Abstract
The K-Cl cotransporter (KCC) regulates red blood cell (RBC) volume, especially in reticulocytes. Western blot analysis of RBC membranes revealed KCC1, KCC3, and KCC4 proteins in mouse and human cells, with higher levels in reticulocytes. KCC content was higher in sickle versus normal RBC, but the correlation with reticulocyte count was poor, with inter-individual variability in KCC isoform ratios. Messenger RNA for each isoform was measured by real time RT-quantitative PCR. In human reticulocytes, KCC3a mRNA levels were consistently the highest, 1–7-fold higher than KCC4, the second most abundant species. Message levels for KCC1 and KCC3b were low. The ratios of KCC RNA levels varied among individuals but were similar in sickle and normal RBC. During in vivo maturation of human erythroblasts, KCC3a RNA was expressed consistently, whereas KCC1 and KCC3b levels declined, and KCC4 message first increased and then decreased. In mouse erythroblasts, a similar pattern for KCC3 and KCC1 expression during in vivo differentiation was observed, with low KCC4 RNA throughout despite the presence of KCC4 protein in mature RBC. During differentiation of mouse erythroleukemia cells, protein levels of KCCs paralleled increasing mRNA levels. Functional properties of KCCs expressed in HEK293 cells were similar to each other and to those in human RBC. However, the anion dependence of KCC in RBC resembled most closely that of KCC3. The results suggest that KCC3 is the dominant isoform in erythrocytes, with variable expression of KCC1 and KCC4 among individuals that could result in modulation of KCC activity.
Keywords: Anion Transport, Chloride Transport, Erythrocyte, Erythropoiesis, Gene Expression, Membrane Function, K-Cl Cotransporter, Expression Profiling, Sickle Cell Disease
Introduction
The SCL12 family of cation-chloride cotransporters represents a group of widely expressed membrane proteins that mediate the coupled, electroneutral symport of chloride with sodium and/or potassium (1). The Na-Cl and Na-K-Cl cotransporters mediate net salt uptake into cells, whereas K-Cl cotransporters (KCC)3 produce net K-Cl loss. In various cells and tissues, cation-chloride cotransporters proteins participate in transmembrane salt movements, modulate membrane potential by establishing internal ion content, and regulate cell volume.
Four independent KCC genes exist in the mammalian genome (1). KCC1 (SCL12A4) is ubiquitously expressed (2), whereas KCC2 (SCL12A5) is restricted to neuronal tissues (3). KCC3 (SCL12A6) is highly expressed in heart, skeletal muscle, kidney, lung, and placenta (4, 5). Two splicing isoforms of KCC3 utilize alternative first exons; KCC3a is widely expressed, whereas KCC3b predominates in kidney (6). KCC4 (SCL12A7) is also widely distributed, with high expression in kidney, heart, lung, and liver (5, 7).
The first report of KCC activity was the Cl−-dependent K+ efflux stimulated by N-ethylmaleimide in red blood cells (RBC) (8, 9). KCC functions as a volume regulator in RBC and is especially active in reticulocytes. Cell swelling activates the transporter, producing K-Cl and water loss with resultant volume reduction. In RBC containing sickle hemoglobin (Hb S) and several other abnormal hemoglobins, K-Cl activity is high and the regulation of KCC activity by cell volume is abnormal (10–13). The apparent consequence is the dehydration of sickle reticulocytes and mature red blood cells relative to cells with normal hemoglobin, leading to high cellular hemoglobin concentration (12–15). Because the rate of polymerization of Hb S is highly dependent on its concentration in the cell, cellular dehydration via K-Cl cotransport contributes to the pathology of sickle cell disease (16–18). However, the basis of abnormal KCC function in sickle red blood cells (SS RBC) has not been elucidated.
In RBC membranes, KCC1 protein has been detected in human (19, 20), sheep (20, 21), rabbit (20), and mouse (20, 22) cells. KCC3 has been reported in sheep RBC (21), and KCC3b (but not KCC3a) has been identified in mouse RBC (22). We demonstrated the presence of RNA transcripts for KCC1, KCC3a, KCC3b, and KCC4 in human reticulocytes (23). The presence of mRNA for KCC1, KCC3, and KCC4 in erythroid precursors was confirmed by De Franceschi et al. (24) who showed that during differentiation in vitro (7 versus 14 days in culture), message levels were relatively stable for KCC1 and KCC3 and increased 2-fold for KCC4. However, the complement of KCC proteins in human RBC membranes has not been ascertained, and their temporal expression during erythroid differentiation has not yet been fully elucidated. Whether significant differences in KCC expression are manifest in sickle cells is also unknown. Mouse RBC expressing Hb S have been valuable for modeling sickle cell pathology, but whether the interactions of Hb S with the mouse RBC membrane faithfully reproduces the cellular pathology of human disease, especially with respect to volume regulation, depends on the complement of KCC proteins and their behavior in mouse RBC.
Here, we report that KCC1, KCC3, and KCC4 proteins are present in human and mouse RBC. We find that RNA transcripts for KCC1 diminish during erythroid differentiation in both human and mouse, whereas KCC3 message levels remain relatively stable. Their relative levels vary considerably among individuals, and no consistent differences in mRNA levels for KCC isoforms were apparent between sickle and normal reticulocytes. When KCC isoforms were expressed in mammalian cells, their kinetic characteristics were similar to each other and to KCC activity in RBC. Anion dependence of KCC in human RBC was most similar to that of KCC3. Taken together, the data suggest that KCC3 is the dominant isoform in erythrocytes, with variable expression of KCC1 and KCC4 among individuals that could result in modulation of KCC activity.
EXPERIMENTAL PROCEDURES
Isolation of Human and Murine RBC and Reticulocytes
Peripheral blood was collected from normal healthy volunteers and from individuals with sickle cell disease via routine phlebotomy into tubes containing either heparin or EDTA, according to a protocol approved by the Institutional Review Board of Cincinnati Children's Hospital Medical Center. Venous blood was collected from the orbital sinus of anesthetized C57/BL6 mice according to a protocol approved by the Cincinnati Children's Hospital Medical Center Animal Care and Use Committee. Transferrin receptor (TfR)-positive reticulocytes were isolated using anti-TfR antibody-conjugated magnetic beads (Miltenyi Biotec Inc.) as described previously (25). Isolated cells were 96–99% reticulocytes by flow cytometric analysis using thiazole orange (BD Biosciences) staining.
Sorting of Human Bone Marrow for Different Stages of Erythroblast Maturation
To assess KCC expression during erythroid differentiation in vivo, fresh bone marrow cells from healthy donors were immunostained with phycoerythrin-Cy5-conjugated anti-CD71 and phycoerythrin-conjugated anti-GPA antibodies (BD Biosciences) as described previously (26, 27). Concurrent staining with 7-amino-actinomycin D (BD Biosciences) was used to gate out apoptotic cells. Single cell suspensions were analyzed using a FACS Canto Flow Cytometer with FacsDiva software version 6.1 and/or sorted using FACS VantageSE with 70-μm nozzle (BD Biosciences). To monitor erythroid differentiation in the sorted samples, cytospin slides were prepared by centrifugation at 300 rpm for 5 min in a Cytospin® 4 cytocentrifuge (Thermo Shandon Inc.) and counter-stained with Wright stain (Harleco EMD).
Sorting of Murine Bone Marrow for Different Stages of Erythroblast Maturation
To assess mKCC expression during in vivo erythroid differentiation in mice, fresh low density bone marrow cells from C57/BL6 mice were isolated by Ficoll density centrifugation and immunostained with FITC-conjugated anti-CD44 and PECy7-conjugated Ter119 antibodies (BD Biosciences) as described by Chen et al. (28). Single cell suspensions were analyzed using a FACS Canto Flow Cytometer with FacsDiva software version 6.1 and/or sorted using FACS VantageSE with a 70-μm nozzle (BD Biosciences). To monitor erythroid differentiation in the sorted samples, cytospin slides were prepared by centrifugation at 500 rpm for 5 min in a Cytospin® 4 cytocentrifuge and counter-stained with Wright stain. Approximately 150 cells per slide (ranging from 129 to 163) were scored for morphology analysis.
Cell Culture
Murine erythroleukemia (MEL) cells were cultured at a concentration of 0.5–2 × 106/ml in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal bovine serum (FBS) (10%), l-glutamine (2 mm), penicillin (100 units/ml), and streptomycin (100 μg/ml). To induce erythroid differentiation, MEL cells were subcultured at 106/ml in DMEM containing FBS (20%) and hexamethylene bisacetamide (1 mg/ml) with medium changed every other day for up to 9 days. To monitor erythroid differentiation, cells were stained with benzidine/hydrogen peroxide solution, and cytospin slides were prepared at 400 rpm for 5 min using a Cytospin® 4 cytocentrifuge. Human embryonic kidney cells (HEK293) (ATCC) were cultured in DMEM containing 10% FBS, penicillin, streptomycin, and other antibiotics as needed to maintain selection depending on the specific plasmid used. Cultures were maintained at 37 °C in 5% CO2 in air.
Real Time RT-QPCR Analysis of mRNA Copy Numbers for KCC Isoforms
Reverse transcription real time QPCR assays were established for human KCC1, KCC1b, KCC3a, KCC3b, and KCC4 mRNA, as well as murine KCC1, KCC3, and KCC4 using two-step multiplex reactions with TaqMan primer/probes. Isoform-specific primer/probe sets (supplemental Table S1) were designed to cross exon junctions to avoid detection of contaminating genomic DNA as described previously (29, 30) using Primer Express Software version 3.0 (Applied Biosystems). The fluorescence label was 6-carboxyfluorescein for KCC1, KCC3a, and KCC4, as well as mouse KCC1, KCC3, and KCC4; tetrachlorofluorescein labeling was used for splicing isoforms KCC1b and KCC3b. After control experiments to identify optimized primer/probe concentrations, standard curves with high correlation coefficients (0.993–1.00) were established for each of the five human KCC and three murine KCC isoforms using serial dilutions of specific plasmids (supplemental Fig. S2 and supplemental Table S1). The amplification efficiency (E) of a single cycle PCR for each target KCC cDNA was determined with corresponding primer/probe set in a range of 1.90–1.94, toward the presumed ideal two in exponential phase. Detection limits approached 10 copies of target sequence per reaction with 6–7 log-fold of quantification (supplemental Table S1), and cross-isoform QPCRs confirmed the specificity of each primer/probe set using plasmids containing 106 or 107 copies of the other KCC isoforms (see supplemental Fig. S1). As an internal control, we utilized VICTM-labeled probe for human or murine glyceraldehyde phosphate dehydrogenase (GAPDH) (Applied Biosystems), which has been reported as the most suitable housekeeping gene for mRNA normalization in human reticulocytes (31). The standard curve for RNA input was determined from (Ct) values for GAPDH (in multiplex amplification with KCC1) measured on serial dilutions of total RNA from human HEK293 cells (supplemental Fig. S2). Control experiments were performed to optimize multiplex RT-QPCR so that the cycle number at threshold (Ct) values for both targets were unaffected by the presence or absence of the other primer/probe sets.
Total RNA was isolated using RNeasy micro kit (Qiagen). Isolated RNA was quantitatively converted into cDNA with a high capacity cDNA archive kit (Applied Biosystems). The KCC1 cDNA and endogenous GAPDH cDNA sequences were quantified in the same reaction well simultaneously by real time PCR in multiplex reactions. The KCC isoforms were amplified in a 25-μl reaction volume containing 12.5 μl of TaqMan 2× Universal Master Mix (Applied Biosystems), RT product from 0.01 to 40 ng of RNA, 80–100 nm probe(s), and 100–300 nm of isoform-specific primers (concentration was optimized for each primer for lowest Ct and the maximal DeltaRn). Unknown samples were run in triplicate, with a no-RT product control for each setup mixture to monitor potential contamination. The amplification conditions were 2 min at 50 °C and 10 min at 95 °C for the first cycle, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Copy numbers for each isoform were determined from the respective Ct values of each sample and the standard curve for that isoform. These values were normalized to RNA input, determined by Ct values for GAPDH of the same sample using a GAPDH standard curve (supplemental Fig. S2).
Western Blot Analysis of Human and Murine RBC Membranes
RBC membranes were prepared by hypotonic lysis of RBC (32). HEK or MEL cells were lysed in hypotonic buffer (10 mm KCl, 1.5 mm MgCl2, 10 mm HEPES (pH 7.6), plus 0.5 mm DTT, and protease inhibitors) and homogenized with a Dounce homogenizer. Nuclei and intracellular vesicles were removed by successive low speed centrifugations, and the supernatants were spun for 30 min at 65,000 rpm in a TLA 120.2 rotor in a Beckman Optima TLX ultracentrifuge. The crude plasma membrane pellets were solubilized in Laemmli sample buffer for immunoblot analysis.
Solubilized membrane (33) preparations were loaded onto 6% SDS-polyacrylamide gels for electrophoresis. Separated proteins were transferred to PVDF membranes and stained with Ponceau S to confirm equivalent loading. Primary antibodies included the following: a rabbit polyclonal antibody raised against an N-terminal 15-amino acid human/mouse KCC1 peptide (22); a rabbit polyclonal antibody raised against an N-terminal 19-amino acid human/mouse KCC3 peptide common to both KCC3a and KCC3b (33, 34); a rabbit polyclonal antibody raised against an N-terminal 15-amino acid mouse KCC3a peptide (22); a rabbit polyclonal antibody raised against an N-terminal 15-amino acid mouse KCC3b peptide (22); a rabbit polyclonal antibody raised against an N-terminal 19-amino acid mouse KCC4 peptide (35); a goat polyclonal antibody raised against a C-terminal human KCC4 peptide (Santa Cruz Biotechnology); a rabbit polyclonal antibody to a peptide corresponding to residues 409–420 of human Myc (Millipore); and a mouse monoclonal antibody to β-actin conjugated to HRP. HRP-conjugated secondary antibodies were purchased from Pierce. Immunoblots were developed using SuperSignal West Pico chemiluminescent substrate (Pierce), and the chemiluminescent signal was detected by autoradiographic film.
Immunoprecipitations
RBC membrane ghosts were solubilized in IP Lysis Buffer (150 mm NaCl, 5% glycerol, 2.5 mm EDTA, 2.5 mm EGTA, 1% Triton X-100, 50 mm Tris-HCl (pH 7.2), 1 mm DTT, and protease inhibitors), and a commercial goat anti-KCC3 antibody (Santa Cruz Biotechnology) was used to immunoprecipitate KCC3. Immune complexes were captured using protein G-agarose beads (Pierce), and proteins were eluted in SDS-PAGE sample buffer. Rabbit anti-KCC1- (22) and rabbit anti-KCC3 (33)-specific antibodies were used to detect their respective proteins in the eluates.
Stable HEK Cell Lines Overexpressing Myc-tagged KCC Proteins
Full-length cDNA constructs were obtained from the I.M.A.G.E. Consortium for KCC1 (accession number BC021193), KCC3 (accession number BC033894), and KCC4 (accession number BC098390). A deletion in exon 14 of the KCC3a cDNA was corrected before cloning to an expression vector. A Myc expression tag (ATGGAACAAAAACTCATCTCAGAAGAGGATCTG) was added in-frame to the 5′ end of each coding sequence using PCR with the Myc sequence tag included in the forward primer. The resulting Myc-tagged cDNAs were then cloned into the pcDNA 3.1 (Invitrogen) expression vector. Primers and restriction sites used in the subcloning are shown in supplemental Table S2. HEK293 cells were transfected with the linearized expression vectors using Mirus LT-1 transfection reagent. Clones were selected using 1.2 mg/ml G418 (Invitrogen) and then maintained in DMEM containing 10% FBS, penicillin, streptomycin, and 0.6 mg/ml G418.
For coexpression of KCC3 with KCC1 or KCC1Δ117, human KCC3a cDNA was cloned into the tetracycline-inducible pcDNA5/FRT/TO vector and transfected into the Flp-In T-Rex HEK293 cell line (Invitrogen). The resulting clone was then transduced with SF91-eGFP-PRE retroviral vector containing either the KCC1 or KCC1Δ117 cDNA. Control cells were transfected with empty vector. Transfected cells were selected for green fluorescent protein (GFP) by flow cytometry, cultured, and assayed for rubidium influx and protein expression. For induction of KCC3a expression, cells were incubated 48 h in 1 ng/ml doxycycline.
Rubidium Flux Assays
Rb+ uptake was measured in HEPES-buffered (pH 7.4) sodium solutions containing 35 mm Rb+; in some experiments, Rb+ concentrations were varied, or Cl− was replaced by other anions as indicated in figures. All flux media contained 0.1 mm ouabain and 0.01 mm bumetanide. RBC samples were processed as described previously for measurement of Rb+ content by flame photometry and hemoglobin by optical spectroscopy (12, 13). HEK cells on polylysine-coated plates (75–90% confluence) were incubated at 37 °C with flux media containing the indicated concentrations of Rb+. Samples were washed with ice-cold media without Rb+ and cells lysed in 4 mm CsCl, 0.1% NH4OH with 0.1% Acationox detergent. Lysates were analyzed for protein by bicinchoninic acid assay (micro-BCA kit, Pierce) and Rb+ by flame emission. Rb+ content was expressed as millimoles/mg of protein, and Rb+ fluxes were calculated as the change in content with time. Parallel incubations in Cl−-free sulfamate media allowed calculation of the Cl−-dependent Rb+ influx as the difference between influx in Cl− and sulfamate media.
RESULTS
KCC1, KCC3, and KCC4 Proteins Are Present in Both Human and Murine RBC Membranes
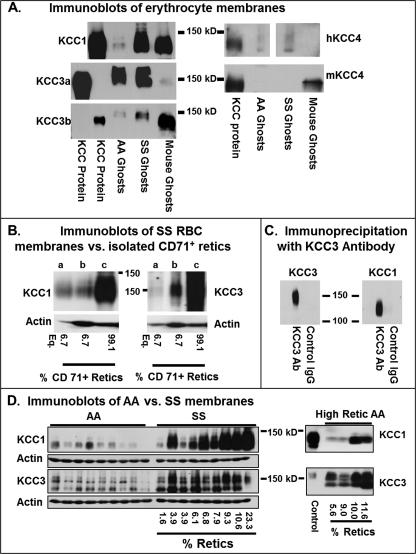
Fig. 1A depicts immunoblots of RBC membranes of healthy volunteers (Hb AA genotype), of individuals with sickle cell disease (Hb SS genotype), and of C57/BL6 mice. Lysates of HEK293 cells expressing KCC1, KCC3a, KCC3b, or KCC4 served as positive controls; isoform specificity of each antibody was demonstrated by lack of binding to membranes from HEK cells overexpressing other isoforms (data not shown). Gels were loaded with equivalent amounts of human AA and SS ghosts and mouse ghost protein (determined by BCA protein assay), permitting semi-quantitative comparisons between AA and SS cells with the same antibody. KCC1, KCC3a, KCC3b, and KCC4 proteins were detected in both AA and SS RBC ghosts and in mouse ghosts (Fig. 1A), and in all cases a stronger signal was obtained in SS ghosts compared with AA ghosts. KCC3a was barely detectable in C57/BL6 mouse ghosts, consistent with a previous study that identified KCC3b as the major KCC3 isoform in mice (22). KCC4 protein (Fig. 1A, right panel) was detected in human AA and SS ghosts using a goat polyclonal antibody raised against a C-terminal human KCC4 peptide. KCC4 was detected in mouse RBC membranes, using a rabbit polyclonal antibody that recognized epitopes in an N-terminal peptide of mouse KCC4. Cross-species reactivity was not apparent with either of these KCC4 antibodies. Rust et al. (22) did not detect KCC4 in mouse RBC membranes using a different antibody to a KCC4 peptide in the C terminus.
FIGURE 1.
KCC protein expression in human and mouse erythrocyte membranes. A, immunoblots of RBC membranes with antibodies specific for KCC isoforms. 150 μg of human AA or SS ghosts or mouse ghosts were loaded per lane. 1–3-μg aliquots of whole cell lysate of HEK293 cells stably expressing human KCC1, KCC3a, KCC3b, or KCC4 were used as positive controls. In the KCC3a and KCC3b blots, HEK293-KCC3a lysate was loaded in the 1st lane and HEK293-KCC3b lysate was loaded in the 2nd lane; note the specificity of antibodies for these splicing isoforms. Subtle differences between erythrocyte membranes and control HEK293 membranes are likely due to differences in post-translational modifications (glycosylation, phosphorylation, etc.) between the HEK cell line and the endogenous red blood cell proteins. B, KCC1 and KCC3 immunoblots of RBC membranes from reticulocytes and whole blood. Reticulocytes were isolated from SS blood using magnetic beads coated with TfR antibody as described under “Experimental Procedures.” Isolated retics (lane c) were 99.1% TfR+ by flow cytometry, and the initial whole blood sample (lane b) was 6.7% TfR+ cells. Equivalent amounts (75 μg) of membrane protein were loaded in lanes b and c. The left-hand lane of both blots (lane a) was loaded with 5 μg of isolated retic membrane protein, representing 6.7% of the protein load of lane c. Actin blots reflect the difference in protein loads. C, immunoprecipitation of RBC membrane proteins. Solubilized membranes from AA RBC were precipitated with antibody to KCC3 (see “Experimental Procedures”), with parallel control IgG incubation. Blots were developed with antibodies specific for KCC3 or KCC1 as indicated. KCC3 and KCC1 antibodies did not cross-react (data not shown). D, KCC protein levels in nine AA and nine SS samples. KCC1 antibody was the same as in A; the KCC3 antibody used here detects both a and b splicing isoforms. Reticulocyte counts for SS samples (CellDyne automated cell counter) are given; AA samples in these blots had normal reticulocytes counts (0.5–1.5%). In the right panel, high reticulocyte AA samples were analyzed. Etiologies of reticulocytosis were (left to right) anti-Rh antibody treatment for immune thrombocytopenia, autoimmune hemolytic anemia, recovery from traumatic blood loss, and treated iron deficiency.
Because KCC activity is known to be greater in reticulocytes, we isolated membranes from TfR+ SS reticulocytes using magnetic beads coated with anti-TfR (anti-CD71) for Western blot analysis. In the example shown in Fig. 1B, the whole blood sample (lane b) contained 6.7% TfR+ reticulocytes by flow cytometry. The isolated TfR+ cells (Fig. 1B, lane c) were 99.1% TfR+ by flow cytometry (data not shown). For Western blot analysis, these lanes (Fig. 1B, lanes b and c) were loaded with equivalent protein amounts (75 μg), as reflected in the actin control; lane a in both blots was loaded with 5 μg of TfR+ retic protein, representing 6.7% of the load of other lanes. If all the KCC protein content resided in TfR+ cells, this lane would be expected to have the same staining intensity as the whole blood lane (Fig. 1B, lane b), which had 6.7% TfR+ cells. This was indeed the case for KCC1. There was intense KCC1 staining in TfR+ retics (Fig. 1B, lane c) compared with the whole blood (6.7% TfR+ cells) as expected, and the diluted sample (lane a) is quite similar in staining intensity to the whole blood sample (lane b), suggesting that most of the KCC1 protein resides in TfR+ cells. For KCC3 the situation is different. The signal is strong in the TfR+ cells (Fig. 1B, lane c) as anticipated, but the signal intensity of the sample diluted (lane a) to reflect the proportion of retics in whole blood is considerably weaker than that of whole blood (lane b). This suggests that the more intense signal in whole blood (Fig. 1B, lane b) results from the presence of KCC3 in cells more mature than the TfR+ population. This is consistent with retention of KCC3 in the membrane beyond the reticulocyte phase. Thus, immature reticulocytes carry the highest complement of KCC1 and KCC3 protein; KCC1 protein appears to be sharply reduced during reticulocyte maturation, whereas KCC3 is retained to some degree in more mature RBC.
Interaction between KCC isoforms has been demonstrated in several experimental systems (38, 39) but has not been heretofore demonstrated in vivo. We performed immunoprecipitation of KCC protein from AA RBC membranes using KCC3 antibody that recognized an epitope not present on KCC1. Subsequent Western blot analysis of precipitated proteins (Fig. 1C) revealed the expected staining with anti-KCC3 antibody (left panel) but also staining of a 130-kDa band by antibody specific for KCC1. KCC4 protein was not detected in the precipitate by the anti-KCC4 antibody that recognized KCC4 in native membranes (data not shown). These studies indicate that KCC3 antibody can immunoprecipitate KCC1 in RBC membranes, suggesting that the two proteins interact directly or indirectly in the native membrane.
KCC1 and KCC3 Western blots comparing AA and SS RBC membranes are shown in Fig. 1D. Both antibodies detected multiple bands, which are well illustrated in the KCC3 blots and which persisted after de-glycosylation (data not shown). KCC1 antibody staining was much stronger for SS than AA samples with normal reticulocyte counts (Fig. 1D, left panel), although the elevated reticulocyte counts and young cell age of SS samples complicate this analysis. It is noteworthy that staining of SS samples was generally, although not rigidly, correlated with reticulocyte count (indicated on the figure) for KCC1. Likewise in high reticulocyte AA samples, KCC1 expression was increased but somewhat variable. With KCC3 antibody that recognized both splicing isoforms but did not cross-react with KCC1, staining of SS membranes was only slightly more intense than that of AA membranes, and it correlated less well with reticulocyte count than KCC1 in both SS and AA RBC. This result is consistent with the immunoblots of TfR+ cells that suggested more complete loss of KCC1 compared with KCC3 with reticulocyte maturation (Fig. 1B). However, reticulocytosis does not explain all of the inter-individual variability observed. For example, two SS samples with the same reticulocyte count (3.9%) had very different content of KCC1 and KCC3; similar large differences in protein staining were apparent in two AA samples with similar reticulocyte counts of 9.0 and 10%. Also apparent are inter-individual differences in the banding pattern for KCC1 and KCC3 and in the apparent ratio of KCC1 to KCC3 in both AA and SS RBC.
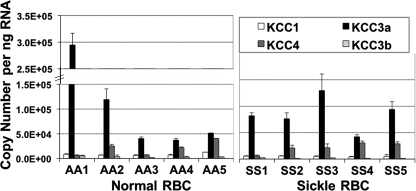
KCC3a Is the Predominant mRNA Species in Human Reticulocytes
Fig. 2 shows KCC RNA levels assessed by multiplex RT-QPCR analysis in human reticulocytes isolated from whole blood of normal controls (AA) or patients with sickle cell disease (SS). The levels of KCC1 message were low and relatively similar among all reticulocyte samples. KCC3a mRNA levels were consistently the highest among all five KCC species, 4–35-fold higher than KCC1 levels in normal AA reticulocytes (n = 5) and 13–26-fold higher than KCC1 in SS reticulocytes (n = 5). The KCC4 mRNA levels varied from 1- to 7-fold of KCC1 levels (n = 10) and approached those of KCC3a in several samples. Message levels for KCC3b were relatively low (20–80% of KCC1), and levels for KCC1b were negligible (1–2% of KCC1; data not shown). Substantial variability in the relative levels of KCC3a and KCC4 message was observed among individual samples, but no pattern was apparent comparing sickle and normal reticulocyte mRNA.
FIGURE 2.
KCC isoform RNA levels in normal and sickle reticulocytes. RT-QPCR analysis for KCC RNAs in AA and SS reticulocytes isolated from whole blood by magnetic separation using anti-TfR-coated beads. Data are shown as means ± S.D. of 5–6 measurements for RBC from five normal individuals (AA) and five sickle patients (SS). The amount of RNA in each sample was determined by simultaneous amplification of GAPDH with KCC1 as multiplex reaction, and interpreted by using standard curves with known initial RNA content from HEK293 cells against Ct of GAPDH amplification.
KCC Expression Profiles Change during Erythroid Maturation in Vivo
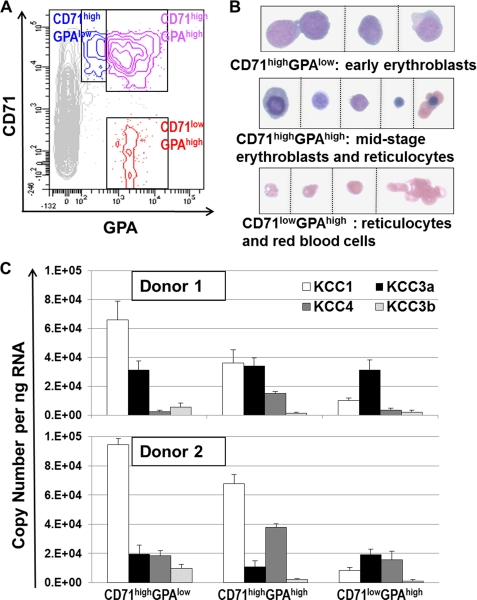
To analyze KCC expression during in vivo erythropoiesis, we isolated three erythroid subpopulations from fresh bone marrow of healthy volunteers based on differential staining for the cell surface markers CD71 (i.e. TfR) and GPA (26), as shown on the flow cytograms in Fig. 3A. The CD71highGPAlow population consisted of early stage erythroblasts, mainly proerythroblasts (26), as confirmed by cytospin imaging (Fig. 3B). The CD71highGPAhigh population contained a mixture of middle and late stage erythroblasts (basophilic, polychromatophilic, and orthochromatic) and few reticulocytes (26). The CD71lowGPAhigh population contained reticulocytes and RBC.
FIGURE 3.
KCC expression during in vivo human erythroid maturation. A, fresh bone marrow cells from normal donors were immunostained with CD71-CyChrome (PECy5) and GPA-PE and sorted into three subpopulations (CD71highGPAlow, CD71highGPAhigh, and CD71lowGPAhigh) as shown in the representative flow cytogram. B, representative morphological images from cytospins slides are shown for the sorted populations that correspond to progressively maturing erythroid precursors. C, RT-QPCR analysis in sorted subpopulations. Two FACS sorting experiments were performed using bone marrow from Caucasian (top panel) or African-American donors (bottom panel), both of whom were Hb AA. Reverse transcription reaction was repeated twice for each RNA sample, followed by QPCRs in triplicate.
The expression levels for KCC1, KCC3a, KCC3b, and KCC4 progressively changed in erythroblasts at these different maturation stages, as shown for two normal donors in Fig. 3C. In early precursors (CD71highGPAlow), KCC1 was the predominant KCC mRNA species, followed by KCC3a. KCC1 levels declined with maturation, whereas KCC3a levels remained relatively constant. KCC4 mRNA levels were relatively low in early stage erythroblasts, increased during differentiation, and then declined in reticulocytes. KCC3b expression was apparent in early stage precursors but decreased with maturation. The KCC expression profile in reticulocytes (CD71lowGPAhigh) isolated from fresh bone marrow was similar to those observed in isolated circulating reticulocytes. Temporal changes in KCC expression during ex vivo erythropoiesis recapitulated the in vivo pattern of KCC isoform expression (see supplemental Fig. S3).
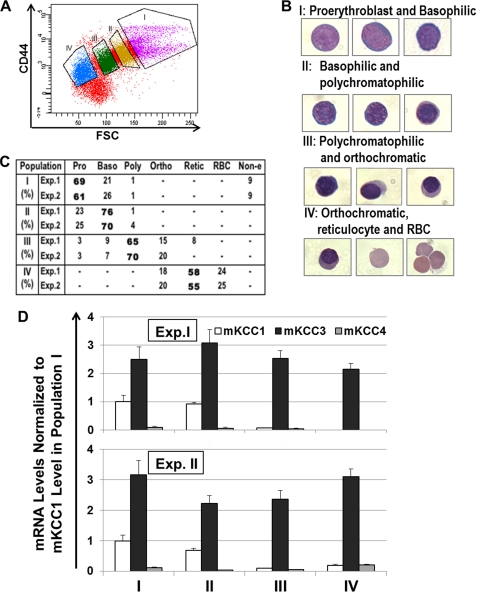
KCC3 mRNA Expression Remains Predominant during in Vivo Erythroid Differentiation in Mouse
Fresh mouse bone marrow was harvested and subjected to cell sorting to isolate different populations of erythroid cells as described by Chen et al. (28). Ter119-positive erythroid cells were sorted on the basis of CD44 staining intensity and forward scatter (Fig. 4A). Populations indicated by the various gates in Fig. 4A contained cells at progressive stages of differentiation, as demonstrated by cytospins in Fig. 4B. The cell type compositions of sorted subpopulations were quantified by morphological analysis, confirming the progression of erythroid differentiation from subpopulation I to IV (Fig. 4C). Analysis of KCC isoform-specific RNA by RT-QPCR (Fig. 4D) revealed predominance of mKCC3 message throughout differentiation. mKCC1 message was present only in populations containing proerythroblasts and basophilic erythroblasts; after the pro-normoblast stage, mKCC1 message was minimal. mKCC4 message was present but remained quite low relative to mKCC3. Nevertheless, mKCC1, mKCC3, and mKCC4 proteins were detected in murine erythrocyte membranes prepared from peripheral blood (Fig. 1A).
FIGURE 4.
KCC expression during in vivo murine erythroid maturation. Fresh low density bone marrow cells from C57/BL6 mice were immunostained with Ter119-PECy7 and CD44-FITC. A, representative flow cytogram indicates sorting gates for four subpopulations (I–IV) that are derived from Ter119+ cells. B, representative morphological images from cytospin slides are shown for corresponding subpopulations. C, summary of cell type composition in corresponding subpopulations by morphological analysis of cytospin images. An average of 150 cells were scored per slide. D, RT-QPCR analysis of murine KCC isoforms in sorted populations. Two FACS sorting experiments were performed, with two reverse transcription reactions for each RNA sample that were followed by two QPCRs in duplicate.
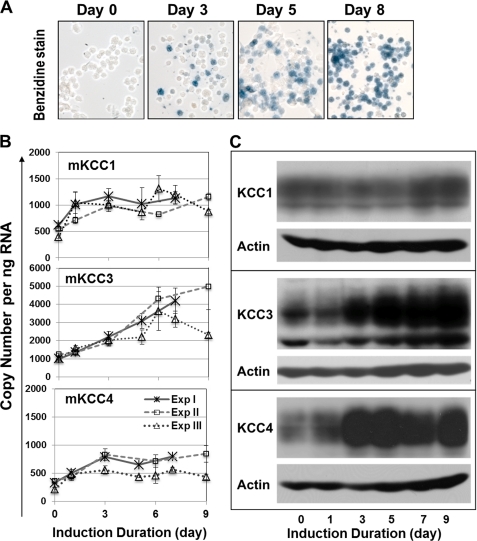
KCC Isoform Expression Increases at Different Rates during in Vitro Erythroid Induction of Murine Erythroleukemia Cells
Flow cytometric separation of erythroid marrow cells produced insufficient numbers of cells for protein analysis with the available antibodies. To determine whether RNA changes during differentiation were associated with increased protein expression, we examined the temporal changes in KCC isoform expression in a MEL cell line induced to differentiate in vitro by treatment with hexamethylene bisacetamide (Fig. 5). Progressive erythroid differentiation during erythroid induction of MEL cells (up to 9 days) was confirmed by morphological evaluation of cytospins demonstrating a gradual reduction of cell size, and by histochemical staining with benzidine, which showed an increasing number of hemoglobin-expressing cells (Fig. 5A). Quantitative PCR analysis (Fig. 5B) demonstrated that mKCC3 was the most abundant mKCC mRNA isoform in uninduced MEL cells, followed by mKCC1 (∼½ of mKCC3) and mKCC4 (∼⅓ of mKCC3). Moreover, mKCC3 mRNA levels increased steadily (by ∼3.5–5-fold) during 8–9 days of erythroid differentiation. Both mKCC1 and mKCC4 levels approximately doubled by day 3 and remained stable thereafter. Temporal changes of message levels were paralleled by changes in KCC protein analyzed by Western blot analysis (Fig. 5C). The mKCC3 protein increased monotonically during the induction period; mKCC1 expression was higher toward the end of the incubation period; and mKCC4 increased and then declined somewhat. These results are consistent with the presence of mKCC1, mKCC3, and mKCC4 isoforms in mouse RBC and support the notion that mKCC3 is the dominant transporter in erythroid cells.
FIGURE 5.
KCC expression in murine erythroleukemia cells during differentiation. MEL cells were induced to differentiate with hexamethylene bisacetamide. A, hemoglobin-expressing cells were detected by histochemical staining with benzidine/hydrogen peroxide solution at indicated days of induction. Representative morphological images from cytospin slides are shown. B, RT-QPCR analysis of mKCC isoforms in induced cells at the indicated days from three independent experiments. The mKCC3 primer/probe set detected both mKCC3a and mKCC3b. Data are shown as mean ± S.D. with two separate RT reactions for each sample, followed by QPCR amplifications in triplicate. C, immunoblot analysis using antibodies against mKCC1, mKCC3, and mKCC4 with anti-actin as loading control.
Physiological Characterization of KCC Isoforms in a Mammalian Expression System
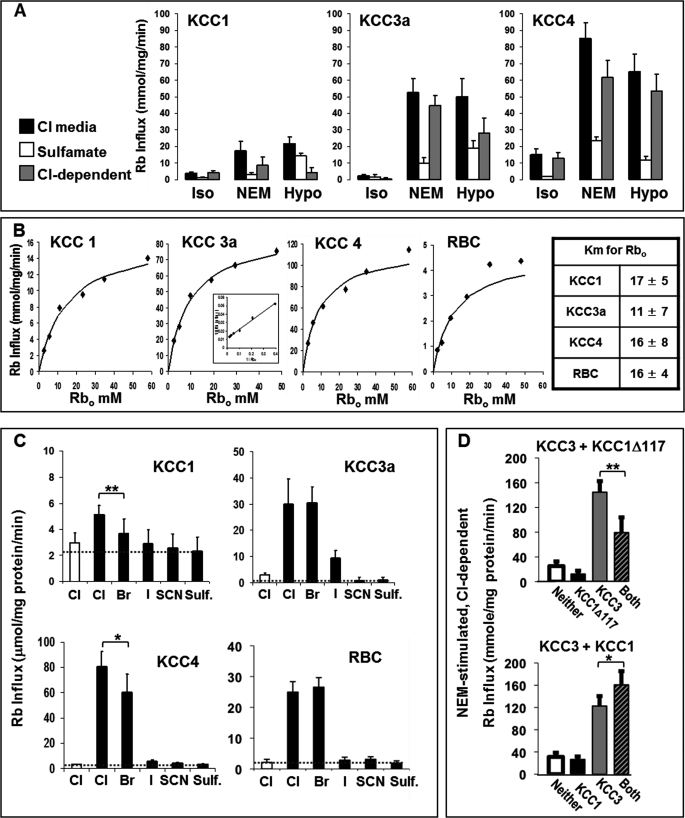
Because direct comparisons of the functional characteristics of the KCC isoforms have been limited to nonmammalian expression systems (34, 36), we evaluated the kinetic properties and anion dependence of human KCC1, KCC3, and KCC4 proteins expressed in human HEK293 cells. We expressed Myc-tagged human KCC1, KCC3a, and KCC4 cDNA in HEK293 cells and selected stably transfected cells by antibiotic resistance. Western blot analysis using anti-Myc antibodies showed similar expression (supplemental Fig. S4). Immunofluorescence analysis revealed expressed protein at the plasma membrane as well as in intracellular vesicles (supplemental Fig. S5); a greater portion of KCC1 was localized to perinuclear vesicles relative to plasma membrane. Nevertheless, KCC-mediated Rb+ uptake could be demonstrated in all of these cell lines, as shown in Fig. 6. Rb+ uptake was measured under isotonic conditions (control), NEM stimulation, and hypotonic incubation. Each condition was paired with a measurement of uptake in sulfamate medium to establish the Cl−-dependent component of uptake. Initial rates of uptake are shown in Fig. 6A, along with the calculated Cl−-dependent components. All three isoforms exhibited Cl−-dependent fluxes stimulated by hypotonic conditions and by NEM. Although KCC fluxes were more robust with KCC3a and KCC4 isoforms compared with KCC1, this could be explained by lower levels of KCC1 protein at the plasma membrane so that direct comparison of activity is not possible.
FIGURE 6.
Physiological characterization of human KCC isoforms expressed in mammalian cells. A, stably transfected HEK293 cells overexpressing different human KCC proteins were maintained in culture with G-418 and grown to 75–90% confluency. Ouabain- and bumetanide-resistant Rb+ influx was measured at 37 °C as described under “Experimental Procedures” in both Cl− and sulfamate media under conditions of isotonicity (Iso), treatment with 1 mm N-ethylmaleimide (NEM), or 220 mOsM hypotonic media (Hypo). For each condition, the flux in sulfamate media was subtracted from that in Cl− media to give the Cl−-dependent flux. B, Rb+ dependence of KCC fluxes. NEM-stimulated, Cl−-dependent Rb+ influx is plotted versus external Rb+ concentration. RBC fluxes represent NEM-stimulated, Cl−-dependent (ouabain- and bumetanide-resistant) Rb+ influx measured at 37 °C as described previously (12). Double-reciprocal plots (inset) were linear and yielded values for Km (mm) shown in the table at right (n = 3). Curves depicted were drawn from the resultant Michaelis-Menten equations. C, anion dependence of KCC fluxes. NEM-stimulated fluxes were measured in isotonic solutions containing various anions. Note different scales. Fluxes higher than those in sulfamate media, indicated by dashed line, reflect KCC-mediated fluxes supported by the indicated anion. Data are shown as mean ± S.D. of three independent experiments. *, p = 0.05; **, p < 0.01. D, KCC activity (NEM-stimulated, Cl−-dependent Rb+ influx) in HEK293 cells coexpressing KCC3 with KCC1 constructs, either N-truncated KCC1Δ117 (upper panel) or full-length KCC1 (lower panel). KCC3a DNA was inserted into a tetracycline-inducible vector integrated into the FlipIn T-REX HEK293 cell line (see “Experimental Procedures”). This KCC3 cell line was then transfected with an SF91-eGFP-PRE vector encoding KCC1Δ117 or full-length KCC1. Control cells were transfected with empty SF91-eGFP-PRE vector. Cells labeled None had empty vector and no induction. KCC1 (or KCC1Δ117) cells were not induced and thus expressed KCC1 (KCC1Δ117) alone. KCC3 cells were transduced with empty vector (no KCC1 or KCC1Δ117 expression) and subsequently induced for 48 h with 1 ng/ml doxycycline prior to flux assay. Cells labeled Both expressed both KCC3+ and KCC1 (or KCC1Δ117) constructs (transduced with KCC1/KCC1Δ117 vector and induced with tetracycline). Bars represent mean ± S.D. of six experiments for KCC1 and three experiments for KCC1Δ117. Paired t test: *, p < 0.006; **, p < 0.001.
The activation of KCC isoforms by external Rb+ (Fig. 6B) conformed to Michaelis-Menten kinetics and double-reciprocal plots (see inset in Fig. 6B) yielded Km values of 11–17 mm. A representative experiment is shown for Rb+ uptake in RBC. Km values did not differ statistically among KCC isoforms and were similar to the value obtained in RBC.
Anion dependence was assessed by measuring NEM-stimulated Rb+ influx in solutions in which chloride was replaced by anions indicated in Fig. 6C. Dashed horizontal lines in Fig. 6C indicate the flux in the presence of sulfamate, assumed to provide minimal anion support for KCC activity, and therefore permit assessment of the degree to which KCC activity is supported by other anions. Similar measurements in RBC are shown for comparison and illustrate similar Rb+ influx in the presence of Cl− and Br− (slight stimulation of RBC KCC activity in Br− media has been reported by others (37)) and minimal support of Rb+ influx by iodide, thiocyanate, or sulfamate. Among the three KCC isoforms expressed in HEK293 cells, only KCC3a shows Rb+ influxes in Cl− and Br− that are comparable with human RBC; Rb+ flux mediated by KCC1 or KCC4 is less in Br− than in Cl− (p < 0.01 and p = 0.05, respectively, by paired t test). Thus, the anion dependence of KCC activity in RBC most closely resembles that of KCC3a.
Our demonstration of in situ interaction between KCC1 and KCC3 raised the question of whether such interactions have functional implications. Previous studies demonstrated a dominant negative effect in Xenopus oocytes lacking the N-terminal domain (KCC1Δ117) when coexpressed with full-length KCC1 or KCC3 (39). We examined the effect of coexpression in HEK cells of KCC3 with either KCC1Δ117 or full-length KCC1. Cells containing KCC3 cDNA in a tetracycline-inducible cassette were transfected with a viral vector containing KCC1 or KCCΔ117 cDNA (linked to green fluorescent protein, GFP) or with a control vector coding for GFP alone. Transfected cells were selected by flow cytometry and cultured and analyzed for KCC activity (NEM-stimulated, Cl−-dependent Rb+ uptake) as shown in Fig. 6D. KCC activity in cells transfected with GFP control vector and not exposed to tetracycline (labeled Neither in Fig. 6D) was slightly higher than that in cells without the tetracycline-inducible KCC3 cassette (data not shown), reflecting a small “leakage” expression of KCC3 in the absence of drug, although no KCC protein was detected in these “un-induced” cells. When KCC1Δ117 was expressed without tetracycline induction (“KCC1Δ117”), KCC activity was slightly reduced (Fig. 6D, upper panel). Tetracycline-induced cells without KCC1Δ117 (“KCC3”) exhibited high KCC activity that was reduced by 45% when KCC1Δ117 was coexpressed (Fig. 6D, Both). Western blots (supplemental Fig. S6) confirmed that expression of KCC1Δ117 did not affect the expression of KCC3 and vice versa, so that the reduced fluxes upon coexpression of KCC3 and KCC1Δ117 were not explained by altered expression of KCC3. This experiment confirms the dominant negative effect of KCC1Δ117 in a mammalian context and demonstrates the capacity of the coexpression system to detect functional interactions between KCC constructs.
When full-length KCC1 was expressed alone in this system (Fig. 6D, lower panel, KCC1), no KCC activity above endogenous levels was detected, consistent with the low levels of KCC activity seen when KCC1 was expressed in HEK cells not containing the tetracycline-inducible cassette (Fig. 6A). When KCC3 was induced (Fig. 6D, KCC3), cells exhibited substantial KCC activity, which was increased by 31% in cells coexpressing KCC1 (KCC3 + KCC1). Immunoblots showed that coexpression of either isoform had no effect on expression of the other (supplemental Fig. S6). These data indicate that KCC1 and KCC3 can interact functionally as well as structurally.
DISCUSSION
We have demonstrated that K-Cl cotransport proteins KCC1, KCC3, and KCC4 are present in human RBC membranes. Because quantitative comparisons of Western blots using different antibodies are not valid, it is not possible to determine the relative amounts of these proteins in RBC membranes. Antibodies specific for KCC3a and -3b detected both splicing isoforms in human RBC membranes. This may account for some of the multiple bands on blots with KCC3 antibody that recognizes both isoforms (Fig. 1D), although there was some overlap between the KCC3a band and the KCC3b band, as can be appreciated in Fig. 1A by comparing the positions of the bands to the 150-kDa marker. KCC proteins are known to be glycosylated (2), but multiple bands were detected after deglycosylation (data not shown), consistent with the known N-terminal heterogeneity of KCC3 transcripts (6).
We confirm the presence of mKCC1 and mKCC3b in mouse RBC membranes (22), and we show for the first time that mKCC4 is also present. A previous study suggested that mKCC4 was absent from mouse RBC (22), and it is not clear whether this discrepancy is due to the use of different antibodies or different strains of mice. In that study, genomic knock-out of KCC1 actually led to an increase in KCC activity because of up-regulation of mKCC3b; KCC fluxes were greatly reduced in KCC3−/− mice (in which no up-regulation of mKCC1 was observed). RBC from double knock-out KCC−/−/KCC3−/− mice exhibited no Cl−-dependent Rb+ uptake, consistent with the absence of other KCC proteins. If the activity of mKCC4 is as robust as hKCC4, it seems unlikely that the protein was present but inactive in the knock-out RBC. The more likely explanation would appear to be differences in the mouse strains, which have been shown to affect KCC activity (40) and which conceivably could involve differences in KCC protein expression.
High levels of KCC1 and KCC3 protein were detected in immature SS reticulocytes expressing transferrin receptor, compared with unfractionated samples. Most of the KCC1 protein appeared to reside in reticulocytes, consistent with loss of KCC1 during reticulocyte maturation. This finding is compatible with the report of Su et al. (20) that KCC1 protein was enriched in low density (high reticulocyte) fractions of SS RBC compared with the high density (low reticulocyte) population of cells. In contrast, although KCC3 protein was abundant in TfR+ cells, the KCC3 content of membranes from whole blood was considerably higher than in membranes of TfR+ cells diluted to reflect their percentage in whole blood, indicating that KCC3 protein persists beyond the reticulocyte stage. These findings are consistent with the tighter correlation of reticulocyte count with membrane content of KCC1 protein compared with KCC3, and with the higher KCC1 content of SS versus AA membranes. In contrast, the difference in KCC3 content between SS and AA membranes was less than for KCC1. The persistence of KCC3 protein in nonreticulocytes is also compatible with previous studies showing KCC-mediated changes in cell volume in the nonreticulocyte fraction of SS RBC (13). Nevertheless, not all of the variation in membrane KCC protein content in both AA and SS samples was a function of reticulocyte count, even in this small sample of patients. Furthermore, the ratio of KCC1 to KCC3 also varied among individuals. Additional work is need to determine whether inter-individual variation in KCC protein expression is a potential source of inter-individual differences in KCC activity and the varying degrees of cellular dehydration observed among patients with sickle cell disease.
Variability in the ratio of KCC isoforms was also apparent at the RNA level in reticulocytes and erythroid precursors. KCC3a was clearly the predominant mRNA species in reticulocytes in both sickle and normal erythrocytes, but comparable levels of KCC1 and KCC4 message were found during erythropoiesis. Messenger RNAs for all three isoforms, as well as the KCC3b splicing isoform, were expressed during human erythroid differentiation in vivo and in erythroid precursors maturing in vitro. KCC3a mRNA was relatively constant during erythroid differentiation and persisted into the reticulocyte stage, becoming the predominant KCC species in terminally differentiated cells. KCC1 mRNA was highest in early erythroblasts, and fell during differentiation. The same trend was seen in KCC3b mRNA, although levels in early erythroblasts were much lower than those of KCC1. Nevertheless, both KCC1 and KCC3b proteins were detected in RBC membranes prepared from peripheral blood. KCC4 message levels first increased and then decreased during differentiation. In mice, the pattern of KCC expression was similar to human; mKCC1 mRNA levels diminished, and mKCC3 mRNA was sustained during in vivo erythroid differentiation. mKCC4 mRNA levels were minimal, but KCC4 protein was nevertheless detected in mouse RBC membranes. In differentiating MEL cells, increases in mKCC1, mKCC3, and mKCC4 mRNA were associated with increases in protein levels in the membrane. These studies provide a foundation for future research on the translational and transcriptional mechanisms regulating KCC gene expression in erythroid cells and on the trafficking and incorporation of KCC proteins into the RBC membrane.
Ouabain- and bumetanide-resistant, Cl−-dependent Rb+ influx, stimulated by NEM and hypotonic conditions, was exhibited by human KCC1, KCC3a, and KCC4 isoforms expressed in HEK293 cells. Affinities of the isoforms for external Rb+ (and presumably K+) were similar to that observed in RBC. Rb+ Km values for KCC3a and KCC4 measured in this study in HEK cells were identical to those measured in oocytes by Mercado et al. (34, 41), whereas Rb+ affinity of KCC1 reported here (Km 17 mm) was somewhat higher than that found in oocytes (26 mm) (41). Anion dependence of NEM-stimulated KCC activity was subtly different among the isoforms. For KCC1 and KCC4, fluxes in Br− were significantly less than those in Cl− media. In contrast, in RBC and HEK cells expressing KCC3a, fluxes in Br− equaled or slightly exceeded those in Cl−. The patterns of Cl−/Br− selectivity reported here for KCC proteins expressed in mammalian HEK cells are similar to those for proteins expressed in Xenopus oocytes (36). Thus, with respect to relative selectivity for Cl− versus Br−, K-Cl cotransport in RBC most closely reflects the behavior of KCC3a expressed in HEK293 cells, suggesting that this isoform is functionally dominant in RBC.
We demonstrate for the first time that KCC1 and KCC3 proteins interact in situ in the RBC membrane, based on the immunoprecipitation of KCC1 by KCC3-specific antibody (Fig. 1C). Interactions between cation-chloride cotransporters have been demonstrated in a variety of ex vivo systems. Simard et al. (38) coexpressed various combinations of KCC proteins in Xenopus oocytes and demonstrated via immunoprecipitation experiments that the KCC proteins had the capacity to form both homodimers and heterodimers with each other via C-terminal interactions. KCC4 interactions with the Na-K-Cl cotransporter (NKCC1) were also demonstrated by immunoprecipitation from Xenopus oocytes and yeast two-hybrid systems and were shown to alter the functional capacity of NKCC1 (38). Casula et al. (39) demonstrated that an N-terminal deletion of KCC1 (KCC1Δ117), which had no KCC activity itself, exhibited a dominant negative effect on the activity of coexpressed full-length KCC1 or KCC3; this functional interaction between the proteins required the C terminus of the truncated protein. We confirm this dominant negative effect of KCC1Δ117 on KCC3 activity in the mammalian HEK293 expression system. Furthermore, we demonstrate an enhancement of KCC activity when KCC3 and full-length KCC1 are coexpressed. It is not clear at present whether this functional effect results from augmented activity of the proteins themselves or from enhanced insertion of transporters into the membrane, but regardless of mechanism, the functional result of KCC3/KCC1 coexpression is increased KCC activity.
The conclusion that KCC3a is the functionally dominant isoform of human RBC is supported by the preponderance of KCC3a message throughout erythroid differentiation, the persistence of KCC3 protein in mature RBC, and the similarity of its anion dependence to KCC activity in RBC. Our findings that KCC1 and KCC3 interact in the native RBC membrane, and that KCC1/KCC3 interactions are capable of producing functional effects, raise the question of the degree to which high content of KCC1 protein in reticulocytes, particularly the stress reticulocytes of sickle cell disease, could play a role in the enhanced KCC activity of these cells via interaction with KCC3. If so, it is possible that the subtle variations noted in the relative expression of KCC1 and KCC3 among individuals could contribute to the striking phenotypic diversity observed in sickle cell disease. Additional studies are required to address these important questions.
Supplementary Material
Acknowledgments
We are grateful to the Clinical Management and Research Support Core of the Division of Hematology/Oncology at Cincinnati Children's Hospital Medical Center for assistance with obtaining blood samples. Services of the Research Flow Cytometry Core and the Translational Trials Development and Support Laboratories were invaluable.
This work was supported, in whole or in part, by National Institutes of Health Grants HL070871 (to C. H. J.), NS064330 (to D. P.), and DK57708 (to D. B. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- KCC
- K-Cl cotransporter
- HEK
- human embryonic kidney
- NEM
- N-ethylmaleimide
- TfR
- transferrin receptor
- GPA
- glycophorin A
- MEL
- murine erythroleukemia.
REFERENCES
- 1. Gamba G. (2005) Physiol. Rev. 85, 423–493 [DOI] [PubMed] [Google Scholar]
- 2. Gillen C. M., Brill S., Payne J. A., Forbush B., 3rd (1996) J. Biol. Chem. 271, 16237–16244 [DOI] [PubMed] [Google Scholar]
- 3. Payne J. A., Stevenson T. J., Donaldson L. F. (1996) J. Biol. Chem. 271, 16245–16252 [DOI] [PubMed] [Google Scholar]
- 4. Hiki K., D'Andrea R. J., Furze J., Crawford J., Woollatt E., Sutherland G. R., Vadas M. A., Gamble J. R. (1999) J. Biol. Chem. 274, 10661–10667 [DOI] [PubMed] [Google Scholar]
- 5. Mount D. B., Mercado A., Song L., Xu J., George A. L., Jr., Delpire E., Gamba G. (1999) J. Biol. Chem. 274, 16355–16362 [DOI] [PubMed] [Google Scholar]
- 6. Mercado A., Broumand V., Zandi-Nejad K., Enck A. H., Mount D. B. (2006) J. Biol. Chem. 281, 1016–1026 [DOI] [PubMed] [Google Scholar]
- 7. Boettger T., Hübner C. A., Maier H., Rust M. B., Beck F. X., Jentsch T. J. (2002) Nature 416, 874–878 [DOI] [PubMed] [Google Scholar]
- 8. Lauf P. K., Theg B. E. (1980) Biochem. Biophys. Res. Commun. 92, 1422–1428 [DOI] [PubMed] [Google Scholar]
- 9. Dunham P. B., Stewart G. W., Ellory J. C. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1711–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brugnara C., Bunn H. F., Tosteson D. C. (1986) Science 232, 388–390 [DOI] [PubMed] [Google Scholar]
- 11. Canessa M., Spalvins A., Nagel R. L. (1986) FEBS Lett. 200, 197–202 [DOI] [PubMed] [Google Scholar]
- 12. Joiner C. H., Rettig R. K., Jiang M., Franco R. S. (2004) Blood 104, 2954–2960 [DOI] [PubMed] [Google Scholar]
- 13. Joiner C. H., Rettig R. K., Jiang M., Risinger M., Franco R. S. (2007) Blood 109, 1728–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brugnara C., Van Ha T., Tosteson D. C. (1989) Blood 74, 487–495 [PubMed] [Google Scholar]
- 15. Franco R. S., Palascak M., Thompson H., Joiner C. H. (1995) J. Clin. Invest. 95, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brugnara C. (2003) J. Pediatr. Hematol. Oncol. 25, 927–933 [DOI] [PubMed] [Google Scholar]
- 17. Nagel R. L., Fabry M. E., Steinberg M. H. (2003) Blood Rev. 17, 167–178 [DOI] [PubMed] [Google Scholar]
- 18. Rinehart J., Gulcicek E. E., Joiner C. H., Lifton R. P., Gallagher P. G. (2010) Curr. Opin. Hematol. 17, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pellegrino C. M., Rybicki A. C., Musto S., Nagel R. L., Schwartz R. S. (1998) Blood Cells Mol. Dis. 24, 31–40 [DOI] [PubMed] [Google Scholar]
- 20. Su W., Shmukler B. E., Chernova M. N., Stuart-Tilley A. K., de Franceschi L., Brugnara C., Alper S. L. (1999) Am. J. Physiol. 277, C899–C912 [DOI] [PubMed] [Google Scholar]
- 21. Lauf P. K., Zhang J., Delpire E., Fyffe R. E., Mount D. B., Adragna N. C. (2001) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130, 499–509 [DOI] [PubMed] [Google Scholar]
- 22. Rust M. B., Alper S. L., Rudhard Y., Shmukler B. E., Vicente R., Brugnara C., Trudel M., Jentsch T. J., Hübner C. A. (2007) J. Clin. Invest. 117, 1708–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crable S. C., Hammond S. M., Papes R., Rettig R. K., Zhou G. P., Gallagher P. G., Joiner C. H., Anderson K. P. (2005) Exp. Hematol. 33, 624–631 [DOI] [PubMed] [Google Scholar]
- 24. De Franceschi L., Ronzoni L., Cappellini M. D., Cimmino F., Siciliano A., Alper S. L., Servedio V., Pozzobon C., Iolascon A. (2007) Haematologica 92, 1319–1326 [DOI] [PubMed] [Google Scholar]
- 25. Cohen R. M., Franco R. S., Khera P. K., Smith E. P., Lindsell C. J., Ciraolo P. J., Palascak M. B., Joiner C. H. (2008) Blood 112, 4284–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loken M. R., Shah V. O., Dattilio K. L., Civin C. I. (1987) Blood 69, 255–263 [PubMed] [Google Scholar]
- 27. Kalfa T. A., Pushkaran S., Zhang X., Johnson J. F., Pan D., Daria D., Geiger H., Cancelas J. A., Williams D. A., Zheng Y. (2010) Haematologica 95, 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen K., Liu J., Heck S., Chasis J. A., An X., Mohandas N. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17413–17418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan D., Gunther R., Duan W., Wendell S., Kaemmerer W., Kafri T., Verma I. M., Whitley C. B. (2002) Mol. Ther. 6, 19–29 [DOI] [PubMed] [Google Scholar]
- 30. Wang D., Zhang W., Kalfa T. A., Grabowski G., Davies S., Malik P., Pan D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19958–19963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Silver N., Best S., Jiang J., Thein S. L. (2006) BMC Mol. Biol. 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bennett V. (1983) Methods Enzymol. 96, 313–324 [DOI] [PubMed] [Google Scholar]
- 33. Pearson M. M., Lu J., Mount D. B., Delpire E. (2001) Neuroscience 103, 481–491 [DOI] [PubMed] [Google Scholar]
- 34. Mercado A., Vázquez N., Song L., Cortés R., Enck A. H., Welch R., Delpire E., Gamba G., Mount D. B. (2005) Am. J. Physiol. Renal Physiol. 289, F1246–F1261 [DOI] [PubMed] [Google Scholar]
- 35. Karadsheh M. F., Byun N., Mount D. B., Delpire E. (2004) Neuroscience 123, 381–391 [DOI] [PubMed] [Google Scholar]
- 36. Mercado A., de los Heros P., Vázquez N., Meade P., Mount D. B., Gamba G. (2001) Am. J. Physiol. Cell Physiol. 281, C670–C680 [DOI] [PubMed] [Google Scholar]
- 37. Kaji D. (1986) J. Gen. Physiol. 88, 719–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simard C. F., Bergeron M. J., Frenette-Cotton R., Carpentier G. A., Pelchat M. E., Caron L., Isenring P. (2007) J. Biol. Chem. 282, 18083–18093 [DOI] [PubMed] [Google Scholar]
- 39. Casula S., Shmukler B. E., Wilhelm S., Stuart-Tilley A. K., Su W., Chernova M. N., Brugnara C., Alper S. L. (2001) J. Biol. Chem. 276, 41870–41878 [DOI] [PubMed] [Google Scholar]
- 40. Armsby C. C., Stuart-Tilley A. K., Alper S. L., Brugnara C. (1996) Am. J. Physiol. 270, C866–C877 [DOI] [PubMed] [Google Scholar]
- 41. Mercado A., Song L., Vazquez N., Mount D. B., Gamba G. (2000) J. Biol. Chem. 275, 30326–30334 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.