Abstract
Ordered nucleosome disassembly and reassembly are required for eukaryotic DNA replication. The facilitates chromatin transcription (FACT) complex, a histone chaperone comprising Spt16 and SSRP1, is involved in DNA replication as well as transcription. FACT associates with the MCM helicase, which is involved in DNA replication initiation and elongation. Although the FACT-MCM complex is reported to regulate DNA replication initiation, its functional role in DNA replication elongation remains elusive. To elucidate the functional role of FACT in replication fork progression during DNA elongation in the cells, we generated and analyzed conditional SSRP1 gene knock-out chicken (Gallus gallus) DT40 cells. SSRP1-depleted cells ceased to grow and exhibited a delay in S-phase cell cycle progression, although SSRP1 depletion did not affect the level of chromatin-bound DNA polymerase α or nucleosome reassembly on daughter strands. The tracking length of newly synthesized DNA, but not origin firing, was reduced in SSRP1-depleted cells, suggesting that the S-phase cell cycle delay is mainly due to the inhibition of replication fork progression rather than to defects in the initiation of DNA replication in these cells. We discuss the mechanisms of how FACT promotes replication fork progression in the cells.
Keywords: Chromatin, DNA Replication, Epigenetics, Histone Chaperone, Nucleosome
Introduction
The mechanisms of DNA replication at the level of naked DNA template in eukaryotes are essentially the same as those in prokaryotes (1, 2). However, eukaryotes must replicate nucleosomes, which are the fundamental units of chromatin (3). The nucleosome is a histone octamer, comprising the histone (H3-H4)2 tetramer and two histone H2A-H2B dimers, which is wrapped by 146 bp DNA (4). The nucleosome structure must be dynamically regulated during each step of DNA replication, including licensing, initiation, and elongation (5–7). Although the molecular mechanisms of these DNA replication processes have been extensively studied (5–7), their relationship to the concomitant changes in chromatin structure remains elusive.
Eukaryotic DNA replication on chromatin template should be regulated by a variety of chromatin-acting factors, such as histone modification enzymes, nucleosome remodeling complexes, and histone chaperones (8–12). Among these factors, histone chaperones are known to facilitate nucleosome assembly and disassembly by promoting specific histone-DNA and histone-histone interactions in an ATP-independent manner (9–13). Especially, at the elongation step of DNA replication, nucleosomes must be disassembled before and reassembled after the passage of replication forks (8–12, 14).
Nucleosome reassembly is well known to be executed by the evolutionarily conserved histone chaperone, chromatin assembly factor-1 (CAF-1),4 which was originally isolated as a nucleosome assembly factor in the SV40 DNA replication system in vitro (15). CAF-1 is also required for the reassembly of nucleosomes into newly synthesized DNA duplexes in the cells (16, 17). Based on functional and physical interactions between CAF-1 and proliferating cell nuclear antigen, a sliding clamp for eukaryotic DNA polymerases δ and ϵ, nucleosome reassembly is reported to be mechanistically coupled to DNA synthesis (18).
Another evolutionarily conserved histone chaperone is CCG1-interacting factor A (CIA) (19), whose budding yeast homologue, anti-silencing function 1 (Asf1), has been shown to possess anti-silencing activity (20), also participates in the nucleosome reassembly reaction on the daughter DNA strands (21). Recently, CIA/Asf1 has been reported to regulate replication fork progression and histone supply and demand in the DNA replication process (22, 23). Furthermore, CIA/Asf1 interacts with a hexameric DNA helicase, MCM2–7 (MCM complex), through histones H3-H4, suggesting that CIA/Asf1 is involved in the transfer of parental histones in DNA replication via an intermediate CIA/Asf1-H3-H4-MCM2–7 complex (22).
Recently, parental histone (H3-H4)2 tetramers have been reported to be transferred as either the tetrameric form or as the histone H3-H4 dimer to daughter strands in a DNA replication-dependent manner in human cells (24). Because CIA/Asf1 has been shown to directly split histone (H3-H4)2 tetramers into histone H3-H4 dimers in vitro (25), CIA/Asf1 is the best candidate for splitting histone (H3-H4)2 tetramers in the cells. Thus, an understanding of the molecular mechanisms of histone transfer from parental to daughter strands and nucleosome reassembly has begun to emerge (10–12, 14). However, the mechanism of parental nucleosome disassembly at the elongation step of DNA replication has not been studied.
In addition to CAF-1 and CIA/Asf1, another evolutionarily conserved histone chaperone, facilitates chromatin transcription (FACT) (26), comprised of Spt16/Cdc18 and structure-specific recognition protein 1 (SSRP1) (27), has also been reported to be involved in DNA replication (28–30). FACT has been shown to directly interact with key DNA replication enzymes and factors such as DNA polymerase α (pol α), replication protein A (RPA), and MCM complex, all of which are essential components for DNA replication (28, 30–33). FACT was also reported to facilitate the DNA helicase activity of the MCM complex on nucleosomal DNA in vitro (31). Furthermore, FACT was shown to be important for proper DNA replication initiation in human cells (31). Despite many studies on the involvement of FACT in DNA replication, the mechanistic roles of FACT in nucleosome disassembly, histone transfer, and nucleosome reassembly at the elongation step of DNA replication remain obscure.
To elucidate the functional roles of FACT during the process of DNA replication on chromatin in the cells, we generated and analyzed chicken DT40 conditional SSRP1 knock-out cells. Here, we provide several lines of evidence to demonstrate that FACT maintains normal DNA replication elongation rates by primarily and preferentially disassembling prereplicative nucleosomes ahead of DNA replication forks.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Gene Disruption
Two SSRP1 disruption constructs were generated from genomic polymerase chain reaction (PCR) products inserted with the puromycine (puro)- or blasticidin (bsr)-selection marker cassette. Chicken SSRP1 cDNA was prepared by reverse transcription PCR, and the FLAG tag was added to its C-terminal end by PCR. FLAG-SSRP1 was inserted into the expression vector carrying the tet-repressible promoter, pUHG 10-3 (34). DT40 cells were successively transfected with puro-SSRP1, FLAG-SSRP1/pUHG 10-3, and bsr-SSRP1. To prepare deletion mutants of SSRP1, SSRP1 fragments were amplified by PCR using appropriate primers on the SSRP1 cDNA template. The DNA fragments obtained were inserted into the vector pAneo (35), which carries the chicken β-actin promoter and the neomycin resistance gene driven by the SV40 promoter. Cells expressing SSRP1 fragments were obtained by subsequent transfection with these vectors.
Western Blotting Assay
Isolation of the chromatin fraction and Western blotting were performed as previously described (36) using antibody against MCM2/4 (a kind gift from Yukio Ishimi, Ibaraki University), Spt16 (29). DNA polymerase α (a kind gift from Fumiko Hirose, University of Hyogo), RPA32 (Cell Signaling), histone H3 (Abcam), histone H2B (Upstate), phospho-Chk1 (Ser345) (Cell Signaling), α-tubulin or FLAG-M2 (Sigma), followed by horseradish peroxidase-conjugated anti-rabbit, anti-rat, or anti-mouse IgG secondary antibody (Cell Signaling). Proteins were visualized using ECL detection reagents (Amersham Biosciences).
Cell Cycle Analysis by Flow Cytometry
Flow cytometry was performed as previously described (17). For two-dimensional cell cycle analysis, cells were cultured in the presence of bromodeoxyuridine (BrdU; BD Biosciences) for 10 min, fixed in 70% ethanol, and stained with FITC-labeled anti-BrdU antibody (BD Biosciences) and propidium iodide.
DNA Fiber Assay
The DNA fiber assay was performed as previously described (37). Fiber lengths were measured using ImageJ, and micrometer values were expressed in kilobases using a conversion factor: 1 μm = 2.59 kb. Measurements were recorded from areas of the slides with untangled DNA fibers to prevent the possibility of recording labeled patches from tangled bundles of fibers.
Molecular Combing Assay
The molecular combing assay was performed as previously described (38). Cells were pulse-labeled for 20 min with 100 μm iododeoxyuridine (IdU), washed with PBS twice, and pulse-labeled for 20 min with 100 μm chlorodeoxyuridine (CldU). To prepare genomic DNA while removing the mitochondrial genome, the nuclei were extracted with buffer A (250 mm sucrose, 20 mm HEPES (pH 7.5), 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA (pH 8.0), 1 mm EGTA (pH 6.8), 1 mm DTT, 0.1 mm PMSF) before resuspension into low-melting point agarose. Combed DNA molecules were heat denatured in 50% formamide and 2× SSC at 72 °C for 12 min. For immunodetection of labeled DNA, denatured DNA molecules were incubated with mouse anti-BrdU monoclonal Ab (1:5) and rat anti-BrdU monoclonal Ab (1:25) for 1 h at 37 °C. After washing with PBS containing 0.05% Tween 20 three times for 5 min each, the DNA molecules were incubated with Alexa Fluor 555-conjugated goat anti-mouse IgG (1:500) and Alexa Fluor 488-conjugated rabbit anti-rat IgG (1:500) for 30 min at 37 °C. All antibodies were diluted in blocking solution (1% (w/v) skim milk and 0.05% Tween 20 in PBS). After washing with PBS containing 0.05% Tween 20 as above, coverslips were mounted using VECTASHIELD (Vector Laboratories). Fluorescent signals were measured by using MetaMorph version 6.1 software (Universal Imaging).
MNase Assay
The MNase assay was performed as previously described (39). Cells were pulse-labeled with 20 μm BrdU for 20 min and harvested. To isolate nuclei, cells were treated with 0.1% Nonidet P-40 in NB buffer (15 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 2 mm magnesium acetate, 2 mm CaCl2, 1 mm DTT, and protease inhibitor mixture (Sigma)). The isolated nuclei were washed twice with NB buffer and digested with 0.0074–0.20 units/ml of MNase (Sigma) at 37 °C for 8 min. Genomic DNA was then isolated using Easy DNA (Invitrogen), electrophoresed in a 2% agarose gel, stained with ethidium bromide, and transferred to Hybond N membrane (GE Healthcare). BrdU-labeled DNA was detected using anti-BrdU antibody.
RESULTS
Generation of Conditionally SSRP1-depleted Cells
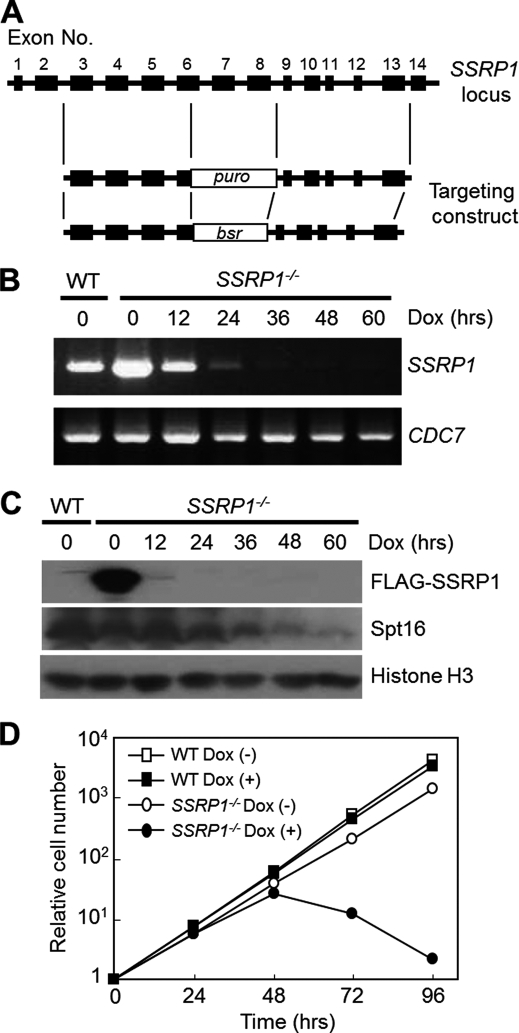
To investigate the functional role of the histone chaperone FACT in DNA replication in the cells, we generated a FACT knock-out cell line by using gene-targeting constructs designed to replace exons 6–8 of the chicken SSRP1 gene, which encodes a small subunit of the FACT complex (27), with a puro or bsr selection marker cassette (Fig. 1A). We presumed that SSRP1 would be essential for the viability of chicken DT40 cells because gene targeting of SSRP1 in mice was lethal (40). To avoid the predicted lethality of SSRP1-deficient cells, we generated SSRP1 conditional knock-out cells. A FLAG-tag conjugated chicken SSRP1 gene under control of a tet-repressible promoter (37) was transfected into DT40 cells after one of the two SSRP1 alleles had been disrupted with the puro-SSRP1 targeting construct. SSRP1−/+ cells expressing FLAG-SSRP1 were selected and subsequently transfected with the bsr-SSRP1 targeting construct to disrupt the other allele, and we obtained “SSRP1−/− + FLAG-SSRP1” (SSRP1−/−) cell lines.
FIGURE 1.
Depletion of SSRP1 causes cell death. A, preparation of SSRP1−/− + FLAG-SSRP1 (SSRP1−/−) cells. Schematic representation of the SSRP1 locus and gene-targeting constructs. Closed boxes indicate exons. puro and bsr indicate the drug resistance genes of puromycin and blasticidin S, respectively. B, suppression of SSRP1 mRNA expression in SSRP−/− cells by Dox. RNA was prepared from SSRP1−/− cells cultured in the presence of Dox for the indicated periods. SSRP1 cDNA was prepared by real-time reverse transcription PCR and electrophoresed in a 2% agarose gel. CDC7 was used as the loading control. It is worth noting that another group independently constructed a similar cell line using DT40 cells (65). C, depletion of the FLAG-SSRP1 protein. Whole cell lysates were prepared from “wild-type (WT)” or SSRP−/− + FLAG-SSRP1 (SSRP−/−) cells cultured in the presence of Dox for the indicated times. FLAG-SSRP1, Spt16, and histone H3 (loading control) were detected by Western blotting. D, growth curves. WT or SSRP−/− cells (1 × 105) were inoculated in 1 ml of medium and passaged daily. Dox was added at time 0.
SSRP1 Is Essential for Cell Viability
As expected, the addition of doxycycline (Dox) to SSRP1−/− cell cultures efficiently suppressed the transgene expression of FLAG-SSRP1. The FLAG-SSRP1 mRNA (Fig. 1B) and protein (Fig. 1C) disappeared after the addition of Dox. Upon depletion of SSRP1, the amount of Spt16, the large subunit of the FACT complex, also gradually decreased (Fig. 1C). SSRP1−/− cells ceased to grow within 72 h and began to die in the presence of Dox (Fig. 1D). These findings suggest that SSRP1 plays an essential role in chicken cell viability.
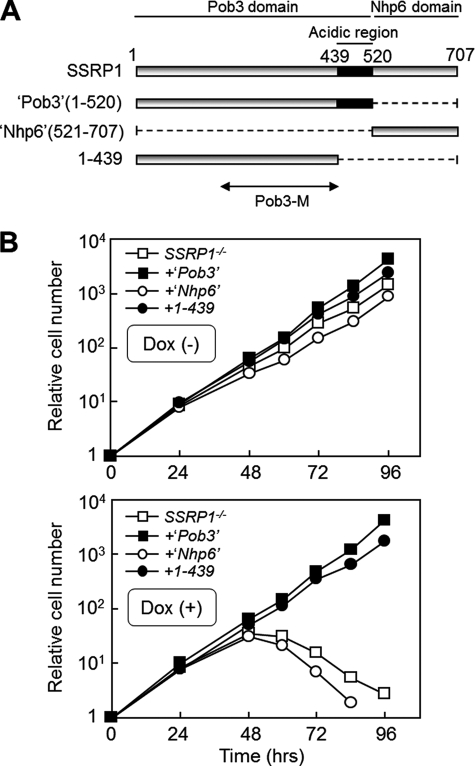
Domain of SSRP1 Required for Cell Viability
Considering the apparent essential role of SSRP1, we attempted to identify the functional domains of SSRP1 required for cell viability. Vertebrate SSRP1 is the bipartite orthologue of yeast Pob3 and Nhp6 (Fig. 2A) (26–30). Histone chaperones including Pob3 are known to usually contain acidic amino acids-rich region(s) (11, 13) (Fig. 2A). To clarify the functional roles of the acidic region of Pob3 and the Nhp6 domain of SSRP1, constructs consisting of the corresponding domains were expressed in SSRP1−/− cells (Fig. 2A). As shown in Fig. 2B, the Nhp6 domain and the acidic region of the Pob3 domain in SSRP1 were not required for cell viability. These results are consistent with the previous findings that Pob3, but not Nhp6, is essential for yeast cell viability (30). Taken together, the domain analysis of SSRP1 revealed that its Pob3 domain without the acidic region was found to be sufficient for cell viability (Fig. 2, A and B).
FIGURE 2.
The Pob3 domain is sufficient for cell viability. A, schematic presentation of SSRP1 deletion mutants. Pob3-(1–520) and –(1–439) are referred to in the text as “Pob3 domain” and “Pob3 domain without the acidic region,” respectively. The “Nhp6 domain” lacks a Pob3 domain. Budding yeast Pob3-M (32), corresponding to the part of the Pob3 domain (1–439) without the acidic region, is described under “Discussion.” B, growth curves. SSRP−/− cells expressing each SSRP1 fragment described in A were cultured in the absence (−) or presence (+) of Dox (upper and lower panels, respectively). Because full-length FLAG-SSRP1 is regulated by Dox, but not the SSRP1 fragments, each SSRP1 fragment is expressed independently of the presence of Dox.
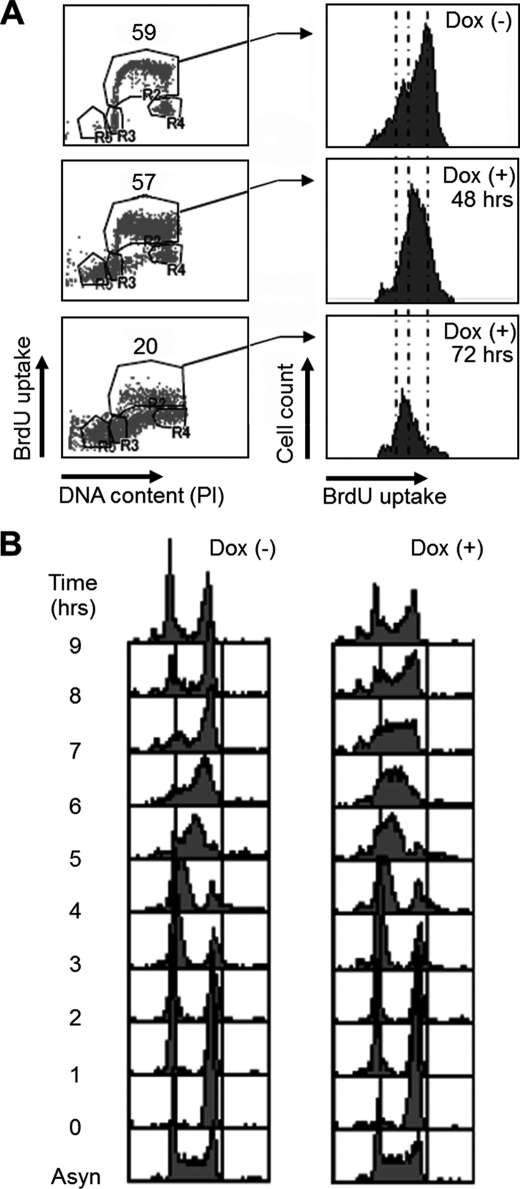
Depletion of SSRP1 Delays the Progression of S Phase
To next address which stages of the cell cycle are perturbed in SSRP1-depleted cells, we characterized the effect of SSRP1 depletion on cell cycle progression by flow cytometry (FACS) using an FITC-conjugated antibody against the thymidine analog BrdU. The proportion of S-phase cells incorporating BrdU decreased within 72 h after the addition of Dox (Fig. 3A, left panels). The amount of BrdU incorporated into individual cells was decreased even 48 h after the addition of Dox (Fig. 3A, right panels). In addition, the G1 and sub-G1 fractions gradually increased in the presence of Dox (Fig. 3A). Considering these results, subsequent experiments were performed using cells cultured for 48 h in the presence or absence of Dox. It was noted that a small amount of Spt16 remained in the SSRP1-depleted cells at that time (Fig. 1C).
FIGURE 3.
Depletion of SSRP1 delays the progression of S phase. A, cell cycle distribution of SSRP1-depleted cells. Cells were cultured in the presence of Dox for the indicated times, pulse-labeled with BrdU for 10 min, and harvested. The cells were stained with FITC anti-BrdU to detect BrdU uptake and with propidium iodide (PI) to detect DNA. Left panels show y axis, BrdU uptake; and x axis, total DNA. Right panels show y axis, cell number; and x axis, BrdU uptake. B, cell cycle progression of SSRP1-depleted cells after release from M-phase block. Cells were cultured in the presence (right) or absence (left) of Dox for 36 h and then cultured in the presence of nocodazole (500 ng/ml) for 8 h. After release from the M-phase block, cells were collected at 1-h intervals, fixed, and stained with PI, and DNA content was analyzed by flow cytometry. Asyn, asynchronous.
After release from a nocodazol block (G2/M arrest), cell cycle progression was further analyzed. Cells cultured in the absence of Dox reached the next G2/M phase 7–8 h after release from the nocodazol block, and SSRP1-depleted cells traversed the S phase of the cell cycle more slowly than those expressing SSRP1 and began to reach G2/M phase at 8–9 h after release (Fig. 3B). It is reported that a mutation of POB3, the yeast SSRP1 homologue, also causes a slower S phase (30). The previous studies and our observations indicate that the functional role of FACT in S-phase cell cycle progression is evolutionarily conserved from unicellular to multicellular eukaryotes.
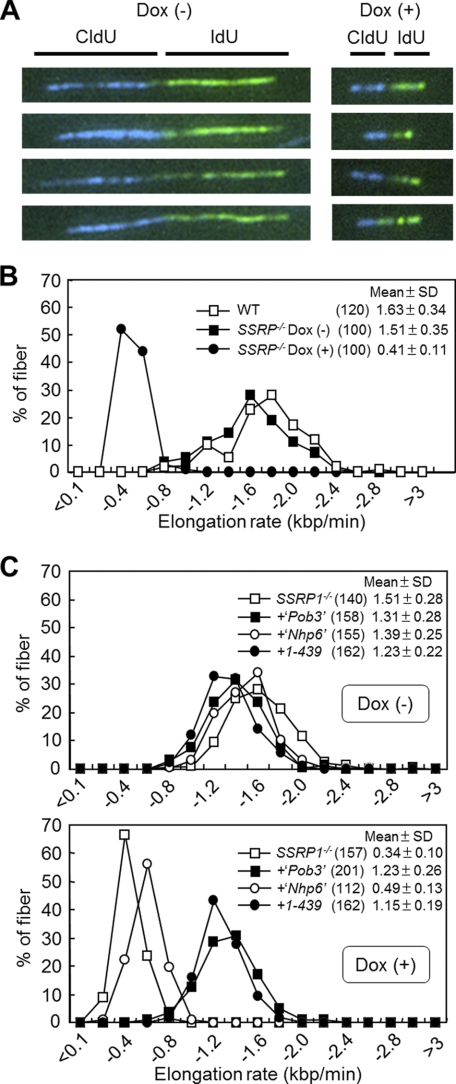
SSRP1 Maintains the Normal Rate of DNA Replication Fork Progression
Delayed S-phase progression could be caused by checkpoint activation or defects in the initiation and/or elongation steps of DNA replication (41). Thus, we attempted to elucidate which of these possibilities caused the S-phase cell cycle defect in SSRP1-depleted cells. No hallmarks of checkpoint activation, such as the phosphorylation of Check1 (Chk1), were observed in SSRP1-depleted cells (supplemental Fig. S1).
To test the other possibilities, the elongation rate of DNA replication (fork rate) and the frequency of DNA replication initiation were measured. The fork rate was examined using a DNA fiber assay (42). In this assay, cells were pulse-labeled with thymidine analogs CldU and IdU for 10 and 15 min, respectively, and the length of labeled DNA replication tracks on DNA fiber spreads were quantified by immunostaining. The fork rate in SSRP1-depleted cells was about one-quarter of that found in wild-type or SSRP1-undepleted cells (Fig. 4, A and B, and supplemental Fig. S2). Interestingly, the Pob3 domain without the acidic region, which supports cell viability (Fig. 2, A and B), is sufficient for replication fork progression (Fig. 4C). Because several components of the eukaryotic DNA replication machinery acting on replication forks (43) have been shown to be required for maintaining a normal replication fork rate (37, 44), FACT could facilitate the fork rate through its concerted actions with DNA replication machinery.
FIGURE 4.
Analysis of DNA replication elongation. A, images of typical DNA fibers. SSRP−/− cells cultured in the presence (+) or absence (−) of Dox for 48 h were successively pulse-labeled with CldU (blue) and IdU (green). DNA fibers were stained as described under “Experimental Procedures.” B, DNA replication elongation rates as determined by the percentage of DNA fibers containing CldU. The lengths of DNA fibers (only CldU tracks that connected with IdU tracks) prepared as shown in A were measured, and DNA replication elongation rates (mean ± S.D.) were calculated as fiber length divided by pulse-labeling time. Results using wild-type cells are also shown. The numbers in parentheses indicate the number of samples tested. C, fork rates in SSRP1−/− cells expressing SSRP1 fragments. Cells were cultured in the presence (+) or absence (−) of Dox for 48 h and successively pulse-labeled with CldU and IdU. Only CldU tracks that connected with IdU tracks were measured. Fork rates were calculated and are displayed as mean ± S.D. The numbers in parentheses indicate the number of samples.
The DNA Replication Initiation May Be Intact in SSRP1-depleted Cells
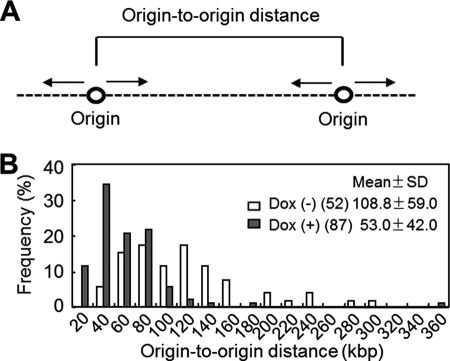
The frequency of DNA replication initiation was next assessed by measuring origin-to-origin distances using a molecular combing assay (45), which allows the visualization and measurement of tethered strands of DNA. An increase or a decrease in the origin-to-origin distance indicates infrequent or frequent DNA replication initiation in a given stretch of DNA, respectively (46). The average of origin-to-origin distance (Fig. 5A and supplemental Fig. S3) measured in DNA from SSRP1-depleted cells decreased to about half of that measured in DNA from SSRP1-undepleted cells (Fig. 5B), indicating that the frequency of initiation of DNA replication was up-regulated in SSRP1-depleted cells. There are two possible explanations for the increase in the initiations of DNA replication found in SSRP1-depleted cells. First, FACT may repress the initiation of DNA replication such that the action of FACT also represses transcription initiation from cryptic sites (47, 48). Alternatively, the decreased fork rate in SSRP1-depleted cells could up-regulate the initiation of DNA replication at the dormant origins, so that slowing the replication fork rate with aphidicolin or hydroxyurea would trigger the initiation of DNA replication at otherwise dormant origins (49–51).
FIGURE 5.
Analysis of DNA replication initiation. A, origin to origin distance. Images of DNA fibers (supplemental Fig. S3) were obtained using a molecular combing assay (“Experimental Procedures”). B, the distances between adjacent origins (in kbp) in clusters of replicons were determined and are displayed as mean ± S.D. The numbers in parentheses indicate the number of samples.
Chromatin Loading of DNA Replication Proteins Was Proficiently Detected in SSRP1-depleted Cells
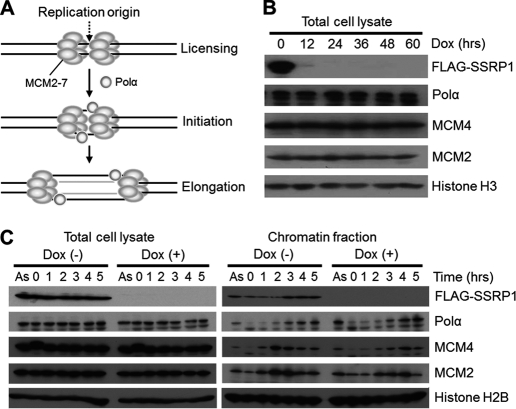
FACT interacts with Polα and the MCM2–7 complex, both of which are essential for DNA replication (28, 31, 32) (Fig. 6A). To determine whether the fork progression defect in SSRP1-depleted cells was caused by a dysfunction of Polα or the MCM2–7 complex, we examined for reductions in the amounts of Polα and two MCM complex components, MCM2 and MCM4, under SSRP1-depletion conditions. The total amounts of these proteins in SSRP1−/− cells, even 60 h after the addition of Dox, was not measurably different from those in SSRP1-undepleted cells (Fig. 6B).
FIGURE 6.
Chromatin binding of MCM and Polα in SSRP1-depleted cells. A, MCM and Polα are known to be involved in both DNA replication initiation and elongation. B, the total amount of MCM and Polα in asynchronous cells. Cells were cultured for the indicated periods in the presence of Dox, and whole cell lysates were prepared. The indicated proteins were detected by Western blotting. C, time course of levels of chromatin-bound MCM and DNA polymerase α (Polα). SSRP−/− cells were cultured in the presence (+) or absence (−) of Dox for 36 h, and then cultured in the presence of nocodazole (500 ng/ml) for 8 h to cause M-phase cell cycle arrest. After release from the M-phase block, cells were collected at 1-h intervals. Cell lysates and chromatin fractions were analyzed by Western blotting.
The chromatin-bound forms of Polα, MCM2, and MCM4 in SSRP1−/− cells were further monitored after the release of cells from nocodazole-induced G2/M arrest in the presence of Dox. The recruitment of Polα, MCM2, and MCM4 onto chromatin was effectively detected in SSRP1-depleted cells (Fig. 6C), although the recruitment of MCM2 and MCM4 seemed slightly delayed. The loaded MCM helicase has been shown to be sufficient for the initiation of DNA replication in SSRP1-depleted cells because the frequency of DNA replication initiation in SSRP1-depleted cells increased instead of decreased (Fig. 5B). This suggested that the fork progression defect in SSRP1-depleted cells was not due to a dysfunction in the chromatin loading of Polα, MCM2, and MCM4.
The Activity of FACT in Nucleosome Assembly on Newly Synthesized Daughter Strands Seems to Be Less Significant
In general, histone chaperones facilitate nucleosome assembly and disassembly (9–13). Indeed, FACT is reported to be involved in both nucleosome disassembly and reassembly in transcription elongation (52). Moreover, nucleosomal histones H2B and H3 are efficiently evicted by RNA polymerase II but do not reform upon inactivation of Spt16, suggesting that Spt16 reassembles nucleosome structure during transcription elongation (53).
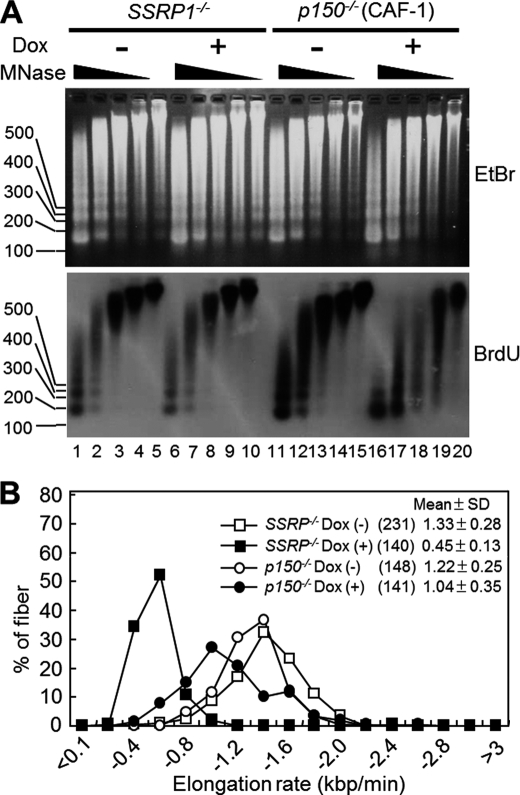
We therefore tried to elucidate whether FACT is involved in nucleosome reassembly during DNA replication. To assess this possibility, a micrococcal nuclease (MNase) digestion assay was performed to monitor nucleosome reassembly on daughter strands. Newly synthesized DNA was pulse-labeled with BrdU and detected with an anti-BrdU antibody. MNase sensitivity was apparently unchanged in SSRP1-depleted cells compared with that in SSRP1-undepleted cells (Fig. 7A, lanes 1–5 versus 6–10). In contrast, p150 (the large subunit of another histone chaperone CAF-1)-depleted DT40 cells, which were previously generated by Takami et al. (17), were more sensitive to MNase (Fig. 7A, lanes 11–15 versus 16–20). This observation is consistent with the previous findings that CAF-1 reassembles nucleosomes on daughter strands during DNA replication both in vitro and in vivo (15–17). These results suggest that the activity of FACT in nucleosome assembly on newly synthesized daughter strands is less significant than that of CAF-1.
FIGURE 7.
Nucleosome assembly and DNA replication elongation rates in SSRP1- versus p150-deficient cells. A, nucleosome reassembly on newly synthesized DNA. After incubation in the presence or absence of Dox for 48 h, SSRP−/− or p150−/− cells were pulse-labeled with BrdU for 45 min. Nuclei were prepared and treated with MNase at 0.20 (lanes 1, 6, 11, and 16), 0.067 (lanes 2, 7, 12, and 17), 0.022 (lanes 3, 8, 13, and 18), 0.007 (lanes 4, 9, 14, and 19), and 0 (lanes 5, 10, 15, and 20) units/ml. DNA was resolved in 2% agarose gels, stained with ethidium bromide (EtBr) (upper panel), transferred onto a Hybond N membrane, and detected with an anti-BrdU antibody (lower panel). B, fork rates in SSRP1−/− versus p150−/− cells. Cells were cultured in the presence (+) or absence (−) of Dox for 48 h and successively pulse-labeled with CldU and IdU. Only CldU tracks that connected with IdU tracks were measured. Fork rates were calculated and are displayed as mean ± S.D. The numbers in parentheses indicate the number of samples.
If the dysfunction of nucleosome reassembly on newly synthesized daughter strands leads to slow DNA replication fork progression, the rate of replication fork progression in p150 (CAF-1)-depleted cells would be expected to be more dramatic than that in SSRP1-depleted cells. However, the deceleration of DNA replication was not as severe in p150-depleted cells as in SSRP1-depleted cells (Fig. 7B). Taken together, the results suggest that the activity of FACT in promoting the rate of DNA elongation is more significant than that of CAF-1, whereas the activity of FACT in nucleosome assembly on newly synthesized daughter strands is relatively weak.
DISCUSSION
A pioneering study using Xenopus egg extracts in a cellular DNA replication system showed that the histone chaperone FACT is required for DNA replication (29). This finding was also supported by S-phase perturbation caused by dysfunction of FACT in the previous (28, 30–33) and present studies (the data in Fig. 3). To further elucidate the functional roles of FACT in DNA replication, we examined the rates of DNA replication elongation (Fig. 4) and initiation (Fig. 5), the amounts DNA replication enzymes/factors (Fig. 6), and nucleosome assembly activity on newly synthesized daughter strands (Fig. 7) under dysfunctional FACT conditions (Figs. 1 and 2). Below, we discuss our observations within the context of previous studies and suggest possible roles for FACT in DNA replication.
Are the DNA Replication Defects Observed in SSRP1-depleted Cells Due to Transcriptional Defects?
Because FACT is known to be involved in transcription elongation (26, 27, 52–55), we first examined the possibility that transcriptional defects caused by FACT dysfunction lead indirectly to DNA replication defects in SSRP1-depleted cells by determining the amounts of several DNA replication factors in both SSRP1-depleted and -undepleted cells. However, the amounts of Polα, MCM2, and MCM4 in SSRP1-depleted cells were not different from those in SSRP1-undepleted cells (Fig. 6). Furthermore, DNA replication factors must have been present in sufficient amounts for DNA replication initiation because origin firing in SSRP1-depleted cells increased compared with that in undepleted cells (Fig. 5). Therefore, inhibition of the rate of replication fork progression in SSRP1-depleted cells (Fig. 4) is probably not caused by a reduction in the transcription of genes encoding DNA replication factors but is related to dysfunction of FACT in DNA replication. This situation is consistent with the previous observation that immunodepletion of FACT from Xenopus egg extracts, in which transcription does not occur, causes defects in DNA replication (29). Taken together, the results suggest that FACT dysfunction directly led to DNA replication defects in SSRP1-depleted cells.
Functional Roles of FACT in DNA Replication Initiation Versus Elongation
FACT is proposed to participate in the initiation stage of DNA replication (31, 33). It has been shown that FACT interacts and coexists with the MCM complex at a particular chromosomal DNA replication origin as determined by chromatin immunoprecipitation analysis (ChIP) in human cells (31, 33). Furthermore, the initiation of DNA replication at a particular site was reduced by the perturbation of the interaction between FACT and the MCM complex (31). Contrary to these observations, in SSRP1-depleted cells, an increase in the frequency of initiation (Fig. 5) and a decrease in the rate of elongation (Fig. 4) were detected by direct analyses of DNA replication products. The apparent differences between the previous (31, 33) and present (Fig. 5) findings on the initiation of DNA replication might be due to different experimental assays and conditions. Further studies, such as chromatin immunoprecipitation analyses of replication enzymes and factors at individual origins in SSRP1-depleted DT40 cells will be needed to clarify the functional roles of SSRP1 in DNA replication initiation. Despite the differences between the results in the previous and present studies, the present results (Fig. 4) suggest that SSRP1 is involved in the elongation stage of DNA replication (Fig. 4).
Functional Roles of FACT in DNA Replication Elongation
Based on the present and previous findings, FACT may act as (i) a factor that stimulates the DNA replication machinery and/or as (ii) a histone chaperone in DNA replication elongation. As described under “Functional Roles of FACT in DNA Replication Initiation Versus Elongation,” FACT interacts with Polα, RPA, and the MCM complex (28, 31, 32) and apparently facilitates replication fork progression via its functional interaction with these replication enzymes/factors. This possibility seems to be supported by a previous study (32) that demonstrated that a point mutation introduced within a conserved surface cluster in the budding yeast Pob3-M, corresponding to the part of the Pob3 domain without the acidic region of the SSRP1 subunit of FACT (Figs. 2 and 4), interferes with DNA replication (30). Because the DNA replication defect in the pob3 point mutant is suppressed by a mutation in RPA, it has been proposed that coordination between the functions of FACT and RPA is important during DNA replication (32). Because the Pob3 domain without the acidic region was sufficient for both cell viability and fork progression (Figs. 2 and 4C) in this study, the defects in cell viability and fork progression would seem to be caused by a dysfunction of RPA in SSRP1-depleted cells.
Histone-based Activities of FACT in DNA Replication Elongation
Because the Pob3 domain without the acidic region was sufficient for both cell viability and DNA replication elongation (Figs. 2 and 4C), it is possible that defects in cell viability and DNA replication elongation are due to a dysfunction in the histone-based activity of the Pob3 domain lacking the acidic region.
Yeast Pob3-M, which corresponds to a part of the Pob3 domain without the acidic region (Figs. 2 and 4), is reported to be genetically linked to the histone H2A C-terminal docking site and to have a function that overlaps with that of the Spt16 N-terminal “peptidase” domain (56). Because the Spt16 N-terminal peptidase domain directly interacts with histones H3 and H4 in vitro (57), the Pob3 domain without the acidic region may regulate chromatin-based DNA replication elongation through its functional interaction with core histones (H2A, H3, and H4). Thus, the histone-based activities of FACT are probably involved in efficient DNA replication elongation.
Functional Roles of FACT in Nucleosome Reassembly during DNA Replication Elongation
Mechanisms for the histone-based activities of FACT have been reviewed very recently (58). In the case of transcription elongation, FACT is proposed to facilitate transcription elongation by assisting in nucleosome disassembly and reassembly ahead and behind, respectively, of RNA polymerase II, which translocates along the DNA template during transcription elongation (26, 27, 52, 54, 55). Another study proposed that FACT promotes a reversible transition between two nucleosome forms that result in unchanged and dramatically increased accessibility to nucleosomal DNA, respectively (59). It is plausible that FACT also plays similar roles in DNA replication elongation.
Because FACT has been proposed to be involved in nucleosome reassembly during DNA replication elongation (32), it is interesting to discuss the implications of our results (Fig. 7A) for the function of FACT in the DNA replication process. In this study, we could not detect a significant requirement for FACT in nucleosome reassembly on daughter strands (Fig. 7A), suggesting that FACT does not seem to be involved in nucleosome reassembly on daughter strands, in contrast to a previous proposal (32). On the other hand, in the case that FACT is involved in nucleosome reassembly on daughter strands as reported, our results (Fig. 7A) could be explained as (i) the reduced fork progression in SSRP1-depleted cells (Fig. 7B) masks the requirement of FACT by compensating for inefficient nucleosome reassembly and (ii) nucleosome reassembly on daughter strands in SSRP1-depleted cells is performed by the remaining pool of Spt16 subunit (Fig. 1C), which has been shown to function alone in DNA replication processes such as in the recovery from DNA replication stress (60). Spt16 alone binds to H2A-H2B dimers and nucleosomes in vitro (52) and redeposits histones during transcription elongation (53). Thus, the remaining pool of Spt16 in SSRP1-depleted cells could redeposit histones, resulting in nucleosome reassembly during DNA replication elongation. The two possibilities may not be mutually exclusive. Taking into account all information described in this paragraph, we cannot fully conclude the degree of importance of FACT in nucleosome reassembly on newly synthesized DNA.
In contrast to FACT, deficiency of CAF-1, which caused a defect in nucleosome reassembly (Fig. 7A) as previously reported (15–17), resulted in an almost normal replication fork rate (Fig. 7B). It suggested that a defect in nucleosome reassembly does not necessarily cause a reduction in replication fork progression. In other words, it seems that the activity of nucleosome reassembly does not significantly facilitate DNA replication elongation.
A Possible Mechanistic Action of FACT in Replication Fork Progression
Prereplicative nucleosomes of simian virus (SV40) minichromosomes disassemble just ahead of replication forks (61, 62). Furthermore, in vivo studies on the dynamics in histone-DNA interactions have suggested that prereplicative nucleosomes are dissolved during the advancement of the replication fork with the release of associated histones in the form of (H3-H4)2 tetramers and H2A-H2B dimers (63). Because MCM helicase complex translocates along DNA and unwinds duplex DNA during the elongation phase of DNA replication (64), it is quite possible that prereplicative nucleosome disassembly might occur ahead of the MCM complex. MCM complex and FACT were reported to be detected in a large replication progression complex, the replisome in budding yeast, using a proteomics approach (43). Furthermore, FACT is known to facilitate the DNA helicase activity of the MCM complex on nucleosomal DNA in vitro (31, 33). Here, we demonstrated that FACT is required for efficient replication fork progression (Fig. 4). Considering all of the results, we favor a model whereby FACT facilitates DNA replication elongation in a concerted action with the MCM complex by promoting efficient prereplicative nucleosome disassembly ahead of the replication fork.
Supplementary Material
Acknowledgments
We thank Drs Y. Ishimi and F. Hirose for gifts of antibodies, and all members of our laboratory for discussions of the manuscript.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CAF-1
- chromatin assembly factor-1
- BrdU
- bromodeoxyuridine
- FACT
- facilitates chromatin transcription
- puro
- puromycine
- SSRP1
- structure-specific recognition protein 1
- MNase
- micrococcal nuclease
- RPA
- replication protein A
- CIA
- CCG1-interacting factor A
- Dox
- doxycycline.
REFERENCES
- 1. Baker T. A., Bell S. P. (1998) Cell 92, 295–305 [DOI] [PubMed] [Google Scholar]
- 2. McHenry C. S. (2003) Mol. Microbiol. 49, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 3. Kornberg R. D. (1974) Science 184, 868–871 [DOI] [PubMed] [Google Scholar]
- 4. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 5. Blow J. J., Dutta A. (2005) Nat. Rev. Mol. Cell Biol. 6, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanaka S., Araki H. (2010) Chromosoma 119, 565–574 [DOI] [PubMed] [Google Scholar]
- 7. Kunkel T. A., Burgers P. M. (2008) Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gruss C., Sogo J. M. (1992) Bioessays 14, 1–8 [DOI] [PubMed] [Google Scholar]
- 9. De Koning L., Corpet A., Haber J. E., Almouzni G. (2007) Nat. Struct. Mol. Biol. 14, 997–1007 [DOI] [PubMed] [Google Scholar]
- 10. Ransom M., Dennehey B. K., Tyler J. K. (2010) Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eitoku M., Sato L., Senda T., Horikoshi M. (2008) Cell Mol. Life Sci. 65, 414–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Groth A., Rocha W., Verreault A., Almouzni G. (2007) Cell 128, 721–733 [DOI] [PubMed] [Google Scholar]
- 13. Philpott A., Krude T., Laskey R. A. (2000) Semin. Cell Dev. Biol. 11, 7–14 [DOI] [PubMed] [Google Scholar]
- 14. Corpet A., Almouzni G. (2009) Trends Cell Biol. 19, 29–41 [DOI] [PubMed] [Google Scholar]
- 15. Smith S., Stillman B. (1989) Cell 58, 15–25 [DOI] [PubMed] [Google Scholar]
- 16. Nabatiyan A., Krude T. (2004) Mol. Cell. Biol. 24, 2853–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takami Y., Ono T., Fukagawa T., Shibahara K., Nakayama T. (2007) Mol. Biol. Cell 18, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shibahara K., Stillman B. (1999) Cell 96, 575–585 [DOI] [PubMed] [Google Scholar]
- 19. Munakata T., Adachi N., Yokoyama N., Kuzuhara T., Horikoshi M. (2000) Genes Cells 5, 221–233 [DOI] [PubMed] [Google Scholar]
- 20. Le S., Davis C., Konopka J. B., Sternglanz R. (1997) Yeast 13, 1029–1042 [DOI] [PubMed] [Google Scholar]
- 21. Tyler J. K., Adams C. R., Chen S. R., Kobayashi R., Kamakaka R. T., Kadonaga J. T. (1999) Nature 402, 555–560 [DOI] [PubMed] [Google Scholar]
- 22. Groth A., Corpet A., Cook A. J., Roche D., Bartek J., Lukas J., Almouzni G. (2007) Science 318, 1928–1931 [DOI] [PubMed] [Google Scholar]
- 23. Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. (2010) Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]
- 24. Xu M., Long C., Chen X., Huang C., Chen S., Zhu B. (2010) Science 328, 94–98 [DOI] [PubMed] [Google Scholar]
- 25. Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007) Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 26. Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. (1998) Cell 92, 105–116 [DOI] [PubMed] [Google Scholar]
- 27. Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D. (1999) Nature 400, 284–288 [DOI] [PubMed] [Google Scholar]
- 28. Wittmeyer J., Formosa T. (1997) Mol. Cell. Biol. 17, 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okuhara K., Ohta K., Seo H., Shioda M., Yamada T., Tanaka Y., Dohmae N., Seyama Y., Shibata T., Murofushi H. (1999) Curr. Biol. 9, 341–350 [DOI] [PubMed] [Google Scholar]
- 30. Wittmeyer J., Joss L., Formosa T. (1999) Biochemistry 38, 8961–8971 [DOI] [PubMed] [Google Scholar]
- 31. Tan B. C., Chien C. T., Hirose S., Lee S. C. (2006) EMBO J. 25, 3975–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., Formosa T. (2006) Mol. Cell 22, 363–374 [DOI] [PubMed] [Google Scholar]
- 33. Tan B. C., Liu H., Lin C. L., Lee S. C. (2010) J. Biomed. Sci. 17, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gossen M., Bujard H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sugawara H., Kurosaki M., Takata M., Kurosaki T. (1997) EMBO J. 16, 3078–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abe T., Ishiai M., Hosono Y., Yoshimura A., Tada S., Adachi N., Koyama H., Takata M., Takeda S., Enomoto T., Seki M. (2008) Cell Signal. 20, 1978–1985 [DOI] [PubMed] [Google Scholar]
- 37. Bermudez V. P., Farina A., Tappin I., Hurwitz J. (2010) J. Biol. Chem. 285, 9493–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katsuno Y., Suzuki A., Sugimura K., Okumura K., Zineldeen D. H., Shimada M., Niida H., Mizuno T., Hanaoka F., Nakanishi M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quivy J. P., Gérard A., Cook A. J., Roche D., Almouzni G. (2008) Nat. Struct. Mol. Biol. 15, 972–979 [DOI] [PubMed] [Google Scholar]
- 40. Cao S., Bendall H., Hicks G. G., Nashabi A., Sakano H., Shinkai Y., Gariglio M., Oltz E. M., Ruley H. E. (2003) Mol. Cell. Biol. 23, 5301–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y., Sanchez Y. (2004) DNA Repair 3, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 42. Jackson D. A., Pombo A. (1998) J. Cell Biol. 140, 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell. Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 44. Petermann E., Helleday T., Caldecott K. W. (2008) Mol. Biol. Cell 19, 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sugimura K., Takebayashi S., Taguchi H., Takeda S., Okumura K. (2008) J. Cell Biol. 183, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hand R. (1975) J. Histochem. Cytochem. 23, 475–481 [DOI] [PubMed] [Google Scholar]
- 47. Kaplan C. D., Laprade L., Winston F. (2003) Science 301, 1096–1099 [DOI] [PubMed] [Google Scholar]
- 48. Mason P. B., Struhl K. (2003) Mol. Cell. Biol. 23, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woodward A. M., Göhler T., Luciani M. G., Oehlmann M., Ge X., Gartner A., Jackson D. A., Blow J. J. (2006) J. Cell Biol. 173, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ge X. Q., Jackson D. A., Blow J. J. (2007) Genes Dev. 21, 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Courbet S., Gay S., Arnoult N., Wronka G., Anglana M., Brison O., Debatisse M. (2008) Nature 455, 557–560 [DOI] [PubMed] [Google Scholar]
- 52. Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., Reinberg D. (2003) Science 301, 1090–1093 [DOI] [PubMed] [Google Scholar]
- 53. Jamai A., Puglisi A., Strubin M. (2009) Mol. Cell 35, 377–383 [DOI] [PubMed] [Google Scholar]
- 54. Kang S. W., Kuzuhara T., Horikoshi M. (2000) Genes Cells 5, 251–263 [DOI] [PubMed] [Google Scholar]
- 55. Belotserkovskaya R., Reinberg D. (2004) Curr. Opin. Genet. Dev. 14, 139–146 [DOI] [PubMed] [Google Scholar]
- 56. VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D. J., Hill C. P., Formosa T. (2008) J. Biol. Chem. 283, 5058–5068 [DOI] [PubMed] [Google Scholar]
- 57. Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., Schffzek K., Ladurner A. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8884–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Winkler D. D., Luger K. (2011) J. Biol. Chem. 286, 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., Formosa T. (2009) Mol. Cell 35, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Donnell A. F., Brewster N. K., Kurniawan J., Minard L. V., Johnston G. C., Singer R. A. (2004) Nucleic Acids Res. 32, 5894–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sogo J. M., Stahl H., Koller T., Knippers R. (1986) J. Mol. Biol. 189, 189–204 [DOI] [PubMed] [Google Scholar]
- 62. Gasser R., Koller T., Sogo J. M. (1996) J. Mol. Biol. 258, 224–239 [DOI] [PubMed] [Google Scholar]
- 63. Jackson V. (1990) Biochemistry 29, 719–731 [DOI] [PubMed] [Google Scholar]
- 64. Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., Berger J. M. (2011) Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Okada M., Okawa K., Isobe T., Fukagawa T. (2009) Mol. Biol. Cell 20, 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.