURE 2.
Heparanase enhances acetylation of histones in myeloma tumors growing in vivo. Subcutaneous tumors in severe combined immunodeficient mice formed by HPSE-low or HPSE-high cells were removed and immunostained for acetylated histone H3 and counterstained with hematoxylin. HPSE-low cells show patchy nuclear staining for acetylated histone H3 in some cells, whereas HPSE-high cells show intense staining for acetylated histone H3 both within the nucleus and nucleolus of the majority of cells. Original magnification, 1300×.
Restoration of High Syndecan-1 Level in Nucleus Decreases HAT Activity in Heparanase-expressing Cells
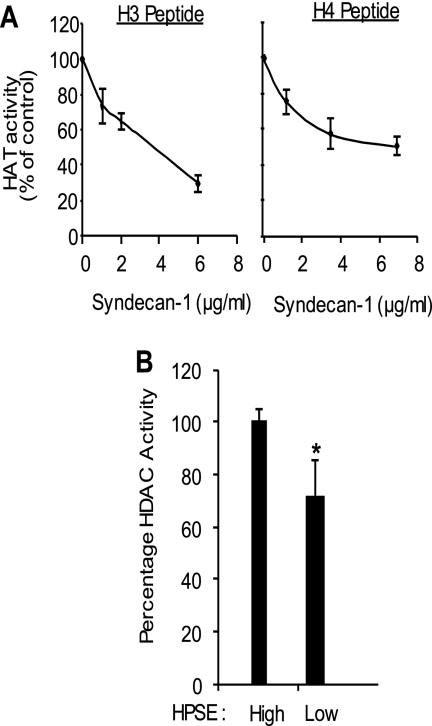
We reported previously that the level of nuclear syndecan-1 (a heparan sulfate-bearing proteoglycan) correlates inversely with the level of heparanase expression and activity (21). When heparanase expression is enhanced in CAG myeloma cells, the level of nuclear syndecan-1 drops by 75% compared with its basal level present in cells expressing low levels of heparanase (21). Because heparan sulfates are potent inhibitors of HAT activity (15), the drop in nuclear heparan sulfate that occurs when heparanase levels are elevated in these cells could be a mechanism for regulating HAT activity. To directly test the influence of syndecan-1 on HAT activity, exogenous syndecan-1 was added to nuclear extracts from CAG cells expressing high levels of heparanase. This decreased HAT activity in a dose-dependent manner (Fig. 3A), demonstrating that the level of syndecan-1 in the nucleus can regulate HAT activity. It also suggests that the enhanced HAT activity in the HPSE-high cells is not due to a decrease in the activity of HDACs, enzymes that remove the acetyl group and balances the activity of HAT. This was confirmed by assaying activities of HDAC present in the nucleus of HPSE-high and HPSE-low cells. Results demonstrated that HPSE-high cells actually have higher levels of HDAC activity than do HPSE-low cells (Fig. 3B).
FIGURE 3.
Exogenous syndecan-1 suppresses HAT activity in HPSE-high cells. A, nuclear extracts from HPSE-high cells, which have low levels of syndecan-1 in the nucleus (21), were subjected to the HAT activity assay utilizing H3 or H4 peptides as substrate in the presence of increasing concentrations of exogenous syndecan-1. Data are expressed as % control from three different experiments. B, levels of HDAC activity present in nuclear extracts from HPSE-high and HPSE-low cells. *, p < 0.001.
Heparan Sulfate Inhibits Activity of HAT in Myeloma Nuclei
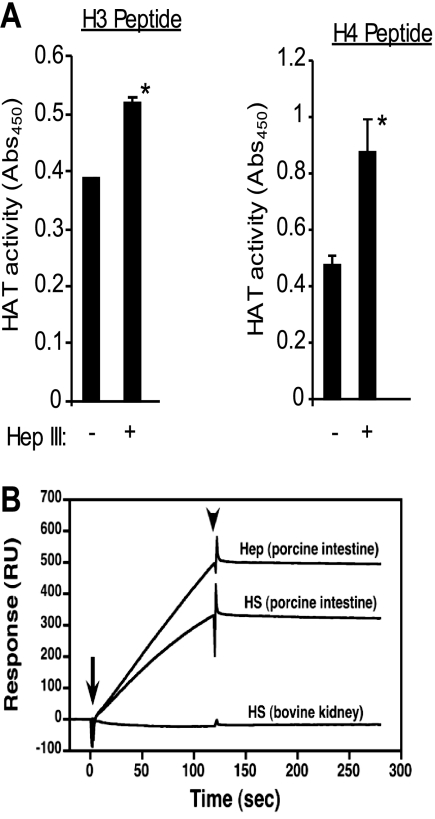
To explore the relationship between heparan sulfate and HAT in myeloma cells, nuclear extracts from wild-type myeloma cells (which have high levels of syndecan-1 in their nucleus) were desalted, and the heparan sulfate chains were degraded using bacterial heparinase III. This resulted in a significant increase in HAT activity as measured by an increase in acetylation of H3 and H4 (Fig. 4A) and points to the ability of heparan sulfate to inhibit HAT activity in these myeloma cells. Using surface plasmon resonance, we also found that the HAT protein p300 can bind directly to heparan sulfate and heparin from porcine intestine (Fig. 4B). Interestingly, p300 did not bind to heparan sulfate from bovine kidney, thus indicating specificity in the interaction between p300 and heparan sulfate. Disaccharide analysis of these heparan sulfates revealed a high level of N-sulfation in heparan sulfate from porcine intestine compared with heparan sulfate from bovine kidney (32% versus 20%, respectively) (supplemental Table 1). This is in accordance with previous work showing that the N-sulfation of heparan sulfate is the strongest requirement for HAT binding (15).
FIGURE 4.
Heparan sulfates in the nucleus of myeloma cells inhibit HAT enzyme activity. A, nuclear extracts were prepared from wild-type CAG cells and purified as described under “Experimental Procedures.” Heparan sulfate chains present within the extract were extensively degraded by treatment with 5 milliunits/ml bacterial heparinase III (Hep III) at 37 °C for 2 h. HAT activity was measured by ELISA. Data are expressed as absorbance ± S.D. *, p < 0.05. B, the HAT enzyme p300 binds to heparan sulfate and heparin. The full-length form of p300 (0.2 μg) was injected over sensor chips containing immobilized heparin from porcine intestine, heparan sulfate from porcine intestine, or heparan sulfate from bovine kidney. The arrow indicates the beginning of the association phase initiated by the injection of the p300, and the arrowhead indicates the beginning of the dissociation phase initiated by the running buffer. Abs, absorbance; RU, response units.
Studies have also shown that heparan sulfate chains can inhibit the biological activity of topoisomerase I, a nuclear enzyme involved in regulating the topological state and transcriptional activity of DNA (30). To determine whether myeloma cells with high levels of heparanase and low levels of nuclear syndecan-1 have high topoisomerase I activity, we compared the activity of topoisomerase I between cells having high and low levels of heparanase. Results demonstrated that in these myeloma cells, the level of heparanase expression did not affect topoisomerase I activity (supplemental Fig. S1D). Interestingly, a recent study showed that topoisomerase I activity was augmented in a breast cancer cell line following epidermal growth factor induced elevation of heparanase in the nucleolus (31). Although the mechanism of heparanase-induced augmentation of topoisomerase I was not examined in this study, it will be interesting to determine if it is related to reduced levels of heparan sulfate present within the nucleolus of these cells.
Increased HAT Activity Up-regulates MMP-9 and VEGF Genes in Heparanase-expressing Cells
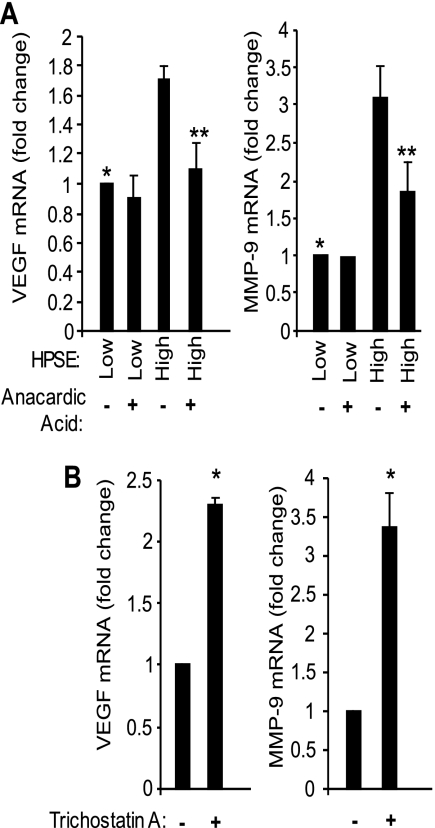
We have demonstrated that elevation of heparanase expression causes both enhanced HAT activity (Fig. 1) and increased transcription of VEGF and MMP-9, two effectors that promote the aggressive behavior of myeloma cells (6, 7). It has been shown that accumulation of acetylated histones leads to transcriptional alteration of specific genes contributing to tumorigenesis (14, 32, 33). Thus, we determined whether the enhanced HAT activity seen in HPSE-high cells contributes to up-regulation of expression of VEGF and MMP-9. CAG HPSE-high and HPSE-low cells were grown in the presence or absence of anacardic acid, a potent inhibitor of HAT (34). In the absence of anacardic acid, the mRNA level of expression of VEGF and MMP-9 were significantly higher in HPSE-high cells compared with HPSE-low cells (Fig. 5A), consistent with our previous findings (6, 7). Treatment of cells with anacardic acid significantly decreased the mRNA levels of VEGF and MMP-9 in HPSE-high cells, indicating that their high level of expression is related to high HAT activity. Treatment of HPSE-low cells with anacardic acid had no significant effect on mRNA levels of MMP-9 and VEGF.
FIGURE 5.
Inhibition of HAT activity suppresses MMP-9 and VEGF expression in heparanase-high cells. A, CAG HPSE-high or HPSE-low cells were treated with 30 μm of HAT inhibitor anacardic acid for 3 h, and the expression of MMP-9 and VEGF was assessed by real time PCR and normalized to 28 S rRNA levels. Data are from three separate experiments ± S.D. *, p ≤ 0.05 versus HPSE-high without anacardic acid; **, p ≤ 0.01 versus HPSE-high without anacardic acid. B, suppression of HDAC activity enhances the expression of VEGF and MMP-9. Wild-type CAG cells were treated with 1 μm of HDAC inhibitor, trichostatin A, for 5 h, and the expression of MMP-9 and VEGF was assessed by real-time PCR and normalized to 28 S rRNA levels. Data are from three separate experiments ± S.D. *, p < 0.01 versus trichostatin-untreated cells.
To further confirm the role of HAT in regulating the expression of MMP-9 and VEGF, trichostatin A was introduced to wild-type CAG cells in culture. Trichostatin A is a potent inhibitor of HDAC, and when added to cells, causes hyper-acetylation of histones (35). Exposure of CAG cells to 1 μm concentration of trichostatin for 5 h significantly enhanced the expression of VEGF and MMP-9 in wild-type CAG cells (Fig. 5B), confirming that acetylation of histone proteins is important for the enhanced expression of these genes in CAG myeloma cells.
DISCUSSION
The ability of heparanase to act as a potent promoter of an aggressive tumor phenotype is due at least in part to its influence on gene expression. In the present work, we demonstrate for the first time that heparanase controls gene expression by up-regulating the activity of HAT, a nuclear enzyme that regulates gene transcription (14). Transfection of heparanase into myeloma cells or addition of exogenous heparanase led to enhanced nuclear HAT activity resulting in increased acetylation of histones in myeloma cells and in tumors growing in vivo. Exposure of cells to the heparanase inhibitor SST0001 significantly inhibited HAT activity. Importantly, expression of two genes up-regulated by heparanase (VEGF and MMP-9), was inhibited by anacardic acid, a potent inhibitor of HAT activity. Mechanistically, we found that the ability of heparanase to increase HAT activity was linked to the decrease in levels of nuclear syndecan-1 that occurs when heparanase expression is elevated, consistent with a previous report that heparin and heparan sulfate inhibit HAT activity (15). Together, these results reveal a novel mechanistic pathway driven by heparanase expression, which leads to decreased nuclear syndecan-1, increased HAT activity, and up-regulation of transcription of multiple genes that drive an aggressive tumor phenotype.
The relationship between heparanase and syndecan-1 localization has emerged as an important regulatory element in myeloma and perhaps other cancers. We previously demonstrated that heparanase enhances syndecan-1 shedding in both myeloma and breast cancer cells and that elevated levels of syndecan-1 in the myeloma tumor microenvironment drives tumor angiogenesis, metastasis and growth (7, 36, 37). In addition to enhancing syndecan-1 shedding, elevation of heparanase diminishes localization of syndecan-1 in the nucleus (21). Interestingly, heparanase also localizes to the nucleus (10), but it is not known how heparanase is transported to the nucleus. One possibility is that heparanase and syndecan-1 are transported together as a complex through an interaction between syndecan-1 heparan sulfate chains and heparan sulfate binding domains known to be present within heparanase. Studies utilizing immunofluorescence microscopy indicate that in some tumor types, syndecan-1 transport to the nucleus is tubulin-dependent and that nuclear translocation requires the RMKKK sequence present within the syndecan-1 core protein (20, 38). If bound to syndecan-1, heparanase could be transported via this same route assuming that the tubulin-mediated transport route is active in myeloma cells.
Elevation of heparanase levels within the nucleus could lead to enhanced degradation of nuclear syndecan-1 heparan sulfate resulting in loss of the degraded syndecan-1 from the nucleus. Conversely, elevation of heparanase could, via unknown mechanisms, lead to poor transport of syndecan-1 into the nucleus resulting in a dramatic reduction in total levels of nuclear syndecan-1. Whatever the mechanism at play, our finding here that loss of syndecan-1 in the nucleus results in up-regulation of HAT activity and gene transcription clearly defines an important new function of heparanase in regulating cell behavior.
We have previously demonstrated that heparanase is present and active in the plasma taken from the bone marrow of myeloma patients (23), and it has also been shown that both myeloma cells and non-tumor cells within the myeloma bone marrow can express heparanase (39). Our finding now that exogenous heparanase can cause up-regulation of HAT activity by myeloma cells (Fig. 1) presents an important and novel mechanism whereby gene expression can be regulated by crosstalk between cells within a tumor. For example, heparanase released from myeloma cells could alter HAT activity and influence gene expression in adjacent tumor cells or in host cells not expressing heparanase. Alternatively, heparanase released by host cells could impact gene expression in tumor cells. This may be particularly important in cancers such as myeloma, which are highly dependent on the tumor microenvironment for their survival.
It will be important to determine how heparan sulfate is able to inhibit HAT activity within the nucleus of cells. We show here that heparan sulfate can bind directly to purified p300 (Fig. 4B), and it was previously shown that another HAT, P300/CBP-associated factor, bound to a heparin affinity column (15). Thus, heparan sulfates may inhibit HAT by binding directly to the enzyme. Alternatively, it is known that heparan sulfate can bind directly to histone proteins, and it has been speculated that heparan sulfate-histone interactions interfere with HAT-mediated acetylation of histone tails (40, 41). Importantly, there does appear to be specificity in heparan sulfate inhibition of HAT because inhibition is dependent on the pattern of sulfation and on the length of the heparan sulfate chain (15). This suggests that modifications to heparan sulfate structure within a cell could fine tune its ability to block HAT activity. Moreover, it may be possible to design heparan sulfate or mimics of heparan sulfate that could be used to block HAT activity therapeutically. In support of this notion, it was recently shown that tumor cells can be primed to express glycosaminoglycans that cause a decrease in acetylation of histone H3 and inhibit tumor cell proliferation (16).
We have recently shown that treatment of animals with the heparanase inhibitor SST0001 down-regulates tumor expression of HGF, MMP-9, and VEGF (26). Exposure of heparanase-high cells to SST0001 failed to directly inhibit HAT activity, likely because SST0001 lacks N-sulfation, a requirement for heparan sulfate inhibition of HAT (15, 42). This indicates that SST0001 inhibition of HAT occurs through its ability to block heparanase activity and provides further information as to the in vivo mechanism of action of this anti-heparanase compound.
Taken together, the findings presented here suggest a model whereby syndecan-1 heparan sulfate present within the nucleus blocks HAT activity, perhaps by binding directly to the enzyme. This results in hypoacetylated histones and suppression of gene transcription. As the level of heparanase rises during tumor progression, the level of syndecan-1 heparan sulfate proteoglycan in the tumor cell nucleus drops. This results in a significant increase in HAT activity that drives histone acetylation and active transcription of genes that promote the aggressive tumor phenotype. This new insight into the mechanism of heparanase-enhanced gene expression provides further understanding of how heparanase is able to wield such a powerful impact on driving tumor progression. Moreover, these results underscore the potential of heparanase inhibitors as anti-cancer drugs and the possibility that they could be used to reverse the aggressive phenotype of some tumors.
Supplementary Material
Acknowledgments
We thank Dr. Israel Vlodavsky (Technion, Haifa, Israel) for providing recombinant heparanase and Dr. Steven Rosen (Northwestern University) for MM.1S cells. We acknowledge Ed Phillips (University of Alabama at Birmingham, High Resolution Imaging Facility) for assistance with fluorescence microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants CA135075 and CA138340 (to R. D. S.). This work was also supported by the Future Drug Discovery and Medical Care Innovation Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (to K. S.). Dr. Claudio Pisano is an employee of sigma-tau Industrie Farmaceutiche Riunite S.p.A.; this company is developing SST0001 for clinical use.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Table 1, and Fig. 1.
- HGF
- hepatocyte growth factor
- HPSE
- heparanase
- HAT
- histone acetyltransferase
- RANKL
- receptor activator of nuclear factor kappa-B ligand.
REFERENCES
- 1. Ilan N., Elkin M., Vlodavsky I. (2006) Int. J. Biochem. Cell Biol. 38, 2018–2039 [DOI] [PubMed] [Google Scholar]
- 2. Barash U., Cohen-Kaplan V., Dowek I., Sanderson R. D., Ilan N., Vlodavsky I. (2010) FEBS J. 277, 3890–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gingis-Velitski S., Zetser A., Flugelman M. Y., Vlodavsky I., Ilan N. (2004) J. Biol. Chem. 279, 23536–23541 [DOI] [PubMed] [Google Scholar]
- 4. Zetser A., Bashenko Y., Edovitsky E., Levy-Adam F., Vlodavsky I., Ilan N. (2006) Cancer Res. 66, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 5. Fux L., Ilan N., Sanderson R. D., Vlodavsky I. (2009) Trends Biochem. Sci. 34, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Purushothaman A., Chen L., Yang Y., Sanderson R. D. (2008) J. Biol. Chem. 283, 32628–32636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purushothaman A., Uyama T., Kobayashi F., Yamada S., Sugahara K., Rapraeger A. C., Sanderson R. D. (2010) Blood 115, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramani V. C., Yang Y., Ren Y., Nan L., Sanderson R. D. (2011) J. Biol. Chem. 286, 6490–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang Y., Ren Y., Ramani V. C., Nan L., Suva L. J., Sanderson R. D. (2010) Cancer Res. 70, 8329–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schubert S. Y., Ilan N., Shushy M., Ben-Izhak O., Vlodavsky I., Goldshmidt O. (2004) Lab. Invest. 84, 535–544 [DOI] [PubMed] [Google Scholar]
- 11. Hebbes T. R., Thorne A. W., Crane-Robinson C. (1988) EMBO J. 7, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loidl P. (1994) Chromosoma 103, 441–449 [DOI] [PubMed] [Google Scholar]
- 13. Turner B. M., O'Neill L. P. (1995) Semin Cell Biol. 6, 229–236 [DOI] [PubMed] [Google Scholar]
- 14. Roth S. Y., Denu J. M., Allis C. D. (2001) Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 15. Buczek-Thomas J. A., Hsia E., Rich C. B., Foster J. A., Nugent M. A. (2008) J. Cell. Biochem. 105, 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nilsson U., Johnsson R., Fransson L. A., Ellervik U., Mani K. (2010) Cancer Res. 70, 3771–3779 [DOI] [PubMed] [Google Scholar]
- 17. Ishihara M., Fedarko N. S., Conrad H. E. (1986) J. Biol. Chem. 261, 13575–13580 [PubMed] [Google Scholar]
- 18. Richardson T. P., Trinkaus-Randall V., Nugent M. A. (2001) J. Cell Sci. 114, 1613–1623 [DOI] [PubMed] [Google Scholar]
- 19. Hsia E., Richardson T. P., Nugent M. A. (2003) J. Cell. Biochem. 88, 1214–1225 [DOI] [PubMed] [Google Scholar]
- 20. Brockstedt U., Dobra K., Nurminen M., Hjerpe A. (2002) Exp. Cell Res. 274, 235–245 [DOI] [PubMed] [Google Scholar]
- 21. Chen L., Sanderson R. D. (2009) PLoS ONE 4, e4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deepa S. S., Umehara Y., Higashiyama S., Itoh N., Sugahara K. (2002) J. Biol. Chem. 277, 43707–43716 [DOI] [PubMed] [Google Scholar]
- 23. Kelly T., Miao H. Q., Yang Y., Navarro E., Kussie P., Huang Y., MacLeod V., Casciano J., Joseph L., Zhan F., Zangari M., Barlogie B., Shaughnessy J., Sanderson R. D. (2003) Cancer Res. 63, 8749–8756 [PubMed] [Google Scholar]
- 24. Yang Y., Macleod V., Bendre M., Huang Y., Theus A. M., Miao H. Q., Kussie P., Yaccoby S., Epstein J., Suva L. J., Kelly T., Sanderson R. D. (2005) Blood 105, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 25. Vreys V., Delande N., Zhang Z., Coomans C., Roebroek A., Dürr J., David G. (2005) J. Biol. Chem. 280, 33141–33148 [DOI] [PubMed] [Google Scholar]
- 26. Ritchie J. P., Ramani V. C., Ren Y., Naggi A., Torri G., Casu B., Penco S., Pisano C., Carminati P., Tortoreto M., Zunino F., Vlodavsky I., Sanderson R. D., Yang Y. (2011) Clin. Cancer Res. 17, 1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fedarko N. S., Conrad H. E. (1986) J. Cell Biol. 102, 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vempati R. K., Jayani R. S., Notani D., Sengupta A., Galande S., Haldar D. (2010) J. Biol. Chem. 285, 28553–28564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mahlknecht U., Hoelzer D. (2000) Mol. Med. 6, 623–644 [PMC free article] [PubMed] [Google Scholar]
- 30. Kovalszky I., Dudás J., Oláh-Nagy J., Pogány G., Töváry J., Timár J., Kopper L., Jeney A., Iozzo R. V. (1998) Mol. Cell Biochem. 183, 11–23 [DOI] [PubMed] [Google Scholar]
- 31. Zhang L., Sullivan P., Suyama J., Marchetti D. (2010) Mol. Cancer Res. 8, 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Isharwal S., Miller M. C., Marlow C., Makarov D. V., Partin A. W., Veltri R. W. (2008) Prostate 68, 1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dekker F. J., Haisma H. J. (2009) Drug Discov. Today 14, 942–948 [DOI] [PubMed] [Google Scholar]
- 34. Balasubramanyam K., Swaminathan V., Ranganathan A., Kundu T. K. (2003) J. Biol. Chem. 278, 19134–19140 [DOI] [PubMed] [Google Scholar]
- 35. Tóth K. F., Knoch T. A., Wachsmuth M., Frank-Stöhr M., Stöhr M., Bacher C. P., Müller G., Rippe K. (2004) J. Cell Sci. 117, 4277–4287 [DOI] [PubMed] [Google Scholar]
- 36. Yang Y., Macleod V., Miao H. Q., Theus A., Zhan F., Shaughnessy J. D., Jr., Sawyer J., Li J. P., Zcharia E., Vlodavsky I., Sanderson R. D. (2007) J. Biol. Chem. 282, 13326–13333 [DOI] [PubMed] [Google Scholar]
- 37. Yang Y., Yaccoby S., Liu W., Langford J. K., Pumphrey C. Y., Theus A., Epstein J., Sanderson R. D. (2002) Blood 100, 610–617 [DOI] [PubMed] [Google Scholar]
- 38. Zong F., Fthenou E., Wolmer N., Hollósi P., Kovalszky I., Szilák L., Mogler C., Nilsonne G., Tzanakakis G., Dobra K. (2009) PloS One 4, e7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mahtouk K., Hose D., Raynaud P., Hundemer M., Jourdan M., Jourdan E., Pantesco V., Baudard M., De Vos J., Larroque M., Moehler T., Rossi J. F., Rème T., Goldschmidt H., Klein B. (2007) Blood 109, 4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watson K., Gooderham N. J., Davies D. S., Edwards R. J. (1999) J. Biol. Chem. 274, 21707–21713 [DOI] [PubMed] [Google Scholar]
- 41. Villeponteau B. (1992) Biochem. J. 288, 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Naggi A., Casu B., Perez M., Torri G., Cassinelli G., Penco S., Pisano C., Giannini G., Ishai-Michaeli R., Vlodavsky I. (2005) J. Biol. Chem. 280, 12103–12113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.