Abstract
Mitochondria are central organelles in cellular energy metabolism, apoptosis, and aging processes. A signaling network regulating these functions was recently shown to include soluble adenylyl cyclase as a local source of the second messenger cAMP in the mitochondrial matrix. However, a mitochondrial cAMP-degrading phosphodiesterase (PDE) necessary for switching off this cAMP signal has not yet been identified. Here, we describe the identification and characterization of a PDE2A isoform in mitochondria from rodent liver and brain. We find that mitochondrial PDE2A is located in the matrix and that the unique N terminus of PDE2A isoform 2 specifically leads to mitochondrial localization of this isoform. Functional assays show that mitochondrial PDE2A forms a local signaling system with soluble adenylyl cyclase in the matrix, which regulates the activity of the respiratory chain. Our findings complete a cAMP signaling cascade in mitochondria and have implications for understanding the regulation of mitochondrial processes and for their pharmacological modulation.
Keywords: Adenylate Cyclase (Adenylyl Cyclase), Cyclic AMP (cAMP), Cyclic GMP (cGMP), Mitochondria, Mitochondrial Metabolism, Phosphodiesterases
Introduction
Mitochondria play central roles in cellular energy metabolism, as well as in the regulation of cell cycle progression, apoptosis, and aging processes (1, 2). Despite their importance, signaling into, from, and within mitochondria is still not well understood. Emerging signaling mechanisms in mitochondria and between the organelle and its environment include reversible protein deacetylation (3, 4), redox regulation and reactive oxygen species formation (5–7), and cyclic adenosine monophosphate (cAMP) signaling (8, 9).
cAMP-dependent effects and proteins of cAMP signaling systems, such as cAMP-responsive element-binding protein (CREB), protein kinase A (PKA), and A-kinase anchoring proteins (AKAPs),3 have been described in mitochondria (10–12). In addition to these effector proteins, a complete cAMP signaling microdomain requires enzymes for synthesis and degradation of the second messenger. Although an intramitochondrial cAMP source has been identified recently (8), there is no known cAMP-degrading enzyme in this organelle. Cyclic AMP is formed inside mitochondria by soluble adenylyl cyclase (sAC) (8), a member of Class III of the nucleotidyl cyclase family, which also comprises the G-protein-regulated transmembrane adenylyl cyclases (13). Unique from transmembrane adenylyl cyclases, sAC is activated by bicarbonate (14), and it appears to act as a metabolic sensor (15), whose mitochondrial form(s) seems to modulate PKA-mediated regulation of respiration (8) and apoptosis (16).
The opponents of the cyclic nucleotide-forming cyclases are cyclic nucleotide monophosphate (cNMP)-degrading phosphodiesterases (PDEs). Mammalian cells contain a varying subset of members of the classical PDE family, which comprises 11 PDE gene families (PDE1–11) (17, 18) and non-generic PDEs such as the protein human Prune (19, 20). The isoforms of the generic PDEs comprise homologous catalytic domains, fused to varying regulatory domains, making them sensitive to a variety of signals such as calmodulin or cNMP binding (17). This regulated degradation of cNMPs, together with their regulated synthesis, determines local cNMP concentrations and thus signal strength and duration. PDE1, for example, contributes to formation of cytosolic and possibly nuclear cNMP signals that can be modulated by Ca2+/calmodulin (18), and the cGMP-activated, specifically cGMP-hydrolyzing PDE5 can contribute to the formation of cGMP signal spikes in platelets (21). Most PDE genes encode several splice variants, such as PDE2A1, PDE2A2, and PDE2A3 (22–24), resulting in more than 100 differently expressed PDEs, which can further contribute to differential modulation and localization of cNMP signals (17).
Although PDE activity has been observed in mitochondria (8), the identity of the mitochondrial PDE remained unknown. Such a mitochondrial PDE should reside inside the matrix to ensure that local sAC-generated cAMP signals are temporally and spatially controlled. PDE4 co-localization with mitochondria, apparently through an interaction with the protein “disrupted in schizophrenia 1” (DISC1), was previously reported (25), but a localization of this, or any other, PDE inside mitochondria has not been shown. Identifying mitochondrial PDEs would reveal specifically localized, well “druggable” targets for treatment of mitochondrial diseases (26) because PDEs are established as excellent targets for pharmacological inhibition. Specific compounds are available for most isoforms, including clinically used drugs for PDE3 and PDE5, and several isoforms serve as therapeutic targets in current drug development efforts (27).
Here, we show that PDE2A isoform 2 is specifically targeted to the mitochondrial matrix, where it forms a local signaling system with sAC regulating the respiratory chain. Our findings complete the cAMP signaling system in mitochondria and have implications for understanding and pharmacologically targeting mitochondrial processes and specific PDE isoforms.
EXPERIMENTAL PROCEDURES
Preparation of Liver and Brain Mitochondria
For the import experiments and initial characterization of lysates in PDE activity and inhibition assays, mitochondria from rat liver and brain were enriched using differential centrifugation. Homogenized liver or brain cells in ice-cold isolation buffer (2 mm Hepes, pH 7.4, 220 mm mannitol, and 70 mm sucrose) were disrupted mechanically. To remove cell debris and nuclei, lysates were centrifuged at 500 × g for 3 min at 4 °C. The supernatant was centrifuged two more times. Mitochondria were collected by centrifugation at 26,000 × g for 10 min at 4 °C. The pellet was washed in isolation buffer and recentrifuged twice, and the mitochondria were finally resuspended in 2 ml of isolation buffer.
For Western blot analysis and functional assays on mitochondria, highly pure mitochondria were prepared from mouse liver and brain mitochondria as described (28). Isolated mitochondria were incubated with sAC or PDE inhibitors in MAITE medium (10 mm Tris-HCl, pH 7.4; 25 mm sucrose; 75 mm sorbitol; 100 mm KCl; 10 mm K2HPO4; 0.05 mm EDTA; 5 mm MgCl2; 1 mg/ml BSA) in the presence of a mixture of phosphatase inhibitors (Sigma-Aldrich). The following conditions were used for all experiments: 1 mm 8Br-cAMP (Sigma-Aldrich), 15 μm erythro-9-(2-hydroxyl-3-nonyl)adenine (EHNA) (Tocris), 20 nm BAY60-7550 (Tocris), 1 μm rolipram (Sigma-Aldrich), and 25 μm KH7 for 10 min.
Preparation of Mitochondrial Subcompartments and Proteinase K Protection Assays
For protein localization, pure brain mitochondria were isolated using a Percoll gradient as described previously (29). 500 μg of mitochondria (10 mg/ml) were resuspended in MS-EGTA (225 mm mannitol, 75 mm sucrose, 5 mm HEPES, 1 mm EGTA, pH 7.4). Water (1/10 volume) and digitonin (1 mg of digitonin/5 mg of mitochondrial protein) were added, and the mixture was incubated on ice for 45 min. Then, KCl (150 mm) was added followed by incubation for 2 min on ice and centrifugation at 18,000 × g for 20 min at 4 °C. The pellet containing the mitoplast fraction was resuspended at 1 mg/ml in 300 mm Tris-HCl, 10 μm CaCl2, pH 7.4. The supernatant containing the post-mitoplast fraction was precipitated with 12% TCA and centrifuged at 18,000 × g for 15 min at 4 °C. The pellet was resuspended in 500 μl of acetone and centrifuged at 18,000 × g for 15 min at 4 °C.
For protease protection assays, 20 μg of mitochondrial protein were treated with 20 μg/ml proteinase K (PK) for 20 min on ice. Then, PK was inactivated with 2 mm phenylmethanesulfonyl fluoride (PMSF) for 10 min on ice. Prior to PK treatment, one aliquot of mitochondria was solubilized with 1% Triton X-100 (Sigma-Aldrich) for 15 min on ice.
Western Blot Analysis
For Western blot analyses of mitochondria and mitoplast samples, 25 μg of protein were separated by 12.5% SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto PVDF filters (Bio-Rad). For protein detection, the following antibodies were used: cytochrome c oxidase (COX) subunit I (Invitrogen), Hsp60, grp75, and cytochrome c (StressGen); PDE2A (K-20, Cell Signaling); Tim23 (BD Transduction Laboratories); and β-actin (Sigma-Aldrich).
PDE Activity Assay and cAMP Measurements
For the characterization of the PDE activity in mitochondrial lysates, cAMP- and cGMP-degrading activities were determined in a radioactive assay, using 10 μm [32P]cAMP or [32P]cGMP as substrate and 10-min incubations, as described before (30). Measurements of mitochondrial cAMP levels after treatment of whole organelles were performed according to manufacturer's instructions using the Direct Correlate-EIA cAMP kit (Assay Designs Inc.). 100 μl of sample were measured. If necessary, samples were diluted to bring the cAMP level of the sample within the linear range of the assay.
In Vitro Test for Protein Interaction with Mitochondria and Confocal Microscopy for Cellular Localization of GFP Fusions
For the in vitro test, fragments coding for PDE2A2(1–210) and PDE2A2(18–210) of mouse PDE2A2 (NM_001143849.1, designated as transcript variant/isoform 3 in contrast to the original designation PDE2A2 (gene identifier 706929), see Refs. 24, 31, and 32) were cloned using BamHI and XhoI into pCDNA3.1zeo(+) with two additional methionine residues at the C terminus. For cloning PDE(1–17)-Map(45–203), an additional SacII restriction site was introduced into PDE2A2 by site-directed mutagenesis, resulting in an insertion encoding an additional proline and arginine. After restriction, the fragment for the PDE2A2 N terminus was ligated into a cleaved pCDNA3.1zeo(+)-Map(45–203) plasmid. Radiolabeled proteins were synthesized in reticulocyte lysate (TnT T7 coupled reticulocyte lysate system, Promega) containing [35S]Met. Rat liver mitochondria were isolated as described before (33). The mitochondria (30 μg of mitochondrial protein) were incubated with 5 μl of reticulocyte lysate in 50 μl of 250 mm sucrose, 80 mm KCl, 20 mm potassium phosphate, 2 mm NADH, 2 mm ATP, 10 mm MOPS/KOH, pH 7.2, containing 3% (w/v) bovine serum albumin. After incubation at 25 °C for 10 min, the mitochondria were reisolated by centrifugation (10 min 16,000 × g). To remove residual reticulocyte lysate, the mitochondria were resuspended in 100 μl of 250 mm sucrose, 150 mm NaCl, 10 mm MOPS/KOH, pH 7.2, and again reisolated. Mitochondria-associated proteins were separated by SDS-PAGE and analyzed using a BAS-1800 II imager (Fujifilm).
For the in vivo localization studies, the gene fragment coding for the N terminus (amino acids 1–216) of PDE2A isoform 2 (NM_001143849.1, see above) was cloned using EcoRI and BamHI into pEGFP-N3 (Clontech) to yield a construct with C-terminally fused GFP. For comparison, we used similar constructs for PDE2A isoforms 1 and 3 fused to CFP as described before (34). For expression, each PDE fusion construct was co-transfected with pDsRed-mito (Clontech), which codes for a fusion of Dicosoma sp. red fluorescent protein (DsRed) with the mitochondrial targeting sequence of human cytochrome c oxidase subunit VIII, into HEK-293 cells using FuGENE 6 (Roche Applied Science) and standard procedures. PDE isoform constructs and DsRed-mito were expressed under control of the immediate early promoter of cytomegalovirus, and cells from three independent expression experiments were visualized using a Zeiss LSM 510 Meta system and a Plan-Neofluar 100×/1.3 objective in sequential scan mode using the 458 (CFP), 488 (EGFP), and 564 nm (DsRed) laser lines with corresponding band-path filters. Image recordings were optimized using the LSM 510 software with images representing single focal planes with a pixel resolution of 0.09 × 0.09 μm. Images were imported into ImageJ, background was subtracted, and regions of interest representing >100 cells for each condition were subjected to intensity correlation analysis (35). Pearson's correlation coefficients were calculated with a value of 1 representing perfect correlation; −1 represents perfect exclusion, and zero represents random localization. Because values close to zero have to be critically evaluated, we calculated in addition the intensity correlation quotient (ICQ) with random staining: ICQ ∼0; segregated staining: 0 > ICQ ≥ −0.5; dependent staining: 0 < ICQ ≤ +0.5. Significance values were calculated using an unpaired Student's t test with p > 0.01.
Assays for COX Activity and Mitochondrial ATP Synthesis and Oxygen Consumption
Oligomycin-sensitive mitochondrial ATP synthesis was measured in isolated mitochondria (15–25 μg of protein) in the presence of ADP (0.1 mm) and malate plus pyruvate (1 mm each) as substrates and of the adenylate kinase inhibitor diadenosine pentaphosphate (0.15 mm), using a kinetic luciferase-luciferin detection system in a recording luminometer, as described previously (36). These conditions yielded linear ATP synthesis rates, which were expressed as rate (nmol/min) of ATP production/mg of mitochondrial protein.
Pyruvate/malate-driven oxygen consumption was measured in isolated mitochondria using an Oxygraph system equipped with a Clark electrode (Hansatech; 100 μg of protein) in the presence of ADP, as described previously (37). Mitochondrial respiration was expressed as μmol of O2 consumed/min/mg of mitochondrial protein. COX activity in isolated mitochondria (2–5 μg of mitochondrial protein) was measured spectrophotometrically as described (38).
RESULTS
Characterization of PDE Activity in Mitochondria from Liver and Brain
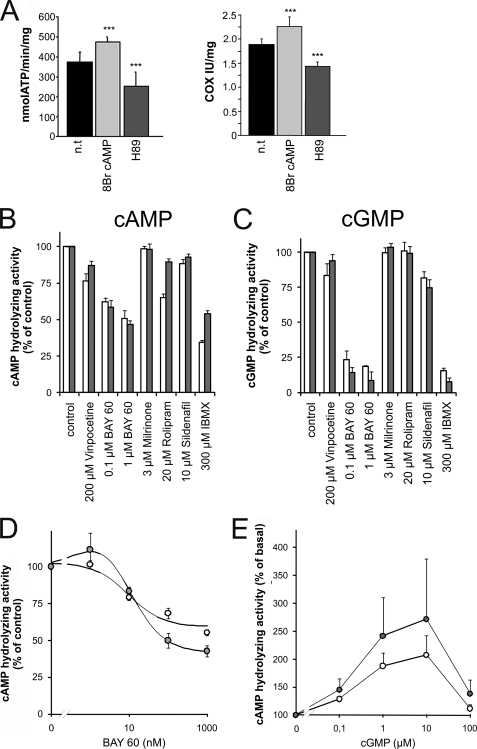
We recently showed that sAC-dependent cAMP formation in the matrix of liver mitochondria regulates respiration (8). We now demonstrate that cAMP also increases ATP production in mitochondria isolated from brain via stimulation of the electron transport chain (Fig. 1A). This form of regulation thus exists in liver and brain, and we assume that it will prove to be a general mitochondrial mechanism. We also showed that cAMP does not pass through the inner mitochondrial membrane (8); thus, this signaling pathway requires a mitochondrial cAMP-degrading PDE for switching off the signal. As a first step toward identifying candidate isoforms for such a mitochondrial PDE, we analyzed lysates of isolated mitochondria from rat liver and brain for cAMP- and cGMP-degrading activities in the absence and presence of different PDE inhibitors (Fig. 1, B and C). Mitochondrial homogenates from both tissues contained readily detectable cAMP- and cGMP-degrading activities. The cAMP-degrading mitochondrial activities from liver and brain were inhibited by ∼50 and 70%, respectively, by the non-selective PDE inhibitor 3-isobutyl-1-methylxanthin (IBMX; Fig. 1B). In contrast, cGMP-degrading activity found in mitochondria from both tissues was almost completely inhibited by this non-selective PDE inhibitor (Fig. 1C).
FIGURE 1.
cAMP-dependent respiratory chain regulation in brain mitochondria and enzymological and pharmacological characterization of mitochondrial PDE activity. A, cAMP-dependent regulation of oxidative phosphorylation in mitochondria isolated from mouse brain. Treatment with the membrane-transmissible cAMP analog 8-Br-cAMP increased the rate of ATP formation (left) and the activity of COX (right). The PKA inhibitor H89, in contrast, decreased both activities. n.t., non-treated. Error bars represent S.E. values. ***, p < 0.0001. B and C, sensitivities of the mitochondrial cAMP-hydrolyzing (B) and cGMP-hydrolyzing (C) activities to a panel of PDE inhibitors. White bars, brain mitochondria; gray bars, liver mitochondria. D, concentration-response curves for inhibition of mitochondrial cAMP-degrading activity from brain (white circles) and liver (gray circles) by BAY60. E, concentration-response curves for stimulation of mitochondrial cAMP-degrading activity from brain (white circles) and liver (gray circles) by cGMP. Error bars represent S.E. values.
To further characterize the PDE isoforms responsible for these activities, isoform-specific inhibitors of PDEs 1 through 5 were employed. The inhibitors of PDE1 (vinpocetine), PDE3 (milrinone), and PDE5 (sildenafil) had little to no effect on cAMP- or cGMP-degrading activities in mitochondrial homogenates from either tissue (Fig. 1, B and 1C). In contrast, the inhibitor BAY60-7550 (“BAY60”), which is specific for PDE2A, a PDE that can degrade both cAMP and cGMP, inhibited the cAMP-degrading activities significantly (Fig. 1B). BAY60 inhibited the activity from liver mitochondria to an extent comparable with IBMX and inhibited activity from brain mitochondria slightly less than IBMX. To evaluate the specificity of BAY60, concentration-response curves for inhibition of mitochondrial cAMP-degrading activity by this compound were recorded (Fig. 1D). The obtained IC50 of ∼10 nm for the BAY60-sensitive activity is in good agreement with the IC50 for BAY60 inhibition of PDE2 (4.7 nm (39)) and confirms specificity of the BAY60 effect. BAY60 also inhibited almost all cGMP-degrading activity present in mitochondrial homogenates from both tissues, comparable with the nonspecific inhibitor IBMX (Fig. 1C). Thus, PDE2A appears to be present in brain and liver mitochondrial fractions and to be responsible for all (liver) or most (brain) of the IBMX-sensitive PDE activity in mitochondria from these tissues. Rolipram, the inhibitor of PDE4, had no significant effect on cAMP-degrading activity from liver mitochondria but showed some inhibition of the activity from brain, albeit to a lesser extent than BAY60 (Fig. 1B). Thus, lysates from brain mitochondria seem to contain PDE4 in addition to PDE2A, and IBMX inhibition of brain mitochondrial PDE activity seems to correspond to the sum of the effects of the PDE4-selective rolipram and the PDE2A-selective BAY60.
To further substantiate the identity of the mitochondrial IBMX-sensitive PDE as PDE2A, stimulation of this activity by cGMP was analyzed (Fig. 1E). In accordance with its known effects on PDE2 (40, 41), cGMP stimulated cAMP degradation with an EC50 of ∼0.3 μm, and it inhibited at higher concentrations. cGMP stimulated cAMP degradation 2–3-fold, which is lower than reported (40, 41) and might reflect the presence of an additional cAMP-degrading PDE, as suggested by the incomplete suppression of cAMP-degrading activity by IBMX or BAY60 described above. In conclusion, our results strongly suggest that brain and liver mitochondria contain PDE2A and a yet to be identified, IBMX-insensitive, cAMP-specific PDE (see “Discussion”).
PDE2 Is Localized in the Mitochondrial Matrix
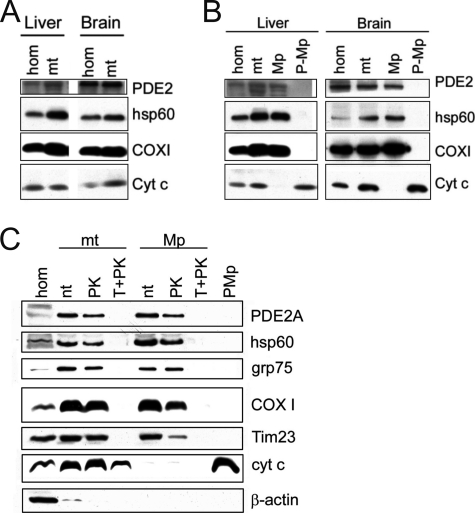
To confirm the presence of PDE2A in mitochondria and to elucidate the PDE2-containing subcompartment, we tested highly purified mitochondria and fractionated mitochondrial subcompartments with a PDE2A-specific antibody. Western blots of whole mitochondria isolated from liver and brain tissue showed positive bands of the expected molecular mass (∼105 kDa; Fig. 2A). Tissue homogenates from brain produced a stronger signal than those from liver, consistent with the previously demonstrated prominent expression of PDE2A in brain (22, 34, 42); however, the PDE2A-specific signal in mitochondria from brain and liver was more comparable, revealing that differences in the levels of the mitochondrially localized PDE2A population are less pronounced.
FIGURE 2.
Identification of PDE2A in the mitochondrial matrix. A, Western blot of mouse liver and brain homogenates (hom) and crude mitochondria (mt). B, Western blot of mitochondrial compartments derived from purified mitochondria from mouse brain and liver. Mitochondria and mitoplasts (Mp; inner mitochondrial compartment) contain a PDE2A species, whereas it is absent from the post-mitoplast fraction (P-Mp; contains intermembrane space proteins). hsp60, marker for mitochondrial matrix proteins; COXI, marker for mitochondrial inner membrane proteins; cyt c, marker for intermembrane space proteins. C, mitochondria and mitoplasts are shown non-treated (n.t.) or after being subjected to PK or Triton plus proteinase K (T+PK) treatment. The β-actin control shows that the small cytoplasmic contamination present in the initial mitochondrial fraction is removed after PK treatment of mitochondria and during mitoplast preparation. hsp60 and grp75, markers for mitochondrial matrix proteins; COXI and Tim23, markers for mitochondrial inner membrane proteins; cyt c, marker for intermembrane space proteins. n.t., non-treated. Blots are representative of three independent experiments.
We next determined which mitochondrial subcompartment contains PDE2A using pure mitochondria from brain. Our Western blots revealed PDE2A in mitochondrial fractions and in the mitoplast fractions, but not in lysates after mitoplast removal (post-mitoplast; Fig. 2B). Furthermore, PDE2A was protected during proteinase K treatment of mitoplasts, as were hsp60 and grp75, two mitochondrial matrix proteins (Fig. 2C). Because post-mitoplast supernatants contain intermembrane space and outer membrane proteins and proteinase K treatment would digest accessible proteins, we conclude that PDE2A is enclosed in the mitochondrial matrix. Consistently, PDE2A was not protected from proteinase K digestion anymore when the mitoplast membrane (inner mitochondrial membrane) was solubilized through the addition of Triton (Fig. 2C). Thus, PDE2A is co-localized with the cAMP source sAC in the mitochondrial matrix.
The N Terminus of PDE2A Isoform 2 Promotes Mitochondrial Localization
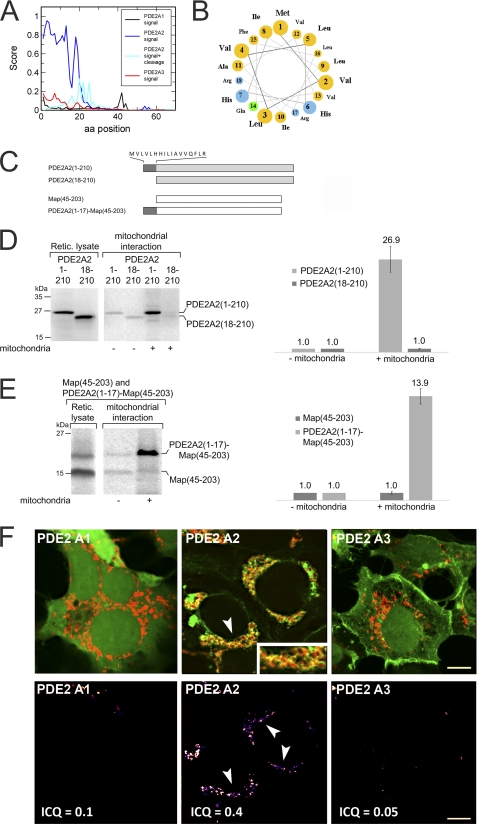
Mitochondrial transport systems can import proteins that contain different types of localization signals, but most matrix proteins contain an N-terminal sorting signal, which is often proteolytically removed during import (43, 44). Three PDE2A isoforms have been described, PDE2A1, PDE2A2, and PDE2A3, which are encoded by a single mammalian PDE2A gene and differ in their N-terminal sequences due to alternative splicing (22–24). The isoform 3 N terminus serves as a myristoylation site for membrane anchoring (34), but no function is known for the N termini of isoforms 1 and 2. We analyzed the N-terminal sequences for the presence of potential mitochondrial localization signals using MitoProtII (45) and SignalP (46) servers. The SignalP neuronal network predicted a high overall probability (72% probability) for the PDE2A2 N terminus to contain a mitochondrial localization signal (Fig. 3A). MitoProtII also predicted that the PDE2A2 N terminus contains a mitochondrial targeting sequence (54%; data not shown). In contrast, much lower probabilities were found for isoforms 3 (MitoProtII: 21%; SignalP: 9%) and 1 (MitoProtII: 2%; SignalP: 14%). Likely signal peptide cleavage sites were also uniquely predicted for isoform 2. Cleavage between residues 19 and 20, in an RRG-QQ motif, had the highest probability score, with two additional putative cleavage sites located between residues 20–28 (Fig. 3A). A helical wheel representation of the N-terminal 19 residues shows that this region can form a positively charged amphipathic α-helix (Fig. 3B), consistent with it functioning as a signal sequence for matrix transport (43). We thus predict that PDE2A2 is targeted to the mitochondrial matrix through recognition of its N-terminal sequence, which might get cleaved at amino acid 19 during this process.
FIGURE 3.
The PDE2A2 N terminus acts as a mitochondrial localization signal. A, probability scores, calculated with SignalP, for the N termini of PDE2A isoforms 1 through 3 to act as mitochondrial localization sequences. A score combining this probability with the likelihood of a position to serve as a mitochondrial processing site (signal + cleavage) identifies three sites with a high probability to serve as proteolytic processing sites during import. aa, amino acid. B, a helical wheel representation of the 19 N-terminal residues of PDE2A2 reveals that these residues can form the amphipathic helix (top, hydrophobic; bottom, hydrophilic) typical for mitochondrial localization sequences. C, constructs used in the mitochondrial interaction experiments shown in panels D and E. Protein parts from the mitochondrial matrix protein Map are symbolized as white bars, residues from human PDE2A2 are symbolized as gray bars, and the 18 N-terminal residues unique for this PDE2A isoform (sequence on top) are symbolized as dark gray bars. D, in vitro tests for interaction with mitochondria. Radiolabeled samples (left) of PDE2A2(1–210) and PDE2A2(18–210) were incubated with rat liver mitochondria for 10 min at 25 °C. Mitochondria were reisolated, and associated proteins were analyzed by SDS-PAGE and autoradiography using a BAS-1800 II imager (left). For quantitation (right), the amount of protein in the absence of rat liver mitochondria was set to a value of 1 (control). Retic. lysate, reticulocyte lysate. E, in vitro tests for the interaction of Map(45–203) and PDE2A2(1–17)-Map(45–203) with mitochondria. The experiments were carried out as described in panel D. Error bars in panels D and E represent S.D. values. F, upper panel, confocal images of HEK cells overexpressing a mitochondrial marker (DsRed-mito, red) and the N termini of PDE2 isoforms fused to EGFP or CFP (green). The three PDE2A isoforms show distinct subcellular distributions, and considerable overlap with the mitochondrial marker, visible in yellow (see for example the arrow), is specifically observed for isoform 2 (see inset). Lower panel, graphical presentation of the intensity correlation analysis highlighting substantial overlap of the PDE2A2 isoform with DsRed-mito. Scale bar: 3 μm.
To test whether the PDE2A2 N terminus is indeed responsible for targeting this PDE2A isoform to mitochondria, we first tested the interaction of different protein constructs (Fig. 3C) with isolated mitochondria in vitro. The PDE2A N terminus (PDE2A2(1–210)) and a truncated version PDE2A2(18–210) lacking the 17 N-terminal residues, which are unique for this isoform, were synthesized in reticulocyte lysate in the presence of [35S]methionine. The radiolabeled proteins were incubated with freshly isolated rat liver mitochondria, and the mitochondria were then reisolated. The PDE2A2(1–210) protein, which harbors the entire PDE2A2 N terminus, bound efficiently to mitochondria, whereas the truncated PDE2A2(18–210) construct was unable to interact with the organelles (Fig. 3D). To further confirm that the 17 unique N-terminal residues of PDE2A isoform 2 are essential and sufficient for targeting a protein to mitochondria, we tested a fusion of this N terminus with the mitochondrial protein Map (47). The intrinsic mitochondrial targeting sequence of Map (residues 1–44) was removed and substituted by the N-terminal 17 residues of PDE2A2. A Map construct covering amino acids 45–203 (Map(45–203)) showed no association with mitochondria, but a Map fusion construct starting with the 17 N-terminal residues of PDE2A2 (PDE2A2(1–17)-Map(45–203)) bound efficiently to mitochondria (Fig. 3E). We thus conclude that PDE2A isoform 2 indeed interacts with mitochondria through its unique N terminus.
To confirm that the PDE2A2 N terminus is responsible for the specific targeting of this isoform to mitochondria in a physiological environment, we analyzed the cellular localization of fusion proteins composed of the N terminus of PDE2A1, PDE2A2, or PDE2A3, respectively, attached to a fluorescent protein (Fig. 3F). Fusion proteins were expressed in HEK-293 cells co-transfected with an expression vector for a fluorescent mitochondrial marker protein (DsRed-mito). Confocal microscopy revealed that the fusion protein harboring the N terminus of isoform PDE2A2 showed significant co-localization with mitochondria, as indicated by the yellow color resulting from an overlap of PDE2A2-N terminus-GFP (green) and mitochondrial DsRed (Fig. 3F, red; overlap indicated by arrows). In contrast, the fusion proteins containing the N terminus of isoform PDE2A1 or PDE2A3 did not show significant co-localization with the mitochondrial marker. This observation was substantiated by co-localization analysis (35); Pearson's correlation coefficients (Rr) and ICQ both demonstrate a significantly (p < 0.01) increased overlap with mitochondria specifically for PDE2A isoform 2. ICQ values are 0.31 + 0.05 (PDE2A2; Rr = 0.60 + 0.11) > 0.19 + 0.06 (PDE2A1; Rr = 0.27 + 0.09) > 0.14 + 0.06 (PDE2A3; Rr = 0.36 + 0.13). We thus conclude that only the isoform-specific N terminus of PDE2A2 can promote targeting to mitochondria in a cellular environment.
PDE2A Regulates Mitochondrial cAMP Levels and Respiration
We next investigated whether mitochondrial PDE2A acts as the cAMP-degrading enzyme counteracting CO2/bicarbonate-responsive sAC in the PKA-containing, cAMP signaling microdomain in the mitochondrial matrix regulating the respiratory chain (8). To test this functional coupling of PDE2A and sAC activities, we analyzed the effects of selective inhibitors on mitochondrial cAMP levels and oxidative phosphorylation. Consistent with PDE2A counteracting sAC activity, we observed an increase in cAMP levels when brain mitochondria were treated with EHNA or BAY60, two specific PDE2A inhibitors (Fig. 4A). In contrast, rolipram inhibition of PDE4, which we found to be a minor component of the PDE activity associated with brain mitochondria (Fig. 1B), did not cause any changes in cAMP levels as compared with uninhibited mitochondria. Thus, the cAMP generated in intact mitochondria is regulated by PDE2A and not by PDE4. KH7, a specific sAC inhibitor, prevented EHNA and BAY60 from elevating cAMP to the levels observed in the absence of KH7 (Fig. 4A), suggesting that both KH7 and BAY60/EHNA are acting on the same cAMP pathway. The level of cAMP in mitochondria treated with KH7 plus EHNA/BAY60 was still moderately elevated as compared with mitochondria treated with KH7 alone (Fig. 4A), presumably due to delayed or diminished degradation of residual mitochondrial cAMP. This moderate elevation was not observed in the presence of rolipram, supporting our conclusion that PDE2A, but not PDE4, acts on the same cAMP pool as sAC.
FIGURE 4.
Mitochondrial PDE2A regulates the respiratory chain. A, cAMP levels in intact mouse brain mitochondria after short treatment (10 min) with inhibitors for PDE2 (EHNA and BAY60), PDE4 (rolipram), and sAC (KH7) (n = 6). nt, non-treated. B, pyruvate plus malate-driven ATP synthesis in intact mouse brain mitochondria after treatment with the agonist 8-Br-cAMP or sAC and PDE inhibitors (n = 9). Values are represented as the percentage of control non-treated mitochondria. C, pyruvate plus malate-dependent respiration of isolated brain mitochondria after treatment with the agonist 8-Br-cAMP or sAC and PDE inhibitors (n = 2). Error bars represent S.E. values. *, p < 0.01; **, p < 0.001; ***, p < 0.0001.
To further confirm the functional coupling of sAC and PDE2A, we analyzed their role in regulating respiratory chain activity. To monitor respiration, we first measured ATP synthesis as a readout of overall function of oxidative phosphorylation. Consistent with the coupling observed when measuring cAMP levels, respiratory chain activity was stimulated by the PDE2A inhibitors EHNA and BAY60, but not by the PDE4 inhibitor rolipram (Fig. 4B). This effect was blocked by inclusion of the sAC inhibitor KH7, confirming that PDE2A modulates the activity of the electron transport chain via the CO2/bicarbonate-regulated intramitochondrial sAC microdomain (8). Analogous changes to cAMP levels and mitochondrial function were observed with mouse liver mitochondria treated with BAY60/EHNA (data not shown), affirming the generality of a functional coupling between mitochondrial sAC and PDE2A2. Consistently, respiration in isolated brain mitochondria monitored directly by measuring oxygen consumption was specifically stimulated by the PDE2A inhibitors EHNA or BAY60 (Fig. 4C). We thus conclude that PDE2A and sAC comprise a general intramitochondrial cAMP signaling cascade that determines matrix cAMP levels, which modulate respiratory chain activity (Fig. 5).
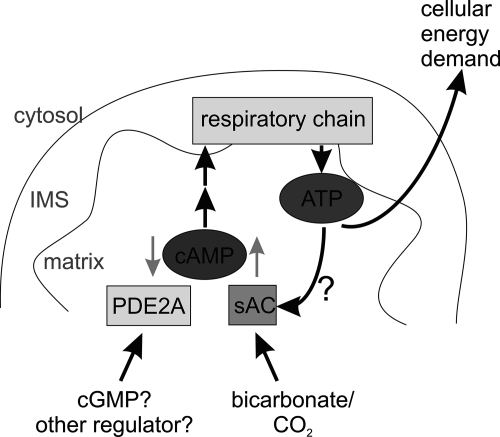
FIGURE 5.
Model for the mitochondrial cyclic nucleotide signaling system. cAMP is formed inside the mitochondrial matrix by a sAC isoform and stimulates the respiratory chain. The cAMP signals are terminated through degradation by PDE2A isoform 2. Thus, formation of cAMP is stimulated by bicarbonate, which activates sAC, and degradation is stimulated by cGMP or a yet to be identified regulatory ligand for the PDE2A2 GAF domains. IMS, intermembrane space.
DISCUSSION
Cyclic nucleotide signaling systems consisting of cyclases, PDEs, and effectors are long known to be highly tissue- and cell type-specific (18). For example, a selective expression and localization of PDE isoforms was reported in neurons (48, 49), and accumulating evidence indicates that defined subcellular cNMP signaling domains exist within a cell (17, 50). The more than 100 PDE isoforms resulting from differential expression and splicing of 11 PDE gene families (17) likely contribute to such specialized function and localization, but details are not established for most of the isoforms. For example, fractionation of rat brain (51) revealed the highest PDE activity in the plasma membrane fraction, but significant activity was also found in the mitochondrial and microsomal fractions. However, the responsible isoforms and their exact locations remain unknown. More recently, AKAP distribution throughout cells (52) and the observation that sAC and cAMP effector proteins reside inside the nucleus and in mitochondria (8, 10, 12, 53) have highlighted the importance of cyclic nucleotides inside cellular organelles. Our finding that a specific PDE isoform, PDE2A2, co-localizes with these proteins in the mitochondrial matrix now confirms that a complete cAMP signaling system exists in this cellular subcompartment.
PDE2A2 was previously suggested to be associated with membranes through its unique N terminus and thus to be responsible, together with PDE2A3, for the PDE2A activity in the particulate fraction of brain homogenates (24). In the case of isoform 3, the membrane association is due to myristoylation, targeting the isoform to synaptosomes (34). Our results indicate that PDE2A2 is found in the particulate fraction due to its import into the mitochondrial matrix. Thus, alternative splicing of PDE2A directs distinct subcellular localization, enabling assembly of independent, localized cNMP signaling domains. During import, PDE2A2 might be proteolytically processed, as suggested by a putative processing site RRG-QQ and the resulting N-terminal Gln, which is the third most abundant N-terminal residue resulting from mitochondrial processing (44). Consistent with the N terminus being more flexible, as typically observed for localization sequences, the PDE2A fragment whose structure was recently solved contained the whole protein except for the N terminus (54).
PDE2A2 forms a mitochondrial signaling domain with sAC, regulating the activity of the respiratory chain (Fig. 5). The stimulatory allosteric effect of bicarbonate on sAC (14, 55) makes oxidative phosphorylation dependent on the activity of the major upstream pathway, the citric acid cycle. PDE2A contains in its regulatory portion tandem GAF domains (first identified in cGMP-regulated PDEs, adenylyl cyclases from Anabaena, transcription factor FhlA), which would allow other signals to modulate this system and hence ATP production. GAF domains are a diverse and widespread family of sensor domains, but in mammals, they occur solely in PDEs and bind cyclic nucleotides (56). In the case of PDE2A, cGMP binding to a GAF domain activates the enzyme (40, 57). Due to its ability to degrade cAMP and cGMP, PDE2A thus can act as a node in the cross-talk between the two cyclic nucleotides (e.g. in the heart (58)). However, to our knowledge, neither occurrence nor functions for cGMP in mitochondria have ever been postulated. Therefore, it remains possible that the GAF domains in mitochondrial PDE2A2 are regulated by other ligands. In fact, cNMP-mediated regulation of the GAF-containing PDE10A and PDE11A has not been reported since their discovery almost 10 years ago, despite extensive efforts (59). Furthermore, a variety of other ligands have been reported for GAF domains in proteins other than PDEs (56), fostering such a speculation on other small molecule PDE regulators acting through the GAF domains. On the other hand, cGMP was long assumed not to exist in prokaryotes, but it was recently identified in cyanobacteria (60, 61), which are assumed to have a common progenitor with mammalian mitochondria (62). It is tempting to speculate that both second messengers, cGMP and cAMP, might be conserved in this organelle, but it remains an exciting question. Does mitochondrial cGMP exist? Also, are there other ligands for the regulatory domains of mitochondrial PDE2A2?
We identified PDE2A2 as a novel regulator of mitochondrial cAMP signals that modulate oxidative phosphorylation. Several other mitochondrial processes also appear to involve cNMPs, such as stress responses and apoptosis initiation (12, 16). It is likely that further mitochondrial PDEs exist, providing additional regulatory domains to influence the cNMP signals that regulate these processes. Our results that cGMP stimulated mitochondrial PDE activity only 2–3-fold and that mitochondrial PDE activity was only partially suppressed with IBMX and BAY60 suggest that additional PDE isoforms exist in mitochondria. For PDE4, attachment to insoluble parts of cell homogenate was reported previously (63), and our inhibition results indicate that PDE4 might partially contribute to the IBMX-sensitive PDE activity in brain mitochondria lysates. Our functional data (Fig. 4, B and C), however, show that PDE4 does not contribute to the cAMP signaling microdomain that regulates respiration. PDE4 shows a unique intracellular distribution through interaction with anchoring proteins, such as AKAPs (18) and the cytosolic domain of the mitochondrial outer membrane DISC1 complex (25), and we thus speculate that PDE4 is located on the outside of brain mitochondria rather than inside the matrix. However, our data indicate that yet another PDE was present in mitochondrial lysates from both brain and liver. The broad spectrum PDE inhibitor IBMX (64) effectively suppressed mitochondrial cGMP-hydrolyzing activity but failed to inhibit 30–50% of cAMP-hydrolyzing activity. These data indicate the presence of an IBMX-insensitive and a cAMP-specific isoform, a characterization that strongly hints at PDE8. However, localization within mitochondria for PDE8 or any other generic PDE or a non-generic cNMP-degrading enzyme (17, 19, 20) remains to be shown. Identifying additional PDEs would reveal further inputs modulating the emerging mitochondrial cNMP signaling network, and it will be interesting to see whether these PDEs contribute to regulation of respiration or other mitochondrial functions.
Many established cytosolic signaling mechanisms, such as protein deacetylation and cNMP signaling, are now also identified in mitochondria, revealing novel drug targets for modulating mitochondrial function (9). Mitochondrial deacetylases, for example, contribute to physiological and pathological processes (3, 65, 66) and are considered attractive therapeutic targets for metabolic and aging-related diseases (67). Modulation of PDEs by using small molecules is even better established and PDEs are widely used as drug targets (27, 31). Our findings that PDE2A2 localizes to the mitochondrial matrix and that its inhibition stimulates oxidative phosphorylation suggest an exciting inroad for generating specific drugs modulating mitochondrial functions by targeting existing PDE inhibitors, through appropriate modifications (68), to mitochondria. Such compounds would help in the exciting challenge to further characterize the mitochondrial cNMP signaling system, its connection to other systems, and its potential therapeutic use.
Acknowledgments
We gratefully acknowledge the technical assistance of Arkadius Pacha and Gina Boanca.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM088999 (to G. M.) and R01 GM62328 and NS552555 (to L. R. L. and J. B.). This work was also supported by grants from the Muscular Dystrophy Association (to G. M.), the United Mitochondrial Disease Foundation (to R. A.-P.), Deutsche Forschungsgemeinschaft (Grant Ko1157) (to M. R.), and Forschungsreferat Ruhr-Universität Bochum Medizin (Grant F630-2008) (to M. R. and C. S.).
- AKAP
- A-kinase anchoring protein
- COX
- cytochrome c oxidase
- EHNA
- erythro-9-(2-hydroxyl-3-nonyl)adenine
- PDE
- phosphodiesterase
- PK
- proteinase K
- Map
- mitochondrial associated protein
- SAC
- soluble adenylyl cyclase
- ICQ
- intensity correlation quotient
- IBMX
- 3-isobutyl-1-methylxanthin
- 8-Br-cAMP
- 8-bromoadenosine-3′, 5′-cyclic monophosphate
- Rr
- Pearson's correlation coefficient.
REFERENCES
- 1. Balaban R. S., Nemoto S., Finkel T. (2005) Cell 120, 483–495 [DOI] [PubMed] [Google Scholar]
- 2. Goldenthal M. J., Marín-García J. (2004) Mol. Cell. Biochem. 262, 1–16 [DOI] [PubMed] [Google Scholar]
- 3. Schlicker C., Hall R. A., Vullo D., Middelhaufe S., Gertz M., Supuran C. T., Mühlschlegel F. A., Steegborn C. (2009) J. Mol. Biol. 385, 1207–1220 [DOI] [PubMed] [Google Scholar]
- 4. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 5. Gertz M., Fischer F., Wolters D., Steegborn C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5705–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gertz M., Steegborn C. (2010) Antioxid. Redox Signal. 13, 1417–1428 [DOI] [PubMed] [Google Scholar]
- 7. Paulsen C. E., Carroll K. S. (2010) ACS Chem. Biol. 5, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Acin-Perez R., Salazar E., Kamenetsky M., Buck J., Levin L. R., Manfredi G. (2009) Cell Metab. 9, 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lakshminarasimhan M., Steegborn C. (2011) Exp. Gerontol. 46, 174–177 [DOI] [PubMed] [Google Scholar]
- 10. Sardanelli A. M., Signorile A., Nuzzi R., Rasmo D. D., Technikova-Dobrova Z., Drahota Z., Occhiello A., Pica A., Papa S. (2006) FEBS Lett. 580, 5690–5696 [DOI] [PubMed] [Google Scholar]
- 11. Cammarota M., Paratcha G., Bevilaqua L. R., Levi de Stein M., Lopez M., Pellegrino de Iraldi A., Izquierdo I., Medina J. H. (1999) J. Neurochem. 72, 2272–2277 [DOI] [PubMed] [Google Scholar]
- 12. Ryu H., Lee J., Impey S., Ratan R. R., Ferrante R. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13915–13920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamenetsky M., Middelhaufe S., Bank E. M., Levin L. R., Buck J., Steegborn C. (2006) J. Mol. Biol. 362, 623–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y., Cann M. J., Litvin T. N., Iourgenko V., Sinclair M. L., Levin L. R., Buck J. (2000) Science 289, 625–628 [DOI] [PubMed] [Google Scholar]
- 15. Zippin J. H., Levin L. R., Buck J. (2001) Trends Endocrinol. Metab. 12, 366–370 [DOI] [PubMed] [Google Scholar]
- 16. Kumar S., Kostin S., Flacke J. P., Reusch H. P., Ladilov Y. (2009) J. Biol. Chem. 284, 14760–14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conti M., Beavo J. (2007) Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 18. Omori K., Kotera J. (2007) Circ. Res. 100, 309–327 [DOI] [PubMed] [Google Scholar]
- 19. Middelhaufe S., Garzia L., Ohndorf U. M., Kachholz B., Zollo M., Steegborn C. (2007) Biochem. J. 407, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. D'Angelo A., Garzia L., André A., Carotenuto P., Aglio V., Guardiola O., Arrigoni G., Cossu A., Palmieri G., Aravind L., Zollo M. (2004) Cancer Cell 5, 137–149 [DOI] [PubMed] [Google Scholar]
- 21. Mullershausen F., Russwurm M., Thompson W. J., Liu L., Koesling D., Friebe A. (2001) J. Cell Biol. 155, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosman G. J., Martins T. J., Sonnenburg W. K., Beavo J. A., Ferguson K., Loughney K. (1997) Gene 191, 89–95 [DOI] [PubMed] [Google Scholar]
- 23. Sonnenburg W. K., Mullaney P. J., Beavo J. A. (1991) J. Biol. Chem. 266, 17655–17661 [PubMed] [Google Scholar]
- 24. Yang Q., Paskind M., Bolger G., Thompson W. J., Repaske D. R., Cutler L. S., Epstein P. M. (1994) Biochem. Biophys. Res. Commun. 205, 1850–1858 [DOI] [PubMed] [Google Scholar]
- 25. Millar J. K., Pickard B. S., Mackie S., James R., Christie S., Buchanan S. R., Malloy M. P., Chubb J. E., Huston E., Baillie G. S., Thomson P. A., Hill E. V., Brandon N. J., Rain J. C., Camargo L. M., Whiting P. J., Houslay M. D., Blackwood D. H., Muir W. J., Porteous D. J. (2005) Science 310, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 26. Acin-Perez R., Salazar E., Brosel S., Yang H., Schon E. A., Manfredi G. (2009) EMBO Mol. Med. 1, 392–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeon Y. H., Heo Y. S., Kim C. M., Hyun Y. L., Lee T. G., Ro S., Cho J. M. (2005) Cell. Mol. Life Sci. 62, 1198–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernández-Vizarra E., López-Pérez M. J., Enriquez J. A. (2002) Methods 26, 292–297 [DOI] [PubMed] [Google Scholar]
- 29. Fischer L. R., Igoudjil A., Magrané J., Li Y., Hansen J. M., Manfredi G., Glass J. D. (2011) Brain 134, 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jäger R., Schwede F., Genieser H. G., Koesling D., Russwurm M. (2010) Br. J. Pharmacol. 161, 1645–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bender A. T., Beavo J. A. (2006) Pharmacol. Rev. 58, 488–520 [DOI] [PubMed] [Google Scholar]
- 32. Juilfs D. M., Soderling S., Burns F., Beavo J. A. (1999) Rev. Physiol Biochem. Pharmacol. 135, 67–104 [DOI] [PubMed] [Google Scholar]
- 33. Domańska G., Motz C., Meinecke M., Harsman A., Papatheodorou P., Reljic B., Dian-Lothrop E. A., Galmiche A., Kepp O., Becker L., Günnewig K., Wagner R., Rassow J. (2010) PLoS Pathog. 6, e1000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russwurm C., Zoidl G., Koesling D., Russwurm M. (2009) J. Biol. Chem. 284, 25782–25790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Q., Lau A., Morris T. J., Guo L., Fordyce C. B., Stanley E. F. (2004) J. Neurosci. 24, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vives-Bauza C., Yang L., Manfredi G. (2007) Methods Cell Biol. 80, 155–171 [DOI] [PubMed] [Google Scholar]
- 37. Hofhaus G., Shakeley R. M., Attardi G. (1996) Methods Enzymol. 264, 476–483 [DOI] [PubMed] [Google Scholar]
- 38. Birch-Machin M. A., Turnbull D. M. (2001) Methods Cell Biol. 65, 97–117 [DOI] [PubMed] [Google Scholar]
- 39. Boess F. G., Hendrix M., van der Staay F. J., Erb C., Schreiber R., van Staveren W., de Vente J., Prickaerts J., Blokland A., Koenig G. (2004) Neuropharmacology 47, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 40. Beavo J. A., Hardman J. G., Sutherland E. W. (1971) J. Biol. Chem. 246, 3841–3846 [PubMed] [Google Scholar]
- 41. Yamamoto T., Manganiello V. C., Vaughan M. (1983) J. Biol. Chem. 258, 12526–12533 [PubMed] [Google Scholar]
- 42. Repaske D. R., Corbin J. G., Conti M., Goy M. F. (1993) Neuroscience 56, 673–686 [DOI] [PubMed] [Google Scholar]
- 43. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. (2009) Cell 139, 428–439 [DOI] [PubMed] [Google Scholar]
- 45. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 46. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 47. Papatheodorou P., Domańska G., Oxle M., Mathieu J., Selchow O., Kenny B., Rassow J. (2006) Cell. Microbiol. 8, 677–689 [DOI] [PubMed] [Google Scholar]
- 48. Juilfs D. M., Fülle H. J., Zhao A. Z., Houslay M. D., Garbers D. L., Beavo J. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3388–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bender A. T., Beavo J. A. (2004) Neurochem. Int. 45, 853–857 [DOI] [PubMed] [Google Scholar]
- 50. Bundey R. A., Insel P. A. (2004) Sci. STKE 2004, pe19. [DOI] [PubMed] [Google Scholar]
- 51. Azila N., Kuppusamy U. R., Ong K. K. (1989) Biochem. Int. 19, 1077–1085 [PubMed] [Google Scholar]
- 52. Welch E. J., Jones B. W., Scott J. D. (2010) Mol. Interv. 10, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zippin J. H., Farrell J., Huron D., Kamenetsky M., Hess K. C., Fischman D. A., Levin L. R., Buck J. (2004) J. Cell Biol. 164, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., Menniti F. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Steegborn C., Litvin T. N., Levin L. R., Buck J., Wu H. (2005) Nat. Struct. Mol. Biol. 12, 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zoraghi R., Corbin J. D., Francis S. H. (2004) Mol. Pharmacol. 65, 267–278 [DOI] [PubMed] [Google Scholar]
- 57. Martinez S. E., Bruder S., Schultz A., Zheng N., Schultz J. E., Beavo J. A., Linder J. U. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zaccolo M., Movsesian M. A. (2007) Circ. Res. 100, 1569–1578 [DOI] [PubMed] [Google Scholar]
- 59. Matthiesen K., Nielsen J. (2009) Biochem. J. 423, 401–409 [DOI] [PubMed] [Google Scholar]
- 60. Ochoa De Alda J. A., Ajlani G., Houmard J. (2000) J. Bacteriol. 182, 3839–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rauch A., Leipelt M., Russwurm M., Steegborn C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hedges S. B., Chen H., Kumar S., Wang D. Y., Thompson A. S., Watanabe H. (2001) BMC Evol. Biol. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jin S. L., Bushnik T., Lan L., Conti M. (1998) J. Biol. Chem. 273, 19672–19678 [DOI] [PubMed] [Google Scholar]
- 64. Wang H., Yan Z., Yang S., Cai J., Robinson H., Ke H. (2008) Biochemistry 47, 12760–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 66. Huang J. Y., Hirschey M. D., Shimazu T., Ho L., Verdin E. (2010) Biochim. Biophys. Acta 1804, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 67. Lavu S., Boss O., Elliott P. J., Lambert P. D. (2008) Nat. Rev. Drug Discov. 7, 841–853 [DOI] [PubMed] [Google Scholar]
- 68. Yousif L. F., Stewart K. M., Kelley S. O. (2009) Chembiochem 10, 1939–1950 [DOI] [PubMed] [Google Scholar]