Abstract
Protein S-nitrosylation is a reversible protein modification implicated in both physiological and pathophysiological regulation of protein function. In obesity, skeletal muscle insulin resistance is associated with increased S-nitrosylation of insulin-signaling proteins. However, whether adipose tissue is similarly affected in obesity and, if so, what are the causes and functional consequences of increased S-nitrosylation in this tissue are unknown. Total protein S-nitrosylation was increased in intra-abdominal adipose tissue of obese humans and in high fat-fed or leptin-deficient ob/ob mice. Both the insulin receptor β-subunit and Akt were S-nitrosylated, correlating with body weight. Elevated protein and mRNA expression of inducible NO synthase and decreased protein levels of thioredoxin reductase were associated with increased adipose tissue S-nitrosylation. Cultured differentiated pre-adipocyte cell lines exposed to the NO donors S-nitrosoglutathione (GSNO) or S-nitroso-N-acetylpenicillamine exhibited diminished insulin-stimulated phosphorylation of Akt but not of GSK3 nor of insulin-stimulated glucose uptake. Yet the anti-lipolytic action of insulin was markedly impaired in both cultured adipocytes and in mice injected with GSNO prior to administration of insulin. In cells, impaired ability of insulin to diminish phosphorylated PKA substrates in response to isoproterenol suggested impaired insulin-induced activation of PDE3B. Consistently, increased S-nitrosylation of PDE3B was detected in adipose tissue of high fat-fed obese mice. Site-directed mutagenesis revealed that Cys-768 and Cys-1040, two putative sites for S-nitrosylation adjacent to the substrate-binding site of PDE3B, accounted for ∼50% of its GSNO-induced S-nitrosylation. Collectively, PDE3B and the anti-lipolytic action of insulin may constitute novel targets for increased S-nitrosylation of adipose tissue in obesity.
Keywords: Adipocyte, Adipose Tissue Metabolism, Lipolysis, Obesity, Oxidative Stress
Introduction
Protein S-nitrosylation, the covalent attachment of NO to cysteine residues in the presence of O2 to form S-nitrosothiol adducts, is increasingly recognized to constitute a prevalent protein modification regulating the function and stability of proteins (1, 2). Like other chemical reactions involving reactive oxygen and nitrogen species, protein S-nitrosylation occurs under normal physiological conditions, but when augmented it can contribute to pathophysiology (3). Being a reversible modification, increased S-nitrosylation occurs as a consequence of increased NO generation by nitric-oxide synthases (NOS), and/or by a diminution of de-nitrosylation processes (4). The inducible NO synthase (iNOS),3 which is a high capacity NOS expressed in inflammatory cells and in other cell types following exposure to inflammatory signals (5), is strongly associated with increased (pathophysiological) levels of protein S-nitrosylation (6, 7). In obesity, a condition now recognized to be associated with increased systemic and local inflammation (8), increased iNOS expression has been documented in insulin-sensitive tissues in both rodents and humans (9–11). In skeletal muscle, the ensuing increase in protein S-nitrosylation, specifically of proteins along the insulin-signaling cascade, was proposed to contribute to the induction of insulin resistance (12). These include the insulin receptor β-subunit (IRβ), insulin receptor substrate 1 (IRS-1), and Akt. S-Nitrosylation of IRβ and Akt in muscle attenuates their kinase activities, and S-nitrosylation of IRS-1 reduces its tissue expression (11, 12). Prevention of S-nitrosylation of these proteins by decreasing iNOS expression improved insulin action (12). Consistently, targeted disruption of iNOS prevents whole-body and skeletal muscle insulin resistance in high fat-fed (HFF) mice (13). Moreover, lipopolysaccharide (LPS)-induced skeletal muscle insulin resistance and increased S-nitrosylation of IRβ and of IRS-1 were also prevented in iNOS−/− mice, suggesting the importance of iNOS-mediated S-nitrosylation in inflammation-induced muscle insulin resistance (14). Finally, direct induction of protein S-nitrosylation by administration of pharmacological NO donors like GSNO or SNAP in nonobese mice also induced muscular insulin resistance, providing compelling evidence for the importance of increased S-nitrosylation as a mediator in the induction of insulin resistance in muscle (14, 15). Whether increased S-nitrosylation occurs in obesity in other major sites of insulin action, like adipose tissue, and what are the sources and functional consequences of S-nitrosylation remain largely unexplored.
Whereas the main physiological function of insulin in skeletal muscle is to induce glucose uptake and utilization, in adipose tissue (where these functions are also observed), the inhibition of lipolysis is particularly important for whole-body metabolic regulation. This effect of insulin is achieved via its classical “metabolic signaling cascade” involving insulin receptor (IRSs), PI3K, and Akt, the latter activating phosphodiesterase 3B (PDE-3B) via phosphorylation on Ser-273 (16). PDE3B then reduces cellular cAMP levels, thereby diminishing the lipolytic activation mediated by PKA-induced regulation of various lipases and lipid droplet-associated proteins, such as the phosphorylation of hormone-sensitive lipase (HSL) and perilipin (17). In obesity, dysregulated lipolysis results in elevated levels of free fatty acids, which in turn contribute to impaired glucose and lipid metabolism in skeletal muscle and liver leading to insulin resistance (18).
In this study, we hypothesized that increased S-nitrosylation occurs in adipose tissue in obesity and may contribute to obesity-associated insulin resistance of adipocytes. Given the importance of dysregulated lipolysis in the pathogenesis of obesity-associated metabolic dysfunction, we assessed whether increased S-nitrosylation in adipocytes specifically targets the anti-lipolytic action of insulin. Our results suggest this is indeed the case and that PDE3B may constitute a specific target of increased S-nitrosylation in adipocytes.
EXPERIMENTAL PROCEDURES
Materials
Tissue culture medium, serum, antibiotic solutions, and recombinant human insulin were obtained from Biological Industries (Beit-Haeemek, Israel). Anti-phosphotyrosine (4G10) antibody was used at a 1:4000 dilution (Transduction Laboratories); anti-Ser(P)-473 PKB/Akt, anti-PKB/Akt, anti-pPKB/Akt substrates, anti-pPKA substrates, anti-Ser(P)-21/9 GSK-3α/β, anti-GSK3α/β, and anti-IRβ antibodies (all from Cell Signaling, Beverly, MA) were used at 1:1000 dilution; anti-iNOS, anti-eNOS, anti-nNOS, and anti-thioredoxin reductase 1 (Abcam, San Francisco, CA) were used at 1:1000 dilution; anti-IRS-1 and anti-IRS-2 antibodies (Upstate Biotechnology, Inc.) were used at 1 μg/ml dilution; anti-β-actin was used at a 1:5000 dilution and anti-FLAG was used at a 1:1000 dilution (both from Sigma); anti-perilipin and anti-HSL antibodies were generated as described previously (19). The NO donor GSNO was from Sigma, and SNAP was purchased from Alexis (San Diego). Murine PDE3B FLAG-tagged wild-type plasmid was kindly provided by Prof. Vincent Manganiello, Translation Medicine Branch, National Institutes of Health, Bethesda. Anti-PDE3B antibody was kindly provided by Prof. Eva Degerman, Lund University, Sweden.
Human Samples
All procedures were approved in advance by the Soroka University Medical Center Institutional Review Committee, and patients gave written informed consent prior to all procedures. Intra-abdominal (omental) fat biopsies were obtained from obese (body mass index (BMI) ≥30 kg/m2) or nonobese BMI ≤26 kg/m2) men and women undergoing elective laparoscopic abdominal surgeries, mainly bariatric surgery or cholecystectomy, at the Soroka University Medical Center (Beer-Sheva, Israel) as described and characterized elsewhere (20–22). Biopsies were immediately transferred to the laboratory and frozen in liquid nitrogen until further analyzed.
Animals and Treatments
The study was approved by Ben-Gurion University Institutional Animal Care and Use Committee and was conducted according to the Israeli Animal Welfare Act following the guidelines of the Guide for Care and Use of Laboratory Animals (National Research Council, 1996). Male C57BL/6 mice and ob/ob mice were purchased from Harlan Laboratories (Rehovot, Israel). Animals were allowed free access to standard rodent chow and water. Diet-induced obesity was achieved by feeding 6-week-old wild-type mice a high fat diet for 19 weeks. The high fat diet consisted of 58.7% calories derived from fat, 25.5% from carbohydrate, and 15% from protein (TD.88137, Harlan Teklad, Madison, WI). At the end of the experiment, mice were killed with CO2. Peri-epididymal fat pads were removed and immediately frozen in liquid nitrogen until further analysis. Acute treatment with GSNO (1.25 mg) was performed by intraperitoneal injection 30 min before insulin was administered. Plasma free fatty acids (FFA) and glucose levels were measured 15 min later, as we described previously (23), using the ACS-ACOD-MEHA method kit (Wako Chemicals GmbH, Neuss, Germany) and FreeStyle glucometer (Abbott), respectively.
Cell Culture and Adipocyte-Macrophage Co-culture
Differentiated mouse preadipocytes (either 3T3-L1 (ATCC, Manassas, VA) or epididymal preadipocyte cell line (24)) were grown and differentiated to mature adipocytes as we described previously (24, 25). After being serum-starved for 24 h, cultured adipocytes were pretreated with or without 0.5 mm SNAP or 1 mm GSNO for an additional 24 h. Insulin stimulation was performed with 100 nm insulin for 10 min. The macrophage cell line RAW264.7 was obtained from ATCC (Manassas, VA) and cultured in RPMI 1640 medium supplemented with l-glutamine (2 mm), heat-inactivated FBS (10%), and antibiotics. Co-culture of adipocytes and macrophages was performed by trypsinizing RAW cells grown on a 15-cm plate onto a 15-cm dish of confluent differentiated epididymal adipocytes. The cells were co-cultured for an additional 24 h and then stimulated (where indicated) with 1 μm LPS for an additional 24 h. Control experiments for the co-culture were performed by mixing lysates from adipocytes and macrophages grown separately, each derived from a 15-cm dish.
Total and Specific Protein S-Nitrosylation
S-Nitrosylation of proteins was detected using the biotin-switch technique as described previously (26), with minor modifications. In this procedure free sulfhydryls are first blocked, followed by reducing S-NO and reacting them with biotin, which was then identified. Fat tissue (∼100–500 mg) or cultured cells (∼1500 μg of protein) were extracted with ice-cold lysis buffer containing 25 mm Hepes, 50 mm NaCl, 1% Nonidet P-40, 0.5 mm PMSF, and protease inhibitors (a 1:1000 dilution of protease inhibitor mixture; Sigma). The extracts were adjusted to 0.5 mg/ml protein with HEN buffer (100 mm Hepes, 1 mm EDTA, 0.1 mm neocuproine) followed by addition of freshly prepared S-methyl methanethiosulfonate (10% v/v in N,N-dimethylformamide) and SDS (25% v/v) to final concentrations of 0.1 and 2.5%, respectively. Following frequent vortexing at 50 °C for 20 min, proteins were precipitated with 3 volumes of acetone at −20 °C for 1 h. The proteins were recovered by centrifugation at 5000 × g for 5 min, followed by gentle rinsing of the pellet with 4× 1 ml of 70% acetone/H2O. The pellets were then resuspended in 240 μl of HEN buffer containing 1% SDS. For labeling, the blocked samples were mixed with 0.1 volume of biotin-HPDP (2.5 mg/ml in Me2SO) and 0.1 volume (20 mm) of freshly prepared sodium ascorbate in HEN buffer. Labeling reactions were performed in the dark at room temperature for 1 h. For direct detection of biotinylated proteins, 40–60 μg of each labeling reaction was resolved by nonreducing SDS-PAGE, followed by immunoblotting with anti-biotin antibody (Sigma, 1:500). To detect an individual S-nitrosylated (SNO) protein from lysates, the labeling reaction was acetone-precipitated as described previously. The washed pellet was resuspended in 250 μl of HEN/10 (HEN diluted 10-fold into H2O) containing 1% SDS, followed by addition of 750 μl of neutralization buffer (25 mm Hepes, 100 mm NaCl, 1 mm EDTA, 1% Triton X-100, pH 7.5). This material was incubated overnight at 4 °C with 50 μl of a streptavidin-agarose slurry. The beads were washed with 4× 1 ml of wash buffer (neutralization buffer + 500 mm NaCl), followed by 2× 1 ml of neutralization buffer. The dried beads were eluted with 50 μl of HEN/10 + 1% β-mercaptoethanol at room temperature for 20 min. The eluted mixture was then analyzed by SDS-PAGE, followed by immunoblotting with specific protein antibody (anti-Akt, IRβ, PDE3B, and FLAG Ab).
To optimize the detection of endogenous PDE3B-SNO, specific minor modifications to the assay were used based on optimization experiments as follows: (i) starting material was 8 mg of total adipose tissue protein; (ii) sodium ascorbate was used at 50 mm instead of 20 mm; and (iii) time of labeling reaction was extended to 3 h.
Site-directed Mutagenesis and Cell Transfection
cDNAs for mutants of FLAG-tagged PDE3B (C275A, C299A, C768A, C777A, and C1040A), in which cysteine was replaced by alanine, were generated by QuikChange II XL site-directed mutagenesis kit (Stratagene, Santa Clara, CA). Human embryonic kidney (HEK) 293T cells (ATCC, Manassas, VA) were grown in DMEM supplemented with 10% heat-inactivated FBS. Cells were transfected using TurboFect in Vitro Transfection Reagent (Fermentas Life Sciences). 48 h later, cells were rinsed three times with PBS and scraped in ice-cold lysis buffer containing 25 mm Hepes, 50 mm NaCl, 1% Nonidet P-40, 0.5 mm PMSF, and protease inhibitors (1:1000 dilution of protease inhibitor mixture; Sigma). The lysates were treated with 0–100 μm GSNO for 30 min in room temperature, and S-nitrosylation of PDE3B was assessed using anti-FLAG Ab, as described above.
Cell Lysates and Western Blot Analysis
After treatments, the cells were rinsed three times with PBS, and proteins were extracted as we described previously (25). Protein concentration was determined using the BCA protein assay (Pierce). Protein samples were resolved on 10% SDS-PAGE and subjected to Western blot, followed by quantification as described previously using ImageGauge software (version 4.0, Fuji Photo Film, Tokyo, Japan) (25).
RNA Extraction and RT-PCR
Total RNA from fat pads was extracted with the RNeasy lipid tissue minikit (Qiagen, Germantown, MD) and quantified with nanodrop. Then 2 μg of RNA were reverse-transcribed with high capacity cDNA reverse transcriptase kit (Applied Biosystems, Foster City, CA). Taqman system (Applied Biosystems) was used for real time PCR amplification. Relative gene expression was obtained after normalization to 18S1 rRNA (Hs0392899_g1) using the formula 2∧ −ΔΔCt. The following primers were used: iNOS (Mm00440502_m1), eNOS (Mm00435217_m1), nNOS (Mm00435175_m1), and Txnrd1 (Mm00443675_m1).
Other Assays
2-Deoxyglucose uptake measurements were performed as described previously (27). Assays were performed for 10 min using 50 μm 2-deoxy[3H]glucose (1 μCi/ml). For lipolysis, adipocytes were stimulated with 5 nmol/liter isoproterenol (Calbiochem) for 1 h as we described previously (25). Insulin (100 nmol/liter) was included 15 min prior to and during the incubation with isoproterenol. Glycerol was measured spectrophotometrically using a glycerol-3-phosphate oxidase Trinder kit (Sigma).
Statistical Analyses
Data are expressed as the means ± S.E. Statistical significance of differences between two groups (treatment versus control) was evaluated using the Student's t test. The criterion for significance was set at p < 0.05. Correlation between S-nitrosylated Akt and IRβ/weight from obese human omental fat explants was done by Pearson's correlation test.
RESULTS
Increased Protein S-Nitrosylation in Adipose Tissue in Obesity
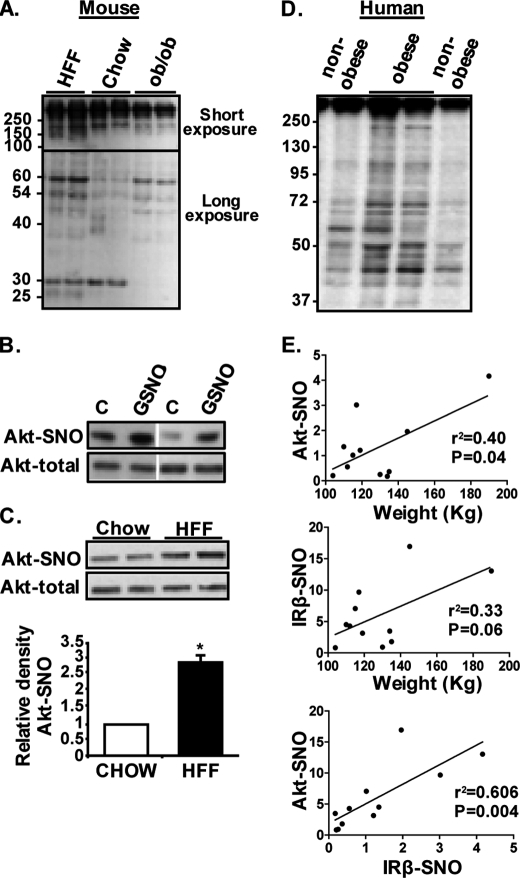
To determine whether obesity is associated with increased protein S-nitrosylation, adipose tissues from nutritional (HFF) and genetic (leptin-deficient, ob/ob) mouse obesity models and human adipose tissue were used. Using the biotin-switch assay (detailed under “Experimental Procedures”), increased total protein S-nitrosylation could be demonstrated in mouse adipose tissue of both HFF and ob/ob models (Fig. 1A). Both models shared highly nitrosylated proteins compared with lean mice (for example, bands at 60, 54, and ∼45 kDa), and occasional bands occurred only in one but not the other model (see as an example a band seen at ∼25 kDa only in HFF mice). The intensity of some bands was decreased in ob/ob mice compared with the other two groups (for example, a band of ∼30 kDa). Specific protein S-nitrosylation could also be detected. Akt was S-nitrosylated (Akt-SNO) in adipose tissue even in control (lean) mice, and its nitrosylation was further increased in response to a single dose of intraperitoneally injected GSNO, an NO donor (Fig. 1B), consistent with reported findings in muscle (12). Moreover, Akt-SNO was ∼2.5-fold higher in adipose tissue of HFF compared with normal chow-fed control mice (Fig. 1C).
FIGURE 1.
Increased protein S-nitrosylation in adipose tissue in obesity. A, total S-nitrosylation in mouse peri-epididymal fat. Shown are representative blots of six normal chow-fed (chow), four HFF, and two 16-week-old ob/ob mice (serving as positive controls). Because higher molecular weight exhibited a stronger signal, shown is a short exposure of the membrane to film for the higher molecular weight (more than ∼100 kDa) and a longer exposure of the same membrane for the lower molecular weight proteins. B, effect of acute administration of the NO donor GSNO on Akt S-nitrosylation in peri-epididymal fat in normal chow-fed mice. Shown are representative blots of two control (vehicle-treated, C) and two GSNO-treated mice. Vertical white line denotes deletion of unrelated bands from the same membrane for clarity of the representation. C, detection of specific S-nitrosylation on Akt from adipose tissue of chow-fed control or HFF mice, using the biotin-switch assay, as detailed under “Experimental Procedures.” Shown are representative blots and quantification of three chow control and six HFF mice. *, p = 0.01. D, total protein S-nitrosylation in human omental adipose tissue biopsies from nonobese (BMI <26 kg/m2, n = 6) and obese (BMI >30 kg/m2, n = 6) assessed by the biotin-switch assay, as described under “Experimental Procedures.” E, S-nitrosylation of specific proteins (Akt or the insulin receptor β-subunit (IRβ)) was detected as in B and quantified by densitometry analysis. Correlations with total body weight and intercorrelations between Akt-SNO and IRβ-SNO are shown.
In human omental adipose tissue, total protein S-nitrosylation was clearly increased in obese compared with nonobese persons (Fig. 1D). Such a notion was evident by both higher intensity of an S-nitrosylation signal from bands detectable in the nonobese or in bands seen only in adipose tissue from obese persons. Consistent with the findings in mouse adipose tissue, S-nitrosylation of Akt could be detected in human adipose tissue (data not shown), and the degree of Akt-SNO correlated with total body weight (Fig. 1E, upper graph). A similar trend was observed with the β-subunit of the insulin receptor, and the amount of Akt-SNO strongly inter-correlated (r2 = 0.606, p = 0.004) with that of IRβ-SNO (Fig. 1E, lower graph), consistent with the total protein S-nitrosylation finding (Fig. 1D).
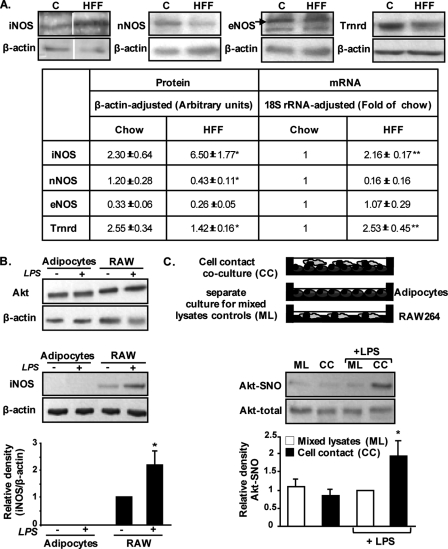
To determine the potential mechanism(s) for increased S-nitrosylation in adipose tissue in obesity, and particularly to assess if it may result from increased NO production and/or diminished de-nitrosylation capacity, we assessed the expression of NOS isoforms and of thioredoxin reductase, respectively. Of the three NOS isoforms, only iNOS was increased at both protein and mRNA levels (Fig. 2A). Thioredoxin reductase, an enzyme that converts oxidized to reduced thioredoxin and thus participates in de-nitrosylation (28), was elevated at the mRNA but decreased at the protein level.
FIGURE 2.
Increased adipose tissue total protein S-nitrosylation in obesity is associated with increased expression of iNOS and decreased expression of thioredoxin reductase 1. A, top panel, representative blots of iNOS, eNOS, nNOS, and thioredoxin reductase 1 (Trnrd) in peri-epididymal fat pads from normal chow (C) or HFF mice (n = 5 per group). Vertical white line denotes splicing of the same membrane for clearer presentation. Bottom panel, results of protein quantification by densitometry analyses of Western blots and of mRNA levels by quantitative real time PCR analyses. Results are means ± S.E. fold change of the respective protein per β-actin over a control sample or relative mRNA levels per 18 S rRNA. *, p < 0.05; **, p < 0.01 versus WT. B, representative blots of Akt and iNOS protein expression in control or LPS (1 μm/ml for 24 h)-treated differentiated epididymal pre-adipocyte cell line or RAW264.7 macrophages. Below the blots are the results of densitometry analyses, expressed as the ratio of iNOS to β-actin, and presented as means ± S.E. of three independent experiments, with a value of 1 given to untreated RAW264.7 cells. *, p < 0.05 versus control. C, effect of LPS and adipocyte-macrophage co-culture on Akt S-nitrosylation (Akt-SNO). RAW264.7 macrophages were seeded on top of cultured adipocyte monolayer, and the contact cell co-culture system (CC) was stimulated as indicated with LPS. As a control, adipocytes and RAW macrophages were grown separately and stimulated separately as indicated with LPS, and lysates prepared from each cell culture were mixed (mixed lysates (ML)), keeping the cell type ratios identical to the CC systems. The amount of Akt-SNO and total Akt was then assessed as described under “Experimental Procedures.” Shown are representative blots and densitometry analyses of four independent experiments. *, p < 0.05 versus LPS-treated mixed lysate controls (ML).
Because adipose tissue is composed of different cell types, the elevated S-nitrosylation observed in the tissue may in fact represent a cross-talk phenomenon between adipocytes and nonadipocytes. In particular, iNOS is shown to be increased in obesity (Fig. 2A) (10, 13) and is a typical hallmark of activated macrophages (29, 30). To address the possibility that increased S-nitrosylation in adipocytes requires activated macrophages, we performed co-culture experiments. When grown separately, both cultured mouse intra-abdominal differentiated pre-adipocytes and RAW264.7 macrophages express Akt, but the expression of iNOS is observed only in the macrophages but not in the adipocyte cell line. In addition, iNOS is further increased by exposing macrophages to LPS (Fig. 2B). As a next step, RAW264.7 macrophages were seeded onto adipocytes (cell contact co-culture, CC). Cells were then stimulated as indicated with LPS, and Akt S-nitrosylation was assessed in lysates of the co-cultured cell population. As a control for CC, cells were grown separately and stimulated as indicated with LPS, and lysates from both cultures were mixed post-cell treatment, as reported previously (31) and as illustrated in Fig. 2C. Notably, a significant 2-fold increase in S-nitrosylated Akt was only observed in adipocyte-macrophage CC culture that was stimulated with LPS (Fig. 2C). The lack of detectable Akt-SNO in the mixed lysates control suggests that stimulated macrophages did not significantly nitrosylate their own Akt. Similar results were obtained in a co-culture apparatus in which macrophages were separated from the adipocytes by a permeable membrane (data not shown), suggesting that a diffusible factor from macrophages is responsible for the increased adipocyte Akt-SNO. Thus, it is reasonable to propose that LPS-induced increase in NO, likely the result of iNOS activation in macrophages, nitrosylated Akt predominantly in the adipocytes. Collectively, these results suggest that increased total protein S-nitrosylation in adipose tissue in obesity is likely contributed by changes in both sides of the S-nitrosylation/de-nitrosylation balance and that cross-talk between macrophages and adipocytes may underlie the augmented S-nitrosylation of specific proteins like Akt.
Increased S-Nitrosylation in Adipocytes Interferes with the Anti-lipolytic Action of Insulin
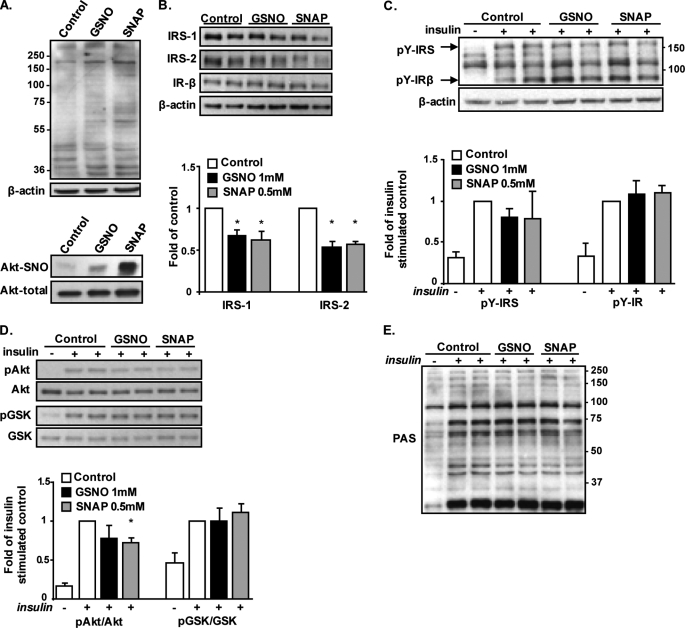
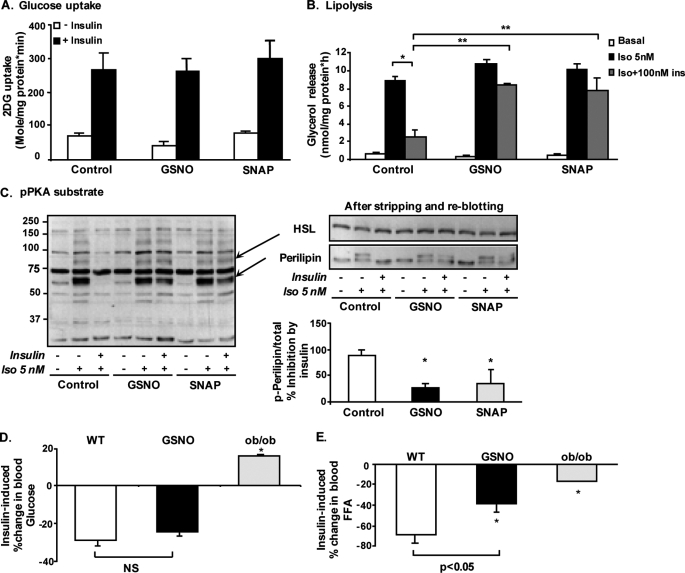
In skeletal muscle increased S-nitrosylation was shown to diminish insulin signaling (12, 15). To determine whether this similarly occurs in adipocytes, we studied the insulin signaling cascade in cultured adipocyte cell lines in which S-nitrosylation was directly induced using the NO donors GSNO and SNAP. Total protein S-nitrosylation and S-nitrosylation of Akt were increased by the two NO donors (Fig. 3A), an effect that was associated with decreased protein contents of IRS-1 and IRS-2 (Fig. 3B). Yet, despite this decrease, insulin-stimulated tyrosine phosphorylation of bands corresponding to the IRSs or to the β-subunit of the insulin receptor was not altered in response to either NO donor (Fig. 3C). Further downstream, the insulin signaling cascade, insulin-stimulated phosphorylation of Akt, was mildly decreased, reaching a statistically significant ∼30% in SNAP-treated adipocytes (Fig. 3D). However, the phosphorylation of GSK3, an Akt substrate involved in the regulation of glycogen synthesis, was unaltered. Moreover, no significant attenuation of Akt-mediated phosphorylation could be observed on additional Akt substrates, as detected by blotting lysates with anti-phospho-Akt substrate antibody, which recognizes phosphorylated proteins on their consensus Akt phosphorylation sequence (RXRXX(S/T) (Fig. 3E) (16, 32). Thus, although mild decreases in IRS protein content and in insulin-stimulated Akt phosphorylation could be observed in adipocytes with increased protein S-nitrosylation, this did not seem to manifest functionally on insulin regulation of several “classical” Akt substrates. Consistently, at the level of metabolic response to insulin, increased S-nitrosylation did not affect the ability of the hormone to stimulate glucose uptake activity (Fig. 4A). Yet, because a major action of insulin in adipocytes is the inhibition of lipolysis, we measured the effect of increased S-nitrosylation also on the anti-lipolytic action of the hormone. As shown in Fig. 4B, isoproterenol, a β-adrenergic agonist, increased lipolysis ∼10-fold in both GSNO- and SNAP-treated cells, as in control adipocytes, and as measured by glycerol release to the medium. Although in control cells insulin inhibited this response by ∼60%, this insulin effect was significantly attenuated by pretreatment of the cells with either GSNO or SNAP. Together, the results indicate that in adipocytes, the increased S-nitrosylation specifically diminishes the anti-lipolytic effect of insulin.
FIGURE 3.
Effect of increased S-nitrosylation by NO donors on insulin signaling in 3T3-L1 adipocytes. Differentiated 3T3-L1 adipocytes were exposed to 1 mm GSNO or 0.5 mm SNAP for 24 h, as detailed under “Experimental Procedures.” A, total S-nitrosylation (top panel) and S-nitrosylation of Akt (bottom panel) were assessed using the biotin-switch assay. B, effect of GSNO or SNAP on protein expression of IRS-1, IRS-2, and the IRβ. Densitometry analyses of IRS-1 and -2 content in five independent experiments are shown and expressed as means ± S.E., with a value of 1 given to the intensity of the band achieved in untreated control cells. *, p < 0.05 versus control. C, following treatments with NO donors, cells were rinsed and incubated in the absence or presence of 100 nm insulin for 10 min. Insulin-stimulated tyrosine phosphorylation was assessed with anti-phosphotyrosine antibody. Densitometry analysis of the phosphotyrosine bands corresponding to the IRS proteins or the insulin receptor β-subunit are shown. D and E, effect of NO donors on insulin-stimulated phosphorylation of Akt and GSK3 (D) or Akt substrates (using PAS antibody, E). Results of densitometry analysis of phosphorylated to total Akt and GSK3 are presented as means ± S.E. of four independent experiments performed in duplicate, with a value of 1 given to the intensity of the band achieved in insulin-stimulated control cells. *, p < 0.05 versus control with insulin.
FIGURE 4.
Increased S-nitrosylation in adipocytes interferes with the anti-lipolytic action of insulin. Differentiated 3T3-L1 adipocytes were exposed to 1 mm GSNO or 0.5 mm SNAP for 24 h. A, following treatments, cells were rinsed and incubated in the absence (□) or presence (■) of 100 nmol/liter insulin for an additional 20 min, after which 2-deoxyglucose (2DG) uptake was measured, as detailed under “Experimental Procedures.” Results are means ± S.E. of three independent experiments, each performed in duplicate. B, following treatments with NO donors, cells were rinsed and incubated in the absence (□) or presence of either 5 nmol/liter isoproterenol (Iso) alone (■) or with 100 nmol/liter insulin ( ) 15 min prior to and then for an additional 1 h in KRBH, after which glycerol release was assayed as described under “Experimental Procedures.” Results are presented as means ± S.E. of three independent experiments performed at least in duplicate. *, p < 0.05 between isoproterenol treated control cells with or without insulin; **, p < 0.005 between GSNO or SNAP pre-treated adipoctyes that were then incubated with insulin + isoproterenol, compared to respective control cells. C, left, representative Western blot analysis of phospho-PKA substrates of three independent experiments. Right, representative Western blot analysis for HSL and perilipin after stripping and re-blotting of the membrane show in the left panel. Results of densitometry analysis of the inhibitory effect by insulin on the intensity of bands representing PKA-mediated phosphorylated perilipin are presented as means ± S.E. of three independent experiments, *, p < 0.05. D and E, C57BL6 mice were injected intraperitoneally with saline (□, n = 7) or GSNO (■, n = 7) 30 min before an additional intraperitoneal injection of 1 IU/kg insulin for 15 min. ob/ob mice served as a positive control for blunted insulin response (
) 15 min prior to and then for an additional 1 h in KRBH, after which glycerol release was assayed as described under “Experimental Procedures.” Results are presented as means ± S.E. of three independent experiments performed at least in duplicate. *, p < 0.05 between isoproterenol treated control cells with or without insulin; **, p < 0.005 between GSNO or SNAP pre-treated adipoctyes that were then incubated with insulin + isoproterenol, compared to respective control cells. C, left, representative Western blot analysis of phospho-PKA substrates of three independent experiments. Right, representative Western blot analysis for HSL and perilipin after stripping and re-blotting of the membrane show in the left panel. Results of densitometry analysis of the inhibitory effect by insulin on the intensity of bands representing PKA-mediated phosphorylated perilipin are presented as means ± S.E. of three independent experiments, *, p < 0.05. D and E, C57BL6 mice were injected intraperitoneally with saline (□, n = 7) or GSNO (■, n = 7) 30 min before an additional intraperitoneal injection of 1 IU/kg insulin for 15 min. ob/ob mice served as a positive control for blunted insulin response ( ). Blood samples were collected before and 15 min after the insulin injection. D, results are presented as means ± S.E. of the percent of change in glucose between time 0 and 15 min after insulin administration. *, p < 0.05 versus saline WT mice. NS, not significant. E, FFA level was determined as described under “Experimental Procedures.” Results are presented as means ± S.E. of the percent of change in FFA with insulin. *, p < 0.05 versus saline WT mice.
). Blood samples were collected before and 15 min after the insulin injection. D, results are presented as means ± S.E. of the percent of change in glucose between time 0 and 15 min after insulin administration. *, p < 0.05 versus saline WT mice. NS, not significant. E, FFA level was determined as described under “Experimental Procedures.” Results are presented as means ± S.E. of the percent of change in FFA with insulin. *, p < 0.05 versus saline WT mice.
The antilipolytic action of insulin in adipocytes occurs by activation of phosphodiesterases, mainly PDE3B, which converts cAMP to AMP (16, 33–35), thereby decreasing PKA-induced phosphorylation that mediates the stimulatory effect of isoproterenol on lipolysis. Using an anti-pPKA substrate antibody (directed against RRX(pS/T)) it was evident that GSNO and SNAP interfered with the ability of insulin to attenuate isoproterenol-induced phosphorylation on PKA substrates (Fig. 4C). By stripping and re-blotting the same membranes, we could verify that this indeed occurred specifically on two major PKA substrates in adipocytes that regulate lipolysis, HSL and the lipid droplet-associated protein perilipin. With the latter, the diminished ability of insulin to negate PKA-induced phosphorylation was also evident by the migration of the perilipin bands, and after treatment with NO donors perilipin still migrated at a higher molecular weight even after insulin stimulation, consistent with its elevated phosphorylation state on PKA sites (Fig. 4C, right panel).
To corroborate these direct effects of increased S-nitrosylation by NO donors in the in vivo setting, we tested whether GSNO blunts insulin-induced anti-lipolysis and/or glucose utilization in wild-type mice. Mice received an intraperitoneal injection of GSNO or vehicle based on a previously published protocol (12), and 30 min later insulin was administered. Glucose and free fatty acid (FFA) levels were measured at times 0 and 15 min of insulin administration, and the percent decrease in the level of either metabolite was calculated. Control mice (wild-type receiving vehicle injection and then insulin) exhibited a nearly 70% drop in FFA (Fig. 4E) and a 30% decrease in base-line glucose levels (Fig. 4D), reflecting insulin's ability to inhibit lipolysis and increase glucose utilization, respectively. In ob/ob mice that develop severe insulin resistance, both metabolic responses to exogenous insulin were markedly impaired. In contrast, wild-type mice who received GSNO prior to insulin administration exhibited a significant resistance to insulin's effect on FFA but not on glucose levels. These results were consistent with the observations in adipocytes in culture, in which GSNO and SNAP inhibited insulin-induced anti-lipolysis but not the stimulation of glucose uptake by the hormone (Fig. 4, A and B). Collectively, these results suggest that the anti-lipolytic action of insulin is specifically targeted in adipose tissue/adipocytes subjected to increased protein S-nitrosylation.
PDE3B May Constitute a Direct Target of S-Nitrosylation in Adipocytes
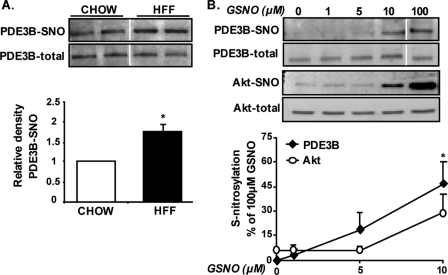
Using the biotin-switch assay, a significant increase in PDE3B-SNO could be detected in adipose tissue of HFF obese mice compared with chow-fed lean controls (Fig. 5A). To further test the likelihood of PDE3B to undergo increased S-nitrosylation in adipocytes, lysates of cultured adipocytes were exposed to increasing doses of GSNO, and the sensitivity of endogenous PDE3B or Akt to SNO modification was determined by the biotin-switch assay. Fig. 5B demonstrates that under basal conditions S-nitrosylation was absent in PDE3B, whereas Akt exhibited a certain level of S-nitrosylation even without exposure to GSNO (consistent with our observations in adipose tissue, Fig. 1, B and C). Moreover, the sensitivity of PDE3B to GSNO-induced S-nitrosylation seemed at least as high as, if not higher, that of Akt; 5 μm GSNO already induced more than 10% of the maximal nitrosylation we observed on PDE3B at higher doses of GSNO, whereas Akt showed no increase above basal level with this concentration. Thus, endogenous PDE3B is more highly nitrosylated in adipose tissue from obese mice, and this protein is at least as sensitive as Akt, a well documented target of increased S-nitrosylation, to this protein modification.
FIGURE 5.
PDE3B is a potential target of S-nitrosylation in adipocytes. A, degree of S-nitrosylation of PDE3B was determined in adipose tissue of HFF obese or chow-fed lean control mice using the biotin-switch assay, with several specific optimizations, as detailed under “Experimental Procedures.” Shown are representative blots and densitometry analysis of results obtained from three chow-fed and six HFF mice, p = 0.04. B, lysates of differentiated epididymal pre-adipocyte cell line were exposed to 0, 1, 5, 10, or 100 μm GSNO for 30 min at room temperature, after which S-nitrosylations of PDE3B and Akt were assessed. Densitometry of PDE3B-SNO and Akt-SNO per their respective total protein is derived from five independent experiments and presented as the percent of intensity achieved with 100 μm GSNO. Vertical white line denotes splicing of the same membrane for clearer presentation.
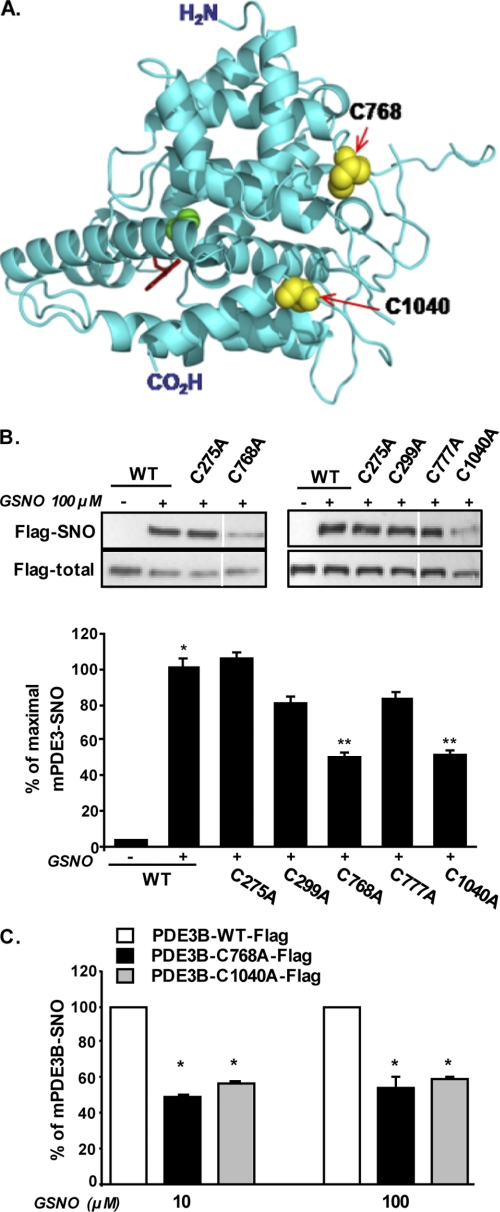
PDE3B has several cysteines in its primary sequence. We chose to assess the contribution of five cysteines to PDE3B S-nitrosylation; Cys-275 and Cys-299 are of interest because they are immediately adjacent to Ser-273 and Ser-296 that are phosphorylation targets of Akt and PKA, respectively (16). Within the catalytic domain, we chose surface-exposed cysteines that were predicted to exist in nonbonding state in the native protein sequence (based on Biocomp.Unibo prediction server) and thus potentially amenable to react with NO. Site-directed mutagenesis was used to individually mutate Cys-275, Cys-299, Cys-768, Cys-777, and Cys-1040 to alanine. Of these residues, Cys-768, Cys-777, and Cys-1040 are the most adjacent to the cAMP (substrate) binding pocket in the catalytic domain of PDE3B (Fig. 6A). HEK 293 cells were transfected with plasmids encoding FLAG-tagged wild-type PDE3B or specific Cys to Ala site mutants, and the level of S-nitrosylation on PDE3B was assessed after exposure to GSNO. As shown in Fig. 6B, wild-type PDE3B showed robust S-nitrosylation when cells were exposed to GSNO, and a similar level of PDE3B-SNO was observed with the C275A mutant. (Fig. 5C). C299A and C777A single site mutants exhibited a small decrease in S-nitrosylation that did not reach statistical significance. Conversely, when either Cys-768 or Cys-1040 was mutated, GSNO-mediated S-nitrosylation of PDE3B was markedly (although not fully) diminished. This effect could be demonstrable with both 10 and 100 μm GSNO (Fig. 6C).
FIGURE 6.
Cys-768 and Cys-1040 are likely major sites for S-nitrosylation of PDE3B. A, ribbon model of the catalytic domain of human PDE3B (Protein Data Bank code 1SOJ (47)), highlighting Cys-768 and Cys-1040 side chains as yellow spheres, and 3-isobutyl-1-methylxanthine molecule at the substrate-binding site is shown by red sticks, and magnesium ions are shown as green balls. (3-Isobutyl-1-methylxanthine is a purine-derived competitive inhibitor of most known phosphodiesterases). B, HEK 293T cells were transfected with plasmids encoding either the wild-type (WT) murine PDE3B or specific cysteine to alanine mutants generated by site-directed mutagenesis as described under “Experimental Procedures.” (C275A, C299A, C768A, C777A, and C1040A). Lysates of transfected cells were exposed to 100 μm GSNO for 30 min at room temperature; S-nitrosylation was performed using the biotin-switch assay, and PDE3B was identified using anti-FLAG antibodies. Results of densitometry analysis are presented as means ± S.E. of five independent experiments, with a value of 100% given to the intensity of the band achieved with lysates of WT-FLAG-PDE3B-expressing cells after GSNO treatment. *, p < 0.05 compared with WT without GSNO. **, p < 0.05 compared with GSNO-treated WT-PDE3B. Vertical white line denotes splicing of the same membrane for clearer presentation. C, HEK 293T cells were transfected with plasmids encoding either the wild-type murine PDE3B or specific site mutants C768A or C1040A. Lysates of transfected cells were exposed to 10 or 100 μm GSNO for 30 min. Results of densitometry analysis are presented as means ± S.E. of five independent experiments, with a value of 100% given to the intensity of the band achieved with lysates of WT-FLAG-PDE3B-expressing cells with GSNO treatment. *, p < 0.05 compared with WT-PDE3B.
DISCUSSION
Increased oxidative stress has been implicated in the pathogenesis of obesity-associated morbidity in general, and recently in adipose tissue dysfunction (36, 37). “Oxidative stress” is a common term for an exceedingly large variety of mediators (reactive oxygen and/or nitrogen species) and targets (proteins, nucleic acids, and lipids) and of different types of chemical alterations (38). Notably, attempts to crudely use antioxidant supplementation as a means of alleviating oxidative stress and thereby improving metabolic functions in obesity and diabetes have proven to be largely disappointing. Better definitions of the specific oxidative and/or nitrative species, their specific cellular target(s), and the specific type of chemical modification(s) such targets undergo are required for more efficient means of alleviating oxidative damage to be designed. This study proposes that PDE3B and/or the Akt-PDE3B interaction are potential novel target(s) for increased S-nitrosylation in adipocytes in obesity, which may interfere with the anti-lipolytic action of insulin and can thus contribute to the development of insulin resistance.
Several oxidative/nitrative modifications of various cellular targets have been shown to be increased in obesity and/or diabetes and to contribute to their pathogenesis, including products of lipid peroxidation (like isoprostanes), DNA oxidation (deoxyguanosine), and protein carbonylation and tyrosine nitration (39–42). Increased S-nitrosylation of proteins is yet an additional specific form of nitrative modification of sulfhydryls. Like several of the modifications mentioned above, reversible S-nitrosylation may constitute a frequent post-translational modification of proteins used to regulate their function under normal physiological conditions, reminiscent of the more established regulation by phosphorylation/de-phosphorylation (4). Indeed, we found that adipose tissue of lean humans and animals, and cells under nonstressed conditions, exhibit a significant degree of total protein S-nitrosylation. Some proteins like Akt have a “basal” level of S-nitrosylation. In such proteins, an increased degree of S-nitrosylation may represent either a larger percentage of molecules nitrosylated on the same cysteine residue(s) or more cysteine residues modified per protein molecule. Indeed, the pattern of S-nitrosylated proteins in adipose tissue of obese humans or mice included bands migrating at the same molecular weight but which exhibit an increased signal. In skeletal muscle, previous studies demonstrated that the degree of S-nitrosylation of Akt regulates its function under both physiological conditions (exercise) and in insulin resistance states (12, 43). Here in adipose tissue and adipocytes, we could show that Akt is nitrosylated basally, but phosphorylation of its classical substrates, a measure of its function, was not markedly diminished in GSNO- or SNAP-treated adipocytes. Consistently, insulin-induced stimulation of glucose uptake that requires proper Akt signaling input was maintained in these cells. However, a different metabolic function of insulin that requires Akt, the inhibition of lipolysis, was indeed impaired by increasing S-nitrosylation. This was either secondary to a modulation of Akt activity toward a specific target (PDE3B) rather than a general inactivation of the enzyme and/or an interference with PDE3B activation by its direct S-nitrosylation. Our findings presented in Fig. 5B (though not Fig. 5A) suggest that PDE3B may not be S-nitrosylated to a significant degree in basal conditions. Indeed, by observing the total pattern of S-nitrosylated proteins in adipose tissue, it is possible to recognize both intensified bands, and bands not observable in lean controls but that appear only under obese conditions. Indeed, by observing the total pattern of S-nitrosylated proteins in adipose tissue, it is possible to recognize both intensified bands, and bands not observable in lean controls but that appear only under obese conditions. In the case of PDE3B, our data implicate two potential cysteine residues, Cys-768 and Cys-1040, as being responsible for a significant portion of the S-nitrosylation capacity of this protein in response to NO donors. Their proximity to the substrate-binding site underscores the theoretical likelihood that their modification may indeed impair PDE3B activity, resulting in blunted anti-lipolytic response of adipocytes to insulin.
The mechanisms for increased S-nitrosylation in adipose tissue in obesity may well involve both increased generation of NO that in turn would modify protein sulfhydryls and diminution of de-nitrosylation processes (44, 45). Of the latter process, thioredoxins are likely major enzymes catalyzing the de-nitrosylation of S-NO moieties on proteins. Their own redox recycling requires thioredoxin reductases, which we show here to be decreased at the protein level in adipose tissue in obesity. Complementarily, processes responsible for increased S-nitrosylation in obesity are more strongly supported, and of the NOS isoforms, iNOS has a severalfold higher capacity to generate NO compared with nNOS and eNOS (46). It is a major gene activated in inflammatory reactions that is now well accepted to be part of the milieu in the obese adipose tissue. We demonstrate here that indeed the combination of adipocytes and macrophages, the latter being the likely contributor of iNOS-mediated NO generation, is required for an LPS-induced increase in Akt S-nitrosylation. iNOS knock-out mice have been shown to be protected from obesity-associated insulin resistance (13). However, given our results that in adipocytes glucose uptake may not be impaired by increasing protein S-nitrosylation, this physiological effect of iNOS knock-out, or of the insulin resistance induced by administering GSNO to mice, is likely the consequence of increased S-nitrosylation (of Akt) in skeletal muscle. Consistent with the unique functions of adipose tissue, our results suggest that the main metabolic consequence of increased S-nitrosylation of adipose tissue proteins in obesity is the impairment of insulin-induced anti-lipolysis. Increased release of FFA from adipose tissue is a hallmark of the obese state and has been thought to play a central role in the pathogenesis of insulin resistance, fatty liver, and dyslipidemia in obesity. Thus, it is tempting to speculate that the means of preventing or reversing increased S-nitrosylation of the Akt-PDE3B axis in adipocytes may assist in alleviating obesity-associated dys-metabolism.
Acknowledgments
We thank Prof. Vincent Manganiello (Translation Medicine Branch, National Institutes of Health, Bethesda) for providing us the plasmid for FLAG-mPDE3B and Prof. Eva Degerman (University of Lund, Sweden) for anti-PDE3B antibody. We also thank Dr. Daniel Konard, University of Zurich, and Dr. Amir Tirosh, Sheba Medical Center, Tel Hashomer, for helpful discussions and critical reading of the manuscript.
This work was supported in part by Israel Science Foundation Grants 118-06 and 1103-09 (to A. R. and N. B.), Israeli Ministry of Health Grant 3/5067, and by the Leslie and Susan Gonda (Goldschmid) Center for Diabetes Research and Education.
- iNOS
- inducible NO synthase
- SNAP
- S-nitroso-N-acetylpenicillamine
- GSNO
- S-nitrosoglutathione
- IRβ
- insulin receptor β
- IRS
- insulin receptor
- BMI
- body mass index
- HSL
- hormone-sensitive lipase
- HFF
- high fat fed
- FFA
- free fatty acid
- eNOS
- endothelial nitric-oxide synthase
- nNOS
- neuronal nitric-oxide synthase
- SNO
- S-nitrosylated
- CC
- cell contact.
REFERENCES
- 1. Stamler J. S., Simon D. I., Osborne J. A., Mullins M. E., Jaraki O., Michel T., Singel D. J., Loscalzo J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stamler J. S., Toone E. J., Lipton S. A., Sucher N. J. (1997) Neuron 18, 691–696 [DOI] [PubMed] [Google Scholar]
- 3. Foster M. W., Hess D. T., Stamler J. S. (2009) Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lane P., Hao G., Gross S. S. (2001) Sci STKE 2001, re1. [DOI] [PubMed] [Google Scholar]
- 5. Nathan C., Xie Q. W. (1994) Cell 78, 915–918 [DOI] [PubMed] [Google Scholar]
- 6. Brüne B., Mohr S., Messmer U. K. (1996) Rev. Physiol. Biochem. Pharmacol. 127, 1–30 [DOI] [PubMed] [Google Scholar]
- 7. Kaneki M., Shimizu N., Yamada D., Chang K. (2007) Antioxid. Redox. Signal. 9, 319–329 [DOI] [PubMed] [Google Scholar]
- 8. Wellen K. E., Hotamisligil G. S. (2005) J. Clin. Invest. 115, 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elizalde M., Rydén M., van Harmelen V., Eneroth P., Gyllenhammar H., Holm C., Ramel S., Olund A., Arner P., Andersson K. (2000) J. Lipid Res. 41, 1244–1251 [PubMed] [Google Scholar]
- 10. Engeli S., Janke J., Gorzelniak K., Böhnke J., Ghose N., Lindschau C., Luft F. C., Sharma A. M. (2004) J. Lipid Res. 45, 1640–1648 [DOI] [PubMed] [Google Scholar]
- 11. Sugita H., Fujimoto M., Yasukawa T., Shimizu N., Sugita M., Yasuhara S., Martyn J. A., Kaneki M. (2005) J. Biol. Chem. 280, 14203–14211 [DOI] [PubMed] [Google Scholar]
- 12. Carvalho-Filho M. A., Ueno M., Hirabara S. M., Seabra A. B., Carvalheira J. B., de Oliveira M. G., Velloso L. A., Curi R., Saad M. J. (2005) Diabetes 54, 959–967 [DOI] [PubMed] [Google Scholar]
- 13. Perreault M., Marette A. (2001) Nat. Med. 7, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 14. Carvalho-Filho M. A., Ueno M., Carvalheira J. B., Velloso L. A., Saad M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E476–E482 [DOI] [PubMed] [Google Scholar]
- 15. Yasukawa T., Tokunaga E., Ota H., Sugita H., Martyn J. A., Kaneki M. (2005) J. Biol. Chem. 280, 7511–7518 [DOI] [PubMed] [Google Scholar]
- 16. Kitamura T., Kitamura Y., Kuroda S., Hino Y., Ando M., Kotani K., Konishi H., Matsuzaki H., Kikkawa U., Ogawa W., Kasuga M. (1999) Mol. Cell. Biol. 19, 6286–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolditz C. I., Langin D. (2010) Curr. Opin. Clin. Nutr. Metab. Care 13, 377–381 [DOI] [PubMed] [Google Scholar]
- 18. Arner P. (2003) Trends Endocrinol. Metab. 14, 137–145 [DOI] [PubMed] [Google Scholar]
- 19. Souza S. C., Yamamoto M. T., Franciosa M. D., Lien P., Greenberg A. S. (1998) Diabetes 47, 691–695 [DOI] [PubMed] [Google Scholar]
- 20. Bashan N., Dorfman K., Tarnovscki T., Harman-Boehm I., Liberty I. F., Blüher M., Ovadia S., Maymon-Zilberstein T., Potashnik R., Stumvoll M., Avinoach E., Rudich A. (2007) Endocrinology 148, 2955–2962 [DOI] [PubMed] [Google Scholar]
- 21. Blüher M., Bashan N., Shai I., Harman-Boehm I., Tarnovscki T., Avinaoch E., Stumvoll M., Dietrich A., Klöting N., Rudich A. (2009) J. Clin. Endocrinol. Metab. 94, 2507–2515 [DOI] [PubMed] [Google Scholar]
- 22. Harman-Boehm I., Blüher M., Redel H., Sion-Vardy N., Ovadia S., Avinoach E., Shai I., Klöting N., Stumvoll M., Bashan N., Rudich A. (2007) J. Clin. Endocrinol. Metab. 92, 2240–2247 [DOI] [PubMed] [Google Scholar]
- 23. Konrad D., Rudich A., Schoenle E. J. (2007) Diabetologia 50, 833–839 [DOI] [PubMed] [Google Scholar]
- 24. Kovsan J., Osnis A., Maissel A., Mazor L., Tarnovscki T., Hollander L., Ovadia S., Meier B., Klein J., Bashan N., Rudich A. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E315–E322 [DOI] [PubMed] [Google Scholar]
- 25. Kovsan J., Ben-Romano R., Souza S. C., Greenberg A. S., Rudich A. (2007) J. Biol. Chem. 282, 21704–21711 [DOI] [PubMed] [Google Scholar]
- 26. Forrester M. T., Foster M. W., Benhar M., Stamler J. S. (2009) Free Radic. Biol. Med. 46, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tirosh A., Potashnik R., Bashan N., Rudich A. (1999) J. Biol. Chem. 274, 10595–10602 [DOI] [PubMed] [Google Scholar]
- 28. Nikitovic D., Holmgren A. (1996) J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 29. Hibbs J. B., Jr., Taintor R. R., Vavrin Z., Rachlin E. M. (1988) Biochem. Biophys. Res. Commun. 157, 87–94 [DOI] [PubMed] [Google Scholar]
- 30. Weinberg J. B., Misukonis M. A., Shami P. J., Mason S. N., Sauls D. L., Dittman W. A., Wood E. R., Smith G. K., McDonald B., Bachus K. E., et al. (1995) Blood 86, 1184–1195 [PubMed] [Google Scholar]
- 31. Suganami T., Nishida J., Ogawa Y. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2062–2068 [DOI] [PubMed] [Google Scholar]
- 32. Kane S., Sano H., Liu S. C., Asara J. M., Lane W. S., Garner C. C., Lienhard G. E. (2002) J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 33. Loten E. G. (1991) Int. J. Biochem. 23, 649–655 [DOI] [PubMed] [Google Scholar]
- 34. Manganiello V. C., Murata T., Taira M., Belfrage P., Degerman E. (1995) Arch. Biochem. Biophys. 322, 1–13 [DOI] [PubMed] [Google Scholar]
- 35. Degerman E., Landström T. R., Wijkander J., Holst L. S., Ahmad F., Belfrage P., Manganiello V. (1998) Methods 14, 43–53 [DOI] [PubMed] [Google Scholar]
- 36. Furukawa S., Fujita T., Shimabukuro M., Iwaki M., Yamada Y., Nakajima Y., Nakayama O., Makishima M., Matsuda M., Shimomura I. (2004) J. Clin. Invest. 114, 1752–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Houstis N., Rosen E. D., Lander E. S. (2006) Nature 440, 944–948 [DOI] [PubMed] [Google Scholar]
- 38. Bashan N., Kovsan J., Kachko I., Ovadia H., Rudich A. (2009) Physiol. Rev. 89, 27–71 [DOI] [PubMed] [Google Scholar]
- 39. Davì G., Falco A., Patrono C. (2005) Antioxid. Redox. Signal. 7, 256–268 [DOI] [PubMed] [Google Scholar]
- 40. Nishikawa T., Sasahara T., Kiritoshi S., Sonoda K., Senokuchi T., Matsuo T., Kukidome D., Wake N., Matsumura T., Miyamura N., Sakakida M., Kishikawa H., Araki E. (2003) Diabetes Care 26, 1507–1512 [DOI] [PubMed] [Google Scholar]
- 41. Rehman A., Nourooz-Zadeh J., Möller W., Tritschler H., Pereira P., Halliwell B. (1999) FEBS Lett. 448, 120–122 [DOI] [PubMed] [Google Scholar]
- 42. Dalle-Donne I., Giustarini D., Colombo R., Rossi R., Milzani A. (2003) Trends Mol. Med. 9, 169–176 [DOI] [PubMed] [Google Scholar]
- 43. Pauli J. R., Ropelle E. R., Cintra D. E., Carvalho-Filho M. A., Moraes J. C., De Souza C. T., Velloso L. A., Carvalheira J. B., Saad M. J. (2008) J. Physiol. 586, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benhar M., Forrester M. T., Stamler J. S. (2009) Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 45. Lillig C. H., Holmgren A. (2007) Antioxid. Redox. Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 46. Nathan C. (1997) J. Clin. Invest. 100, 2417–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scapin G., Patel S. B., Chung C., Varnerin J. P., Edmondson S. D., Mastracchio A., Parmee E. R., Singh S. B., Becker J. W., Van der Ploeg L. H., Tota M. R. (2004) Biochemistry 43, 6091–6100 [DOI] [PubMed] [Google Scholar]