Abstract
We employed budding yeast and skeletal muscle actin to examine the contribution of the actin isoform to myosin motor function. While yeast and muscle actin are highly homologous, they exhibit different charge density at their N termini (a proposed myosin-binding interface). Muscle myosin-II actin-activated ATPase activity is significantly higher with muscle versus yeast actin. Whether this reflects inefficiency in the ability of yeast actin to activate myosin is not known. Here we optimized the isolation of two yeast myosins to assess actin function in a homogenous system. Yeast myosin-II (Myo1p) and myosin-V (Myo2p) accommodate the reduced N-terminal charge density of yeast actin, showing greater activity with yeast over muscle actin. Increasing the number of negative charges at the N terminus of yeast actin from two to four (as in muscle) had little effect on yeast myosin activity, while other substitutions of charged residues at the myosin interface of yeast actin reduced activity. Thus, yeast actin functions most effectively with its native myosins, which in part relies on associations mediated by its outer domain. Compared with yeast myosin-II and myosin-V, muscle myosin-II activity was very sensitive to salt. Collectively, our findings suggest differing degrees of reliance on electrostatic interactions during weak actomyosin binding in yeast versus muscle. Our study also highlights the importance of native actin isoforms when considering the function of myosins.
Keywords: Actin, Myosin, Protein-Protein Interactions, Skeletal Muscle, Yeast
Introduction
Actin is a highly abundant 43 kDa monomeric protein, highly conserved among eukaryotes, that reversibly polymerizes into filamentous actin (F-actin)2 in a spatially and temporally regulated fashion. One of the major functions of F-actin, in both muscle and non-muscle cells, is to interact with the motor protein myosin to generate contractile force to power processes such as muscle contraction, cell movement, intracellular transport, and cytokinesis.
The myosin motors constitute a structurally and functionally complex family of proteins (1). However, a comparison of these proteins reveals a great degree of conservation in the motor domain. This region of the protein contains an ATP binding site, interacts with F-actin, and couples conformational changes with ATP hydrolysis to generate force via F-actin displacement. Formation of the actomyosin complex involves an initial weak ionic interaction, which transitions into a more hydrophobic, strong-bound state that is coupled with force generation.
Actin is a clam-shaped protein with two domains that are joined by a hinge region (2). The smaller outer domain consists of subdomains 1 and 2, while the larger inner domain consists of subdomains 3 and 4 (Fig. 1A). Between the two domains exists a deep cleft in which sit an adenine nucleotide and a divalent cation to help hold the nucleotide in place. This nucleotide and cation are important for stability of the actin monomer, and the phosphorylation state of the nucleotide is an important determinant of filament stability.
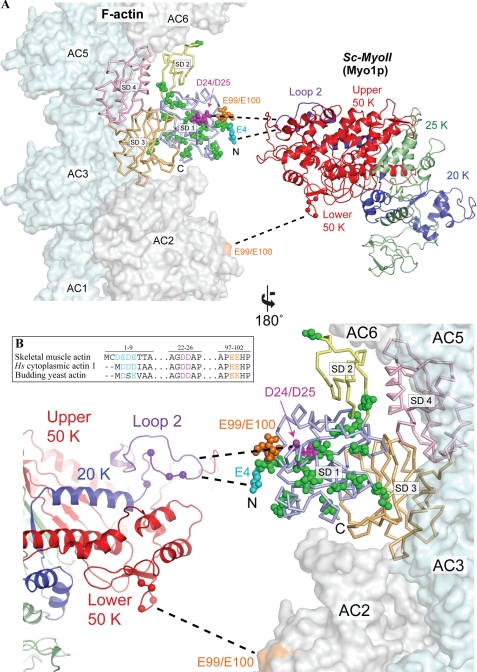
FIGURE 1.
Actin mutations at the predicted actomyosin interface. A, structural model depicting the predicted binding interface between yeast actin and myosin-II. Yeast actin (Act1p, 1YAG) (41) was superimposed onto one of the actin momomers (AC4) in the actomyosin model of Holmes et al. (42). Actin subdomains 1–4 (SD1-SD4) are colored separately. A three-dimensional structure of the yeast myosin-II Myo1p (Sc-MyoII) motor was predicted using the I-TASSER protein structure and function prediction server (43), and colored to indicate the 50K domain (red), 25K domain (green) and 20K domain (blue). The Sc-MyoII motor was superimposed onto the skeletal muscle myosin structure (2MYS) (44) in the orientation predicted for the actomyosin complex, but separated horizontally to expose the binding interface. The cationic loop 2 of Sc-MyoII is indicated in purple. Mutations concentrated in subdomain 1 of the outer domain are shown in space-filling representation for the actin monomer set within the actin filament. Residues relating to the 4Ac (cyan), sub12 (green), D24A-D25A (pink), and E99A-E100A (orange) mutants are shown. For 4Ac actin, of the four glutamates located at the N terminus in this mutant form, only the E4 residue is shown. The sixteen mutations in sub12 are as follows: V5T, A6T, I10C, M16L, C17V, I43V, R68K, V76I, V103T, M110L, S114A, F132M, S135A, T350S, S358T, and H372R. Dashed lines indicate proposed myosin binding sites (which span two adjacent actin monomers): loop 2 to 4Ac, D24-D25 (primary binding site); lower 50K loop to E99-E100 (secondary binding site). The position of positively charged residues within these two Sc-MyoII loops are indicated (balls). B, amino acid sequence homology centered on the negatively charged N termini and D24-D25/E99-E100 amino acid pairs of human skeletal muscle actin, human cytoplasmic actin-1, and budding yeast actin. Alignments were generated using the ClustalX software (45).
A number of studies have led to the conclusion that a major site of interaction of the myosin head with actin involves a substantial portion of actin subdomain 1 (3). One factor believed to be especially important in formation of the initial weak binding state is an ionic interaction between the acidic N-terminal finger of actin with a cationic loop (loop 2) separating the 50 and 20 kDa domains of the myosin head (Fig. 1, A and B). A number of studies support such a model. For example, interaction of these two elements were directly demonstrated by chemical crosslinking (4–6). Blocking the N terminus with an antibody directed against the N-terminal seven residues adversely affects this interaction (7, 8), and substitution of the acidic N-terminal residues in Dictyostelium actin with histidines has the same effect (9).
Other negatively charged surface residues from subdomain 1 of actin are also important for the initial weak-bound actomyosin state. Substitution of either of two conserved charged pairs of residues (D24/D25 and E99/E100 in budding yeast actin, Fig. 1, A and B) with alanines or histidines leads to a loss in skeletal muscle myosin-II activity (10–12). Such substitutions lead to a significant reduction in actomyosin affinity in the presence of ATP, without having any effect on strong actin binding (i.e. in the absence of ATP) (11).
Budding yeast actin will only activate the ATPase activity of skeletal muscle myosin subfragment 1 (S1) about one-tenth as well as skeletal muscle actin (13), despite the fact that their sequences are 87% identical. A difference in the number of N-terminal negative residues between these two actins seems to play a significant role in this activation. Yeast actin has only two N-terminal acidic residues while striated muscle actins have four, and higher eukaryotic non-muscle actins have three (Fig. 1B). Substitution of the yeast N terminus with four acidic residues generated a mutant actin (4Ac) that reduced this 10-fold difference in ATPase activation to 3-fold (13). It was further demonstrated that three negative charges seemed to be as good as four in stimulating this activity (13).
Another question concerning the actomyosin interaction is to what extent, if any, the inner domain of actin can influence the conformation of subdomain 1 (either through the hinge region or the nucleotide bridge), in affecting the interaction of the two proteins. To address this question, we previously generated a hybrid actin (sub12) in which we used site-directed mutagenesis to cumulatively introduce muscle actin-specific residues into the corresponding positions of yeast actin in subdomains 1 and 2 (14) (Fig. 1A). Our results with this actin demonstrated that despite the fact that the outer domain should have constituted a normal binding surface for muscle actin, the hybrid still acted like yeast actin in its ability to activate muscle myosin ATPase activity. This hybrid actin produced an adverse phenotype in yeast expressing it as the only actin in the cell. Furthermore, it has been shown that striated actin is not compatible with yeast viability (15), and that a mammalian non-muscle actin, although it will rescue a lethal actin-null phenotype in yeast, produces at best a very sick cell (15). These results raise the question of to what extent yeast myosins may have evolved to specifically utilize yeast actin and, in particular, how they compensate for the reduced negative N-terminal charge density on yeast actin.
In this study, we have utilized two myosins isolated from budding yeast to address these questions. Myo1p (referred to as Sc-MyoII from here on in) is a conventional class II, double-headed myosin (16–18) that localizes to the division site (bud neck) where it participates in retrograde flow of actin cables (19) and contractile ring function during cytokinesis (20, 21). Myo2p (referred to as Sc-MyoV from here on in) is an unconventional class V, double-headed myosin (22, 23) that transports various cargoes along actin cables toward sites of polarized growth (24). We assessed the activation of Sc-MyoII, Sc-MyoV, and skeletal muscle myosin-II by yeast (wild-type and mutant forms) and skeletal muscle actin in terms of solution ATPase activity and the ability to move actin filaments in the in vitro motility assay. Our results demonstrate markedly different responses of the myosins to the actin variants utilized, implying a significant difference in the modes of actomyosin binding in muscle and yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Standard budding yeast growth conditions and genetic methods were employed (25). Sc-MyoII and Sc-MyoV overexpression strains (GAL1prom-MYO1 and GAL1prom-MYO2) utilized a DNA fragment encoding a nourseothricin (nat) resistance marker and the GAL1 galactose-inducible promoter. The cassette was amplified from pFA6a-natR-GAL1, purified, and integrated into the genome of a wild-type (or MYO2-GFP:HIS3/MYO2-GFP:HIS3 leu2/leu2 ura3/ura3 his3/his3) diploid immediately upstream of MYO1 or MYO2 by homologous recombination (using primers containing 40 bp of flanking DNA specific to the relevant locus) (26). The C-terminal GFP tag favored tethering of active Sc-MyoV motors in motility assays; no difference in activity was observed in ATPase assays for Sc-MyoV-GFP versus untagged Sc-MyoV. nat-resistant strains carrying the desired integration were confirmed by morphological phenotypes associated with Sc-MyoII and Sc-MyoV overexpression and diagnostic PCRs of genomic DNA.
Construction of the pGST-CMD1 calmodulin overexpression plasmid was previously described (27). This plasmid has a pRS316 (URA3) backbone, expressing GST-tagged calmodulin from a GAL1 promoter. CMD1 was replaced with MLC1 or MLC2 to generate pGST-MLC1 and pGST-MLC2. MLC1 primers: 5′ NotI GCGGCCGCATGTCAGCCACCAGAGCCAATAAAG; 3′ SalI GTCGACTCATTGTCTCAAAACATCTTCGATG. MLC2 primers: 5′ NotI GCGGCCGCATGGACCATAGTGAATCGCTAACG; 3′ SalI GTCGACTTAATCGGTGATTGAATCTAAGAAAAGC. The pFA6a-natR-GAL1 plasmid was constructed by replacing the nmt41 promoter of pFA6a-natR-nmt41prom (28) with the GAL1 promoter fragment.
Actin Purification
The yeast haploid strains expressing either 4Ac, sub12, D24A-D25A, or E99A-E100A mutant actins have been described previously (11, 13, 14). Wild-type and mutant yeast actins in the Ca2+ monomeric form were purified by a DNase I affinity and DEAE ion exchange chromatography as described previously (29). G actin concentrations were determined by UV absorbance at 290 nm using the extinction coefficient ϵ = 25.6 mm−1·cm−1, and actin was stored in G buffer (10 mm Tris-HCl pH 7.5, 0.2 mm ATP, 0.2 mm CaCl2, and 1 mm DTT) at 4 °C and used within 5 days. Chicken skeletal muscle actin was purified from acetone powder as previously described (30).
Sc-MyoII and Sc-MyoV Purifications
Four liters of the GAL1p-MYO1 strain (two liters harboring pGST-MLC1, two liters harboring pGST-MLC2) or four liters of the GAL1p-MYO2 strain (two liters harboring pGST-CMD1, two liters harboring pGST-MLC1) were grown in CSM-Ura− with 1% raffinose as the sole carbon source. After the OD595 reached 1.0, cells were induced by the addition of 2% galactose and allowed to grow for an additional 16 h. GAL1p-MYO1 and GAL1p-MYO2 cells were harvested separately via centrifugation and cell pellets were washed in water and resuspended 1:1 in lysis buffer (750 mm KCl, 25 mm Tris-HCl pH 7.4, 4 mm MgCl2, 4 mm ATP, 20 mm EGTA, 1 mm DTT, 2 mm PMSF, 2 mm benzamidine hydrochloride hydrate, and 0.1% Triton X-100) and lysed using a FastPrep-24 (MP Biomedicals). GST-tagged light chains (along with the co-purifying heavy chains) were recovered from lysates using glutathione-Sepharose resin and dialyzed into A15 buffer (500 mm KCl, 10 mm imidazole pH 7.2, 0.375 mm sodium azide, and 1 mm DTT). Light chains over-expressed to greater extent than heavy chains. The concentration of motors was estimated by densitometry of Coomassie-stained heavy chain bands after SDS-PAGE using rabbit skeletal muscle myosin as the standard. Typical yields were ∼150 μg (Sc-MyoII) and ∼50 μg (Sc-MyoV).
Actin-activated ATPase Assays
Actin-activated ATPase assays were carried out at room temperature in 2 mm Tris-HCl, pH 7.2, 10 mm imidazole, 50–175 mm KCl, 0.1 mm CaCl2, 3 mm MgCl2, 2 mm ATP, and 1 mm DTT with 20 nm Sc-MyoII, 3 nm Sc-MyoV, or 50 nm skeletal muscle myosin-II and 0–100 μm actin filaments. Malachite green was used to quantitate Pi release (31). Background ATP hydrolysis from actin (detected in controls lacking myosin) and basal myosin activity (detected in controls lacking actin) were subtracted when deriving actin-activated ATPase rates. Curves were fit to Michaelis-Menten kinetics using the GraphPad Prism software (v5.0b). Myosin heavy chain quantitation (described above) enabled VMAX values to be determined for Sc-MyoII and Sc-MyoV.
In Vitro Motility Assays
We used motility assays (32) with 40 μg/ml of myosin applied to the motility chambers. Fluorescent actin stocks were prepared at 5 μm from G-actin stocks, polymerized by the addition of 50 mm KCl and 1 mm MgCl2 in the presence of 5 μm rhodamine phalloidin (Invitrogen) for 30 min. Sc-MyoII, Sc-MyoV, or skeletal muscle myosin-II were adhered to the surface of a nitrocellulose-coated coverslip for 10 min and the chamber was then washed as follows: (a) three times with motility buffer (25 mm imidazole pH 7.4, 50–150 mm KCl, 1 mm EGTA, 4 mm MgCl2, 2 mm DTT) plus 0.5 mg/ml BSA; (b) three times with motility buffer; (c) twice with motility buffer containing vortexed (1 min) unlabeled actin filaments (1 μm); (d) three times with motility buffer plus 1 mm ATP; (e) twice with motility buffer plus 25 nm rhodamine phalloidin-labeled actin filaments and oxygen scavengers (50 μg/ml catalase, 130 μg/ml glucose oxidase, and 3 mg/ml glucose); (f) twice with motility buffer plus 20 mm DTT, 0.5% methyl-cellulose and oxygen scavengers; and (g) twice with motility buffer plus 20 mm DTT, 0.5% methyl-cellulose, 1.5 mm ATP, and oxygen scavengers. Filaments were observed at room temperature by epifluorescnece microscopy on a Nikon TE2000-E2 inverted microscope with a Plan Apo 60 (1.45 NA) objective (Melville, NY). Fluorescence utilized an EXFO X-CITE 120 illuminator (Nikon). NIS Elements software was used to control the microscope (Nikon), two Uniblitz shutters (Vincent Associates, Rochester, NY), and a Photometrics CoolSNAP HQ2 14-bit camera (Tucson, AZ) were used to record time-lapse images at 2 s intervals for 1 min. ImageJ software with the MTrackJ plug-in was used to calculate filament velocities from the time-lapse series.
RESULTS
Budding yeast actin has only two N-terminal acidic residues compared with the three or four found on higher eukaryotic actins which are believed to be important for the initial weak interaction between actin and myosin. The focus of this paper was to determine, using purified myosins, if myosins from budding yeast compensate for this reduced N-terminal acidic charge density on yeast actin. Previous studies had used small amounts of these myosins isolated in quantities sufficient for in vitro motility assays (19, 22, 33), but this is the first report of a procedure for preparing active Sc-MyoII (Myo1p) and Sc-MyoV (Myo2p) in amounts sufficient for ATPase analysis. We isolated the full-length proteins from yeast extracts (Fig. 2) by pulling them down with overexpressed GST-tagged myosin light chains using glutathione Sepharose resin. As we previously reported for budding yeast myosin-I (27), isolation required co-overexpression of both heavy chain and light chains since isolation of over-expressed light chains (calmodulin, Mlc1p, or Mlc2p) alone failed to co-purify any detectable myosin activity in actin-activated ATPase or motility assays. We also generally observed that overexpression of Sc-MyoII was much better than Sc-MyoV for reasons we do not understand. Both Sc-MyoII and Sc-MyoV samples exhibited robust motor activity as measured by actin-activated ATPase and in vitro motility assays (Table 1). One-step purified myosins were employed since any attempt to further purify these low-yield samples typically resulted in loss of heavy chain and motor activity.
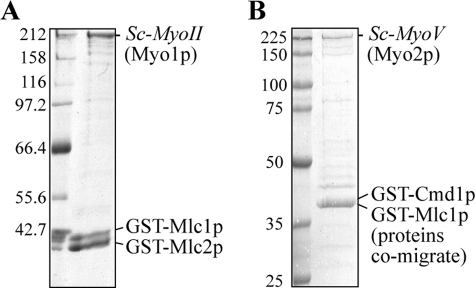
FIGURE 2.
Representative budding yeast Sc-MyoII (Myo1p) and Sc-MyoV (Myo2p) isolations. A, Sc-MyoII and B, Sc-MyoV were isolated from budding yeast lysates using glutathione-Sepharose following co-overexpression of heavy chain and GST-tagged light chains (Mlc1p and Mlc2p for Sc-MyoII; Mlc1p and Cmd1p/calmodulin for Sc-MyoV). Numbers to the left of the SDS-PAGE gel indicate molecular weight markers (kDa).
TABLE 1.
Summary of Sc-MyoII and Sc-MyoV ATPase and motility activities with muscle and yeast actins
|
Sc-MyoII (Myo1p) |
Sc-MyoV (Myo2p) |
|||||||
|---|---|---|---|---|---|---|---|---|
| VMAXa | Kma | Catalytic efficiencyb | Motility ratec | VMAXa | Kma | Catalytic efficiencyb | Motility ratec | |
| s−1 | μm | μm/s | s−1 | μm | μm/s | |||
| Muscle actin | 1.8 ± 0.1 | 11.1 ± 1.5 | 0.17 | 0.87 ± 0.17d (40) | 21.5 ± 0.5 | 6.8 ± 0.6 | 3.17 | 1.51 ± 0.34 (40) |
| Wild-type yeast actin | 3.2 ± 0.2 | 9.6 ± 2.2 | 0.34 | 1.17 ± 0.28 (150) | 39.9 ± 2.6 | 16.1 ± 2.6 | 2.47 | 1.33 ± 0.30 (40) |
| 4Ac yeast actin | 0.93 ± 0.20d (123) | 1.60 ± 0.33 (20) | ||||||
| Sub12 yeast actin | 0.86 ± 0.29d (109) | 1.20 ± 0.27 (20) | ||||||
a VMAX and Km (±S.E.) calculated by fitting actin-activated ATPase data (from Figs. 3A and 4A) to the Michaelis-Menten equation. ATPase activity: molecules of ATP hydrolyzed per motor per second.
b Catalytic efficiency, VMAX/Km.
c Mean in vitro motility rate ± S.D. (n is indicated in parentheses).
d p < 0.001 when compared to yeast actin.
Relative Ability of Yeast and Muscle Actins to Activate the ATPase Activity of Sc-MyoII and Sc-MyoV
Our previous work demonstrated that yeast actin was only one-tenth as active in stimulating skeletal muscle myosin S1 ATPase activity as was muscle actin (13). To determine if this relative ability changed with yeast myosins, we assayed the actin-activated ATPase activity of both Sc-MyoII and Sc-MyoV in the presence of either yeast or skeletal muscle actin. The results demonstrate a number of interesting findings. First, for Sc-MyoII, the VMAX and catalytic efficiency were significantly higher for yeast than for muscle actin (Fig. 3A, Table 1) suggesting co-adaptation of the two proteins for optimal function in yeast. For Sc-MyoV the results were more complicated. There was a significantly higher VMAX with yeast versus muscle actin (Fig. 4A; Table 1). However, it was offset by a higher Km (i.e. a lower apparent actin affinity) for the yeast actin, leading to near equal catalytic efficiencies for the two actins (Table 1). The other observation was that in terms of VMAX and catalytic efficiency, the Sc-MyoV values (with either actin) were >10-fold and 7–20-fold higher, respectively, than those for Sc-MyoII (Table 1).
FIGURE 3.
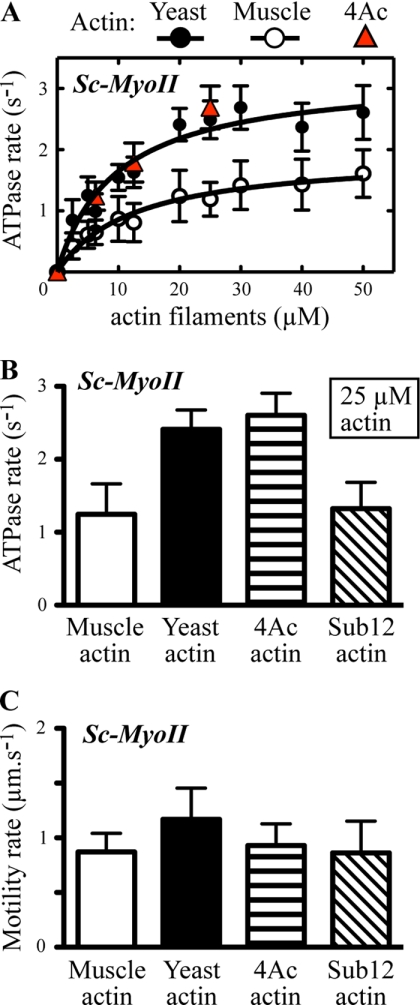
Budding yeast myosin-II motor activities with yeast and muscle actin isoforms. Sc-MyoII motor activity was measured using ATPase and in vitro motility assays with muscle, yeast, and mutant yeast actins. A, actin-activated Mg2+-ATPase activity was measured as a function of actin concentration (yeast, muscle, and the yeast 4Ac mutant). Plots were generated from average values obtained from 3–4 different datasets using at least two independent preparations of Sc-MyoII and the actins. Curves were fit using Michaelis-Menten kinetics. B, actin-activated ATPase data comparing average Sc-MyoII motor activity with 25 μm yeast, muscle, and mutant 4Ac and sub12 yeast actins (n = 3). The difference between the values for wild-type yeast (or 4Ac) and muscle (or sub12) actin are significant (p < 0.0001). C, comparison of the average actin filament gliding rates for Sc-MyoII from in vitro motility assays using yeast, muscle, and mutant yeast actins (n = 40–150).
FIGURE 4.
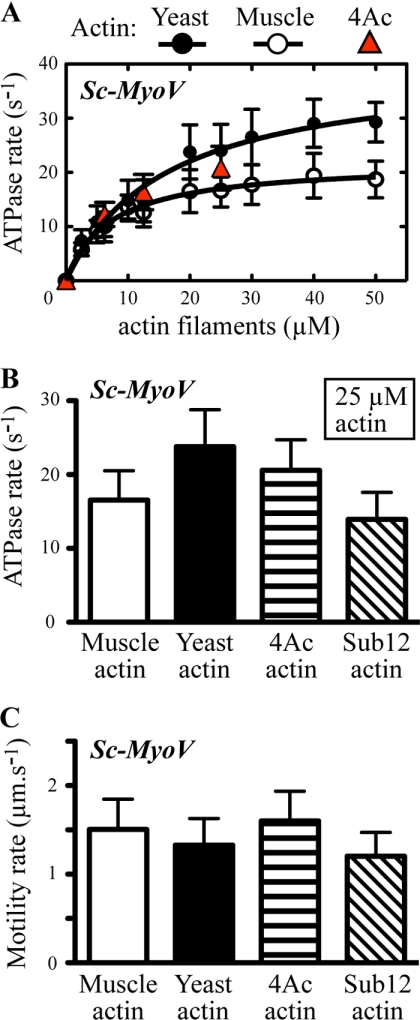
Budding yeast myosin-V motor activities with yeast and muscle actin isoforms. Sc-MyoV motor activity was measured using ATPase and in vitro motility assays with muscle, yeast, and mutant yeast actins. A, actin-activated Mg2+-ATPase activity was measured as a function of actin concentration (yeast, muscle, and the yeast 4Ac mutant). Plots were generated from average values obtained from 3–4 different datasets using at least two independent preparations of Sc-MyoV and the actins. Curves were fit using Michaelis-Menten kinetics. B, actin-activated ATPase data comparing average Sc-MyoV motor activity with 25 μm yeast, muscle, and mutant 4Ac and sub12 yeast actins (n = 3). The difference between the values for wild-type yeast (or 4Ac) and muscle (or sub12) actin are significant (p < 0.0001). C, a comparison of the average actin filament gliding rates for Sc-MyoV from in vitro motility assays using yeast, muscle, and mutant yeast actins (n = 20–40).
Previously, we demonstrated that addition of two negative charges to the N terminus of yeast actin (4Ac actin) increased its ability to activate muscle myosin. With the yeast myosins, however, we observed markedly different results. Because of limitations in available 4Ac actin, we carried out a more limited kinetic analysis preventing calculation of VMAX and Km values. However, for each of the 4Ac actin concentrations tested, the Sc-Myoll ATPase activity was virtually identical to that observed with wild-type yeast actin (Fig. 3, A and B). This result demonstrated a decreased dependence of Sc-MyoII on the negative charge density of the actin N terminus compared with muscle myosin. With Sc-MyoV, the extra negative charge had no significant effect on the ability of yeast actin to activate the myosin (Fig. 4, A and B), again demonstrating a decreased reliance on an ionic interaction between the actin N terminus and myosin in the yeast system.
Behavior of Sc-MyoII and Sc-MyoV in an in Vitro Motility Assay
Another way of assessing actomyosin function is to measure the rate of movement of a fluorescently labeled actin filament over a lawn of myosin heads attached to the surface of a microscope coverslip. We thus applied this assay to the two yeast myosins in the presence of different actins. As shown for Sc-MyoII (Fig. 3C; Table 1), motility with yeast actin is significantly faster than with muscle actin, although the relative difference is less than seen in the solution ATPase assays. Here, the 4Ac actin supported a Sc-MyoII motility rate that was closer to that of muscle (Fig. 3C; Table 1). With Sc-MyoV, the motility observed was basically the same for yeast, muscle, and 4Ac actins (Fig. 4C; Table 1) demonstrating again, a decreased reliance on actin N-terminal negative charge density for yeast myosin activation. However, the relatively slow rate of Sc-MyoV motility (compared with its ATPase) suggests that the duty ratio of this motor probably has a significant impact on functional output in this assay (see “Discussion”).
Activity of Sc-MyoII and Sc-MyoV with a Yeast Muscle Hybrid Actin
We next tested the ability of the mutant yeast actin with muscle-like substitutions in subdomains 1 and 2 (sub12) to support Sc-MyoII and Sc-MyoV activity. As with muscle actin, sub12 actin supported Sc-MyoII and Sc-MyoV ATPase activities that were significantly lower than with wild-type yeast actin (Figs. 3B and 4B). Again, similar to muscle actin, sub12 actin supported a Sc-MyoII motility rate that was significantly lower than wild-type yeast actin (Fig. 3C; Table 1). The motility rate of Sc-MyoV with sub12 actin, if anything, appeared slightly slower than with wild-type yeast, 4Ac, and muscle actins (Fig. 4C; Table 1). Taken together, while the muscle-like 4Ac mutant actin appears more like wild-type yeast actin in its ability to support Sc-MyoII and Sc-MyoV activity, the yeast muscle hybrid sub12 actin appears more like muscle.
Comparing the Properties of Yeast and Skeletal Muscle Actomyosin
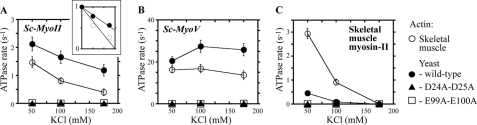
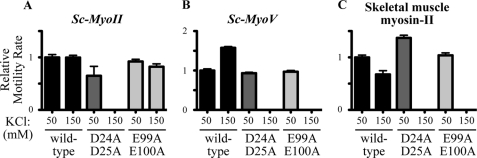
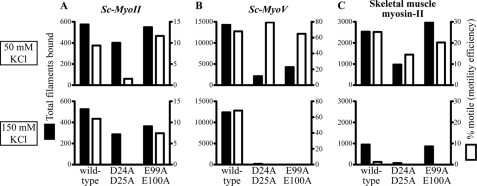
We compared the activity of Sc-MyoII and Sc-MyoV with muscle myosin-II using yeast and muscle actins. Myosin activity was assayed at different salt concentrations to assess the importance of ionic interactions in establishing functional actomyosin complexes. In agreement with our earlier observations (Figs. 3A and 4A) Sc-MyoII and Sc-MyoV showed greater ATPase activity throughout with yeast versus muscle actin (Fig. 5, A and B). The salt sensitivity of Sc-MyoII ATPase activity (Fig. 5A) fell between the insensitivity exhibited by Sc-MyoV (Fig. 5B) and the extreme sensitivity of muscle myosin-II (Fig. 5C). Sc-MyoII activity showed an ∼1.5-fold greater sensitivity to salt with muscle actin (yeast actin IC50: 184 mm KCl, muscle actin IC50: 125 mm KCl; Fig. 5A, inset). In contrast, increasing the salt in motility assays with yeast actin had little impact on Sc-MyoII motility rate (Fig. 6A), filament binding (Fig. 7A), or the percentage of filaments undergoing motility (motility efficiency) (Fig. 7A).
FIGURE 5.
Comparing the salt-sensitivity of Sc-MyoII, Sc-MyoV, and skeletal muscle myosin-II ATPase activities with different actin isoforms. Sc-MyoII (A), Sc-MyoV (B), and muscle myosin-II (C) actin-activated ATPase assays were performed at three different salt concentrations (50, 100, and 175 mm KCl) using four different actins: muscle, wild-type yeast, yeast D24A-D25A, and yeast E99A-E100A (n = 3). All actins were included at a final concentration of 20 μm (except the D24A-D25A mutant which was at a final concentration of 10 μm because of limitations in yield for this particular form). The inset plot in A displays normalized fits comparing the salt-sensitivity of Sc-MyoII ATPase activity for yeast versus muscle actin. The maximum Sc-MyoII activity (which was observed at 50 mm KCl for both actins) was set to 1. These fits allowed the IC50 (half-maximal inhibitory concentration) of KCl to be estimated.
FIGURE 6.
Comparing the salt sensitivity of Sc-MyoII, Sc-MyoV, and skeletal muscle myosin-II motility rates with yeast actins. The average actin filament gliding rates generated by Sc-MyoII (A), Sc-MyoV (B), and muscle myosin-II (C) are compared using three different yeast actins (wild-type, D24A-D25A, and E99A-E100A) at two final salt concentrations (50 and 150 mm KCl). For positive readings, the number of filaments measured (n) were as follows: Sc-MyoII: n = 54–64, except the 50 mm KCl D24A-D25A (n = 6) and 150 mm KCl E99A-E100A (n = 27) values, which were limited by decreased motility efficiency; Sc-MyoV: n = 60; muscle myosin-II: n = 60, except the 150 mm KCl wild-type actin (n = 12) value, which was limited by decreased motility efficiency. Measurements with wild-type yeast actin were generated in parallel with those for D24A-D25A and E99A-E100A mutant actins (i.e. independently of the measurements performed earlier in Figs. 3C and 4C). A uniform concentration of actin filaments (25 nm) was applied to motility chambers throughout. For each myosin, average motility rates (± S.E.) are presented relative to the value measured for wild-type yeast actin at 50 mm KCl (which is set to 1).
FIGURE 7.
Comparing actin filament binding and motility efficiencies for Sc-MyoII, Sc-MyoV, and skeletal muscle myosin-II using the in vitro motility assay. The total number of filaments bound (■), and percentage of bound filaments undergoing motility (□) were quantified for Sc-MyoII (A), Sc-MyoV (B), and muscle myosin-II (C). Filament totals were scored at three set positions (i.e. three 130 μm2 fields) along the motility chamber. As in Fig. 6, yeast actins (wild-type, D24A-D25A, and E99A-E100A) were assayed at two different salt concentrations (50 mm KCl: upper plots; 150 mm KCl: lower plots). 25 nm actin filaments were employed throughout.
With muscle actin, Sc-MyoV ATPase activity showed only a subtle decrease at high salt (Fig. 5B). Surprisingly, with yeast actin, Sc-MyoV ATPase activity increased (and leveled off) when the salt was increased from 50 to 100 (and 175) mm KCl (Fig. 5B). Correspondingly, the Sc-MyoV motility rate increased when the salt was elevated from 50 to 150 mm KCl (Fig. 6B). Increased salt had negligible effects on Sc-MyoV filament binding and motility efficiency in motility assays with yeast actin (Fig. 7B). It is not clear to us why Sc-MyoV activity increases at higher salt, but this effect has been observed previously with other myosin-Vs examined over similar salt concentrations (34, 35).
Consistent with previous findings (13) and in contrast to the yeast myosins, muscle myosin-II ATPase activity was significantly lower with yeast (versus muscle) actin (Fig. 5C). Compared with Sc-MyoII and Sc-MyoV, muscle myosin-II ATPase activity was extremely sensitive to salt (Fig. 5C). This was more pronounced with yeast actin, where activity was essentially lost at 100 mm KCl (Fig. 5C). In motility assays (with yeast actin) this salt sensitivity was reflected by a significant drop in filament gliding rate (Fig. 6C) and filament binding (Fig. 7C), as well as a dramatic reduction in motility efficiency (Fig. 7C).
In summary, yeast and muscle actomyosin function exhibit obvious differences in their actin preference and salt sensitivity. These differences presumably reflect differences at the actomyosin binding interface in yeast versus muscle.
The D24/D25 and E99/E100 Charged Pairs of Actin Residues Are Important for Yeast Actomyosin
Our findings suggest that while muscle actomyosin interactions rely on ionic interactions mediated via the N terminus of actin, yeast actomyosin relies to a greater extent on alternative residues found elsewhere in subdomains 1 and 2. Two pairs of charged residues in subdomain 1 of yeast actin (D24/D25 and E99/E100) are also known to facilitate weak binding with muscle myosin-II (11). Given the lower salt-sensitivity of Sc-MyoII and Sc-MyoV, we tested the degree to which these charged pairs influence yeast actomyosin. As with muscle myosin-II (Fig. 5C) (12), mutant forms of yeast actin lacking either of these charged pairs failed to activate Sc-MyoII or Sc-MyoV ATPase activity at any salt concentration (Fig. 5, A and B). In the case of Sc-MyoV, while both mutant actins show a significant reduction in the total number of filaments undergoing Sc-MyoV-dependent motility at low salt (Fig. 7B), the mutants failed to support any filament binding or motility at high salt (Figs. 7B and 6B), similar to what was seen for muscle myosin-II (Figs. 6C and 7C) (12). On the other hand, while Sc-MyoII motility was clearly sensitive to the D24A-D25A mutant actin (with motility being lost at high salt), the E99A-E100A mutant had no obvious effect on Sc-MyoII motility at low or high salt (Figs. 6A and 7A). We would like to point out the difference in filament numbers bound by the three myosins in Fig. 7 (as shown on the y axes of the plots). The plots here are designed to show the difference in motility behavior for each individual myosin at low versus high salt. In summary, while muscle myosin-II and the yeast myosins exhibit differences in their reliance on the negatively charged N-terminal residues of actin, the weak-binding state of all three myosins rely on negatively charged pairs of residues found elsewhere in subdomain 1.
DISCUSSION
Previous work had suggested that an important aspect of both the binding of myosin to actin and subsequent actin-dependent activation of myosin ATPase activity was an interaction between the negatively charged actin N terminus and a cationic loop (loop 2) separating the 50 kDa and 20 kDa domains in the myosin head (Fig. 1). The focus of this study was to try to gain insight into how, compared with other myosins, yeast myosins function with an actin that has a lower N-terminal negative charge density.
The execution of this work depended on our ability to generate yeast myosins in quantities sufficient for biochemical experimentation. Our successful accomplishment of this goal led to our ability to gain insight into the biochemical differences in the mechanisms that distinguish yeast from mammalian myosins and opens the door to other biochemical avenues of investigation involving these proteins.
We had previously shown that muscle myosin-II ATPase activity is ∼10-fold greater with muscle actin versus budding yeast actin (13) and that increasing the actin N-terminal negative charge density to that of muscle actin decreased this difference to 3-fold. In contrast, we showed here that yeast myosin-II ATPase activity is 2-fold greater with the lower N-terminal negative charge of yeast actin versus muscle actin. These findings suggest that Sc-MyoII somehow compensates for its decreased ability to form an ionic interaction with the yeast actin N terminus. The relatively slow VMAX of Sc-MyoII with muscle actin (1.8 s−1) is 5-fold lower than that of skeletal muscle myosin-II (∼9 s−1) (13, 36), yet Sc-MyoII's VMAX increases with yeast actin (3.2 s−1). This compensation, in terms of an alternative mode of protein-protein interaction, is also evidenced by the fact that Sc-MyoII ATPase activity with yeast 4Ac actin (with its two additional N-terminal negative charges) is no different to wild-type yeast actin.
With Sc-MyoV, however, this actin-dependent difference was not evident. Both yeast and muscle actins displayed equivalent abilities to activate Sc-MyoV ATPase activity suggesting that specific adaptations of the actin molecule are largely inconsequential for some myosins. This difference between the Sc-MyoII and the Sc-MyoV profiles may reflect, in part, the relatively high motor activity of the yeast Sc-MyoV, which, while similar to myosin-Va (37), is ∼10-fold greater than that of Sc-MyoII (with either actin isoform).
The motility rate of Sc-MyoII in filament gliding assays was significantly faster with yeast versus muscle actin (or the 4Ac and muscle-like sub12 yeast actin mutants), although differences in rate were far less striking than those observed in ATPase assays. This smaller difference is understandable since motility rates, while favored by faster ATP hydrolysis cycle times, are rate-limited by the duration of the strong actin-bound myosin ADP state. For example, chicken myosin-Va (and budding yeast Sc-MyoV) ATPase rates are an order of magnitude greater than Sc-MyoII with muscle actin, but this is not reflected by motility rates where the higher duty ratios (percentage time spent in the strong actin-bound state per ATP hydrolysis cycle) of the myosin-Vs probably have a significant influence. Indeed, with all actins tested Sc-MyoV exhibited motility rates that were relatively close to those observed for Sc-MyoII.
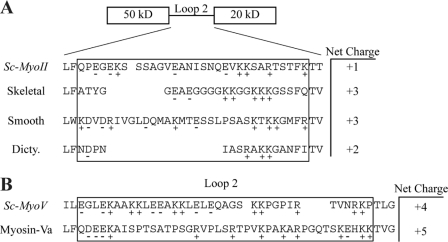
To try to gain insight into the different responses of the class II and class V myosins in yeast to differences in actin N-terminal negative charge density, we investigated the amino acid content of the cationic loops in various myosins. The results, shown in Fig. 8, provided a clue to this difference in behavior. Fig. 8A shows that of the four class II myosins analyzed, Sc-MyoII has by far the lowest net cationic nature with only one positive charge compared with the three found in skeletal and smooth muscle. Thus, muscle actin with its four N-terminal acidic residues would have a stronger ionic interaction with the triple cationic residue muscle myosin loops (compared with yeast). The two negative charges on the yeast actin would have to interact with the one net positive charge on Sc-MyoII, probably requiring another mode of binding between these two proteins. Based on the reduced Sc-MyoII (and Sc-MyoV) ATPase and motility activities with the sub12 mutant yeast actin, such a mode of binding likely relies on other residues in subdomains 1 and 2. This mode of binding may help account for the lower salt-sensitivity of Sc-MyoII and Sc-MyoV motor activities (relative to muscle myosin-II).
FIGURE 8.
Comparing the net charge of myosin-II and myosin-V loop 2 regions. Sc-MyoII and Sc-MyoV loop 2 regions were identified from homology models generated by the Phyre.org server (46) and viewed in Swiss-PdbViewer (47). Alignments using the ClustalX software (45) were used to further refine predicted Sc-MyoII/Sc-MyoV loop 2 regions based on the known structures used to generate the homology models. A, net charge of loop 2 regions from Sc-MyoII, skeletal muscle myosin-II (2MYS (44)), smooth muscle myosin-II (1BR2 (48)), and Dictyostelium discoideum myosin-II (1FMV (49)). B, net charge of loop 2 regions from Sc-MyoV and myosin-Va (1W8J (50)).
On the other hand, as shown in Fig. 8B, both class V myosins, Sc-MyoV and myosin-Va, have cationic loops with greater charge density: +4 charges for the former and +5 for the latter. Furthermore, these are much more evenly distributed across the loop than they are in the class II myosins. This higher positive charge density in the myosins may provide sufficient opportunity for interaction with the two N-terminal negative charges on yeast actin to reach a plateau of stabilization needed for a productive interaction between the two proteins. Once this plateau value is reached, the addition of the two extra negative charges found in muscle actin would produce no additional productive stabilization. This greater charge density and an alternative mode of binding (as discussed above) may explain why Sc-MyoV is less sensitive to salt (cf. Sc-MyoII and muscle myosin-II).
There does appear to be some degree of commonality between muscle and yeast myosins regarding ionic interactions governing the weak actomyosin binding state. The conserved pairs of charged residues (D24/D25 and E99/E100 in yeast) in subdomain 1 that contribute to the weak-bound state of muscle actomyosin (11), are also important for Sc-MyoV ATPase and motility activity. However, while both of these charged pairs of residues were important for Sc-MyoII ATPase, only the D24A-D25A mutant compromised Sc-MyoII motility. Thus, the proposed contact between E99-E100 and the positively charged loop in the lower 50 KDa domain of the myosin motor (Fig. 1A) (38–40) may not be critical for Sc-MyoII motility in the cell.
In summary our results indicate that myosins in muscle have adapted to utilize the higher N-terminal negative charge density of muscle actin. While one can only speculate on how the specialization of muscle myosin-II function might have driven this adaptation, yeast actomyosin reflects an ancestral actin with limited reliance on N-terminal charge. Sc-MyoV, by virtue of its highly positively charged and elongated cationic loop may maximize the opportunity for its interaction with the two acidic residues on the yeast actin N terminus allowing this interaction to reach a threshold needed for activity. In contrast, Sc-MyoII, with only a single net positive charge in the cationic loop, highlights the importance of alternative interactions in achieving functional associations with yeast actin. We conclude that actomyosin binding interfaces are far from universal, and are most likely fine-tuned by constraints imposed by specific endogenous cellular functions.
Acknowledgment
We thank Susan Lowey (University of Vermont, Burlington, VT) for comments and advice regarding Fig. 1 and for providing us with skeletal muscle myosin-II.
This work was supported, in whole or in part, by National Institutes of Health Grant GM33689 (to P. A. R.) and by a New Research Initiative Award (University of Vermont) and a Scientist Development Grant (0835236N) from the American Heart Association (to M. L.).
- F-actin
- filamentous actin.
REFERENCES
- 1. Sellers J. R. (1999) Myosins, Second Ed., Oxford University Press, Oxford [Google Scholar]
- 2. Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. (1990) Nature 347, 37–44 [DOI] [PubMed] [Google Scholar]
- 3. Geeves M. A., Holmes K. C. (1999) Annu. Rev. Biochem. 68, 687–728 [DOI] [PubMed] [Google Scholar]
- 4. Bertrand R., Chaussepied P., Kassab R., Boyer M., Roustan C., Benyamin Y. (1988) Biochemistry 27, 5728–5736 [DOI] [PubMed] [Google Scholar]
- 5. Sutoh K. (1982) Biochemistry 21, 4800–4804 [DOI] [PubMed] [Google Scholar]
- 6. Sutoh K. (1983) Biochemistry 22, 1579–1585 [DOI] [PubMed] [Google Scholar]
- 7. DasGupta G., Reisler E. (1989) J. Mol. Biol. 207, 833–836 [DOI] [PubMed] [Google Scholar]
- 8. DasGupta G., Reisler E. (1992) Biochemistry 31, 1836–1841 [DOI] [PubMed] [Google Scholar]
- 9. Sutoh K., Ando M., Sutoh K., Toyoshima Y. Y. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7711–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johara M., Toyoshima Y. Y., Ishijima A., Kojima H., Yanagida T., Sutoh K. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2127–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller C. J., Reisler E. (1995) Biochemistry 34, 2694–2700 [DOI] [PubMed] [Google Scholar]
- 12. Miller C. J., Wong W. W., Bobkova E., Rubenstein P. A., Reisler E. (1996) Biochemistry 35, 16557–16565 [DOI] [PubMed] [Google Scholar]
- 13. Cook R. K., Root D., Miller C., Reisler E., Rubenstein P. A. (1993) J. Biol. Chem. 268, 2410–2415 [PubMed] [Google Scholar]
- 14. McKane M., Wen K. K., Meyer A., Rubenstein P. A. (2006) J. Biol. Chem. 281, 29916–29928 [DOI] [PubMed] [Google Scholar]
- 15. Karlsson R., Aspenström P., Byström A. S. (1991) Mol. Cell. Biol. 11, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang X., Luo J., Nishihama R., Wloka C., Dravis C., Travaglia M., Iwase M., Vallen E. A., Bi E. (2010) J. Cell Biol. 191, 1333–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watts F. Z., Miller D. M., Orr E. (1985) Nature 316, 83–85 [DOI] [PubMed] [Google Scholar]
- 18. Watts F. Z., Shiels G., Orr E. (1987) EMBO J. 6, 3499–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huckaba T. M., Lipkin T., Pon L. A. (2006) J. Cell Biol. 175, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bi E., Maddox P., Lew D. J., Salmon E. D., McMillan J. N., Yeh E., Pringle J. R. (1998) J. Cell Biol. 142, 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lippincott J., Li R. (1998) J. Cell Biol. 140, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunn B. D., Sakamoto T., Hong M. S., Sellers J. R., Takizawa P. A. (2007) J. Cell Biol. 178, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnston G. C., Prendergast J. A., Singer R. A. (1991) J. Cell Biol. 113, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. (2004) Annu. Rev. Cell Dev. Biol. 20, 559–591 [DOI] [PubMed] [Google Scholar]
- 25. Rose M. D., Winston F., Heiter P. (1990) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 27. Lord M., Sladewski T. E., Pollard T. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8014–8019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sladewski T. E., Previs M. J., Lord M. (2009) Mol. Biol. Cell 20, 3941–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cook R. K., Blake W. T., Rubenstein P. A. (1992) J. Biol. Chem. 267, 9430–9436 [PubMed] [Google Scholar]
- 30. Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 31. Henkel R. D., VandeBerg J. L., Walsh R. A. (1988) Anal. Biochem. 169, 312–318 [DOI] [PubMed] [Google Scholar]
- 32. Kron S. J., Spudich J. A. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6272–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reck-Peterson S. L., Tyska M. J., Novick P. J., Mooseker M. S. (2001) J. Cell Biol. 153, 1121–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodges A. R., Krementsova E. B., Trybus K. M. (2007) J. Biol. Chem. 282, 27192–27197 [DOI] [PubMed] [Google Scholar]
- 35. Wang F., Chen L., Arcucci O., Harvey E. V., Bowers B., Xu Y., Hammer J. A., 3rd, Sellers J. R. (2000) J. Biol. Chem. 275, 4329–4335 [DOI] [PubMed] [Google Scholar]
- 36. Margossian S. S., Lowey S. (1982) Methods Enzymol. 85, 55–71 [DOI] [PubMed] [Google Scholar]
- 37. Krementsov D. N., Krementsova E. B., Trybus K. M. (2004) J. Cell Biol. 164, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Milligan R. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 21–268552606 [Google Scholar]
- 39. Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. (1993) Science 261, 58–65 [DOI] [PubMed] [Google Scholar]
- 40. Volkmann N., Hanein D., Ouyang G., Trybus K. M., DeRosier D. J., Lowey S. (2000) Nat. Struct. Biol. 7, 1147–1155 [DOI] [PubMed] [Google Scholar]
- 41. Vorobiev S., Strokopytov B., Drubin D. G., Frieden C., Ono S., Condeelis J., Rubenstein P. A., Almo S. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5760–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holmes K. C., Angert I., Kull F. J., Jahn W., Schröder R. R. (2003) Nature 425, 423–427 [DOI] [PubMed] [Google Scholar]
- 43. Roy A., Kucukural A., Zhang Y. (2010) Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. (1993) Science 261, 50–58 [DOI] [PubMed] [Google Scholar]
- 45. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 46. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 47. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 48. Dominguez R., Freyzon Y., Trybus K. M., Cohen C. (1998) Cell 94, 559–571 [DOI] [PubMed] [Google Scholar]
- 49. Bauer C. B., Holden H. M., Thoden J. B., Smith R., Rayment I. (2000) J. Biol. Chem. 275, 38494–38499 [DOI] [PubMed] [Google Scholar]
- 50. Coureux P. D., Sweeney H. L., Houdusse A. (2004) EMBO J. 23, 4527–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]