Abstract
FTY720 phosphate (FTY720P) is a high potency agonist for all the endothelial differentiation gene family sphingosine 1-phosphate (S1P) receptors except S1P receptor subtype 2 (S1P2). To map the distinguishing features of S1P2 ligand recognition, we applied a computational modeling-guided mutagenesis strategy that was based on the high degree of sequence homology between S1P1 and S1P2. S1P2 point mutants of the ligand-binding pocket were characterized. The head group-interacting residues Arg3.28, Glu3.29, and Lys7.34 were essential for activation. Mutation of residues Ala3.32, Leu3.36, Val5.41, Phe6.44, Trp6.48, Ser7.42, and Ser7.46, predicted to interact with the S1P hydrophobic tail, impaired activation by S1P. Replacing individual or multiple residues in the ligand-binding pocket of S1P2 with S1P1 sequence did not impart activation by FTY720P. Chimeric S1P1/S1P2 receptors were generated and characterized for activation by S1P or FTY720P. The S1P2 chimera with S1P1 sequence from the N terminus to transmembrane domain 2 (TM2) was activated by FTY720P, and the S1P2(IC1-TM2)S1P1 domain insertion chimera showed S1P1-like activation. Twelve residues in this domain, distributed in four motifs a–d, differ between S1P1 and S1P2. Insertion of 78RPMYY in motif b alone or simultaneous swapping of five other residues in motifs c and d from S1P1 into S1P2 introduced FTY720P responsiveness. Molecular dynamics calculations indicate that FTY720P binding selectivity is a function of the entropic contribution to the binding free energy rather than enthalpic contributions and that preferred agonists retain substantial flexibility when bound. After exposure to FTY720P, the S1P2(IC1-TM2)S1P1 receptor recycled to the plasma membrane, indicating that additional structural elements are required for the selective degradative trafficking of S1P1.
Keywords: G Protein-coupled Receptors (GPCR), Ligand-binding Protein, Lipids, Membrane Trafficking, Receptor Structure-Function, Sphingolipid, FTY720, Gilenya, Sphingosine 1-Phosphate
Introduction
The lipid signaling molecule sphingosine 1-phosphate (S1P)3 is an important regulator of the cardiovascular and immune systems and functions in numerous physiological and pathophysiological conditions (for reviews, see Refs. 1–3). Although S1P has several intracellular targets and can function as a second messenger (4), many of its biological effects are mediated by activation of five members of the endothelial differentiation gene family of G protein-coupled receptors, S1P1–5. The five S1P receptors share high amino acid identity, ranging from 33 to 51% (5), and the individual receptor subtypes couple to distinct as well as overlapping cellular signaling pathways, allowing S1P to mediate its diverse cellular effects (6, 7).
The S1P receptor modulator FTY720 (Gilenya) has been approved as frontline treatment for relapsing/remitting multiple sclerosis (8, 9). FTY720 is phosphorylated in vivo by sphingosine kinase 2 to form the active metabolite FTY720 phosphate (FTY720P), which is a high affinity agonist of all the endothelial differentiation gene family S1P receptors except S1P2 (10–12). This type of receptor selectivity of FTY720P appears to be important to its therapeutic application because activation of S1P2 mediates several undesired responses including pathological angiogenesis, vascular leakiness, vasoconstriction, and increased vascular tone (13–17). Although FTY720P is an agonist of the S1P1 receptor, its therapeutic benefit is derived from its long term down-regulation of S1P1 signaling. Upon stimulation by the natural ligand S1P, S1P1 is internalized in endocytic vesicles, which subsequently recycle S1P1 back to the plasma membrane. In contrast, activation by FTY720P selectively leads to internal sequestration, ubiquitination, and degradation of the S1P1 receptor (18, 19). The immunosuppressive actions of FTY720P are dependent on the down-regulation of S1P1 surface expression on activated T cells, thereby rendering the T cells unresponsive to an S1P gradient in the blood and unable to egress from secondary lymphoid organs (20). Besides its effects on the immune system, the therapeutic benefit of FTY720P in multiple sclerosis might rely on S1P receptor modulation in the central nervous system. The down-regulation of S1P1 signaling in astrocytes appeared to be a primary protective mechanism in an experimental autoimmune encephalomyelitis mouse model (3).
The S1P2 receptor is not activated by para-substituted aromatic ligands such as FTY720P (10, 21). We have applied computational modeling-guided mutagenesis studies for mapping the common and distinguishing features of ligand recognition by endothelial differentiation gene family lysophosphatidic acid and S1P G protein-coupled receptors (22–27). Previously, we have developed and validated a computational model of the ligand-binding pocket of S1P1 that was used to successfully screen the NCI Developmental Therapeutics Library for two non-lipid S1P1 agonists (26). In the present study, we set out to identify the structural basis of the lack of activation of S1P2 by FTY720P. Based on the high degree of sequence similarity between S1P1 and S1P2, we first hypothesized that a computational homology model of the S1P2 ligand-binding pocket derived from the validated S1P1 model might reveal critical interactions with S1P that are lacking with FTY720P.
To test this hypothesis, we generated a library of S1P2 receptor constructs with point mutations to alter the charge, steric, or size properties of residues predicted to line the ligand-binding pocket. We also generated S1P1/S1P2 swap mutation receptors in which we replaced one or more amino acids of S1P2 with the corresponding S1P1 residues. We examined the effects of these mutations on ligand specificity in an effort to uncover the negative selectivity of S1P2 for FTY720P. However, none of these S1P2 mutations could recapitulate S1P1-like activation by FTY720P. This led us to hypothesize that the FTY720P selectivity determinants in the S1P1 and S1P2 receptors might not reside in the ligand-binding pocket; therefore, chimeric receptors were generated with domain exchanges between the S1P1 and S1P2 receptors. Testing the activation of and ligand binding to these receptors using S1P and FTY720P identified a minimal region of S1P1 in the first intracellular loop (IC) and second transmembrane domain (TM) that confers activation by FTY720P in a chimeric S1P2 receptor. Modeling studies indicate that FTY720P binding selectivity is a function of the entropic contribution to the binding free energy rather than enthalpic contributions. Preferred agonists retain substantial flexibility when bound. Using the S1P1/S1P2 chimera, we tested other S1P1-selective analogs including (R)-FTY720 phosphonate (FTY720PN) (21, 28), which also activated this chimera. Interestingly, neither FTY720P nor FTY720PN caused the selective internal sequestration of the S1P1/S1P2 chimera characteristic for S1P1, indicating that additional structural elements are required for the selective trafficking of the S1P1 receptor to the degradative pathway by these types of ligands.
EXPERIMENTAL PROCEDURES
Reagents
S1P and FTY720P were purchased from Avanti Polar Lipids (Alabaster, AL), and 1 mm stock solutions were prepared in PBS containing 1 mm charcoal-stripped bovine serum albumin (BSA). Anti-FLAG M2 monoclonal antibody and cycloheximide were purchased from Sigma-Aldrich. FTY720PN was synthesized (25) and dissolved as described previously (21). [32P]S1P was synthesized enzymatically using recombinant sphingosine kinase 1 as described previously (29); the specific activity was 86 cpm/fmol.
Residue Position Nomenclature
We have used Ballesteros-Weinstein nomenclature in which index positions are in the format X.XX. The first number specifies the TM of the residue, and the second number refers to the position within that TM relative to the most highly conserved residue in the G protein-coupled receptor superfamily that is designated as position 50 (30).
Computational Homology Modeling
Homology models of S1P1 and S1P2 in complex with S1P and FTY720P were used from a previous study (31). The starting structures of the four receptor-ligand complexes were minimized in MOE (Chemical Computing Group, Montreal, Canada, version 2010.10) using a distance-dependent implicit solvent treatment with a dielectric constant of 10 and the MMFF94 force field (32).
Chimeric S1P2 receptors with varying levels of FTY720P selectivity were generated from the S1P2 receptor complexes. One construct, S1P2(a)S1P1, was created by mutating motif a residues ARNS found in S1P2 to the WKTK residues found in S1P1. Hydrogen was added to the mutated residues. The remaining S1P2 chimeric structure, S1P2(a,b,c,d)S1P1, was created in the MOE program by splicing the IC1-TM2 segment of the S1P1 receptor model in place of the corresponding IC1-TM2 segment of the S1P2 receptor after superposition of two residues before and after each splice. Constructs were minimized first with the backbone fixed and then without positional restraints. Chimeric S1P1 receptors with varying levels of FTY720P selectivity were generated from the S1P1 receptor complexes. One construct, S1P1(b)S1P2, was created by mutating motif b residues RPMYY found in S1P1 to the SAMYL residues found in S1P2. Hydrogen was added to the mutated residues. The remaining S1P1 chimeric structure, S1P1(a,b,c,d)S1P2, was created in the MOE program by splicing the IC1-TM2 segment of the S1P2 receptor model in place of the corresponding IC1-TM2 segment of the S1P1 receptor after superposition of two residues before and after each splice. Constructs were minimized first with the backbone fixed and then without positional restraints.
Each receptor-ligand combination was used in molecular dynamics to observe the differences and similarities in structures and dynamics. Molecular dynamics simulations used the same force field and solvent treatment as minimizations. Three or four molecular dynamics simulations were performed for each complex depending on consistency between different simulations. Molecular dynamics simulations used the NVT (mole number, volume, temperature) ensemble and 2-fs time steps with light atom distance restraints with temperature raised to 300 K over 30 ps followed by a production phase of 100 ps at constant temperature. Root mean square fluctuations (RMSF) of ligand atom positions relative to the starting structure were determined from each simulation, and averages for each ligand atom were computed from three or four independent simulations.
Receptor Constructs and Mutagenesis
N-terminal FLAG epitope-tagged human S1P1 and S1P2 receptors in pcDNA3.1 were generous gifts from Dr. Timothy Hla (Weill Cornell Medical College, New York, NY). S1P2 point mutants were generated at specific residues using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA); all constructs were verified by complete sequencing of the inserts.
To generate chimeric receptor constructs, we first constructed a panel of receptors in which the N-terminal sequence of either S1P1 or S1P2 was fused to the C-terminal part of the other receptor to create single splice site chimeras. The splice overlap extension method was used to generate the N-terminal FLAG-tagged chimeric receptors in pcDNA3.1 (33). The single splice site chimeric receptors were used in a second round of splice overlap extension PCR to generate the domain replacement chimeras containing two splice junctions.
Construction of Receptor-Green Fluorescent Protein (GFP) Fusion Constructs
The S1P1, S1P2, or chimeric receptors were fused in-frame at the C terminus to the GFP from Aequorea coerulescens by subcloning the receptors into the pAcGFP-N1 vector (Clontech). The receptors in pcDNA3.1 were amplified using primers engineered to introduce a KpnI restriction site 5′ to the N-terminal FLAG epitope. For receptors with N-terminal FLAG-S1P1 sequence, the forward primer 5′-TCA GAT CGC GGT ACC ACC ATG GAC TAC AAG GAC GAC GAT GAC AGG ACC CAC CAG CG-3′, which also introduced a silent mutation to abolish an ApaI restriction site at the third amino acid of the S1P1 sequence, was used. For receptors with N-terminal FLAG-S1P2 sequence, the forward primer 5′-ACC ATG GGC GCT TGT ACT CGG AGT ACC TGA ACC CCA AC-3′ was used. For receptors containing C-terminal S1P1 sequence, the reverse primer 5′-GAC CGG TGG ATC CCG GGC CCC GGA AGA AGA GTT GAC-3′ was used to introduce an ApaI site to fuse the receptor in-frame at the C terminus to the GFP polypeptide. For receptors containing N-terminal S1P2 sequence, the reverse primer 5′-GAC CGG TGG ATC CCG GGC CCC GAC CAC CGT GTT GC-3′ was used. PCRs were carried out using the Expand High Fidelity PCR system (Roche Applied Science) using the following conditions: denaturation (95 °C for 2 min) and 30 cycles of annealing/extension (94 °C for 1 min, 55 °C for 1 min, and 72 °C for 2 min) followed by an anneal/extension step (72 °C for 7 min). Purified PCR products were digested with KpnI and ApaI (Promega, Madison, WI) and ligated into the pAcGFP-N1 vector. Positive clones were verified by complete sequencing of inserts and splice sites.
Cell Culture and Transfection
McArtl rat hepatoma RH7777 cells (ATCC, Manassas, VA), rat hepatoma HTC4 cells (a kind gift from Dr. Edward Goetzl, University of California, San Francisco), and HEK293 cells (ATCC) were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin, 10 μg/ml streptomycin, and 2 mm glutamine. For cellular Ca2+ assays of the N-terminal FLAG-tagged wild type (WT) and point mutant S1P2 receptors, the constructs were transfected into RH7777 cells using Xfect transfection reagent (Clontech). The receptor constructs were cotransfected with an equal amount of Gαq plasmid to allow the receptors to couple effectively to Ca2+ mobilization. For cellular Ca2+ assays of the N-terminal FLAG-tagged S1P1, S1P2, and chimeric receptors, HTC4 cells were cotransfected with an equal amount of Gα16 plasmid and receptor plasmid using Effectene transfection reagent (Qiagen, Valencia, CA). HEK293 cells were transfected with N-terminal FLAG-tagged S1P receptor constructs fused at the C terminus to GFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The transfected HEK293 cells were selected for stable expression in growth medium supplemented with 1.0 mg/ml G418 (Invitrogen).
Flow Cytometric Analysis
Expression of receptor constructs on the cell surface was measured by flow cytometry using indirect immunofluorescence staining with anti-FLAG M2 antibody as described previously (26).
Radioligand Binding
Ligand binding assays were performed as described previously (27). Briefly, HEK293T cells were plated in 24-well plates (3 × 105 cells/well) and the following day transfected with 0.6 μg of receptor constructs using Lipofectamine 2000 (Invitrogen). Two days later the cells were washed with ice-cold binding buffer (50 mm Tris, pH 7.4, 150 mm NaCl). The cells were then incubated in binding buffer containing 4 mg/ml fatty acid-free BSA and 3 nm [32P]S1P on ice for 45 min in the presence or absence of 3 μm unlabeled S1P or FTY720P as a competitor. The cells were washed twice with cold binding buffer containing 0.4 mg/ml BSA and then lysed in 0.5% SDS, and binding was quantified by scintillation counting. Quadruplicate samples were measured for each condition.
Receptor Activation/Ca2+ Mobilization Assays
To examine the impact of point mutations on receptor activation, N-terminal FLAG-tagged receptor constructs were transiently expressed in RH7777 cells. After 6 h, the cells were detached using HyQtase Cell Detachment Solution, replated into poly-l-lysine-coated 96-well plates (30,000 cells/well), and cultured for 16 h. The culture medium was replaced with Krebs buffer, 4 h later the cells were incubated with Fura-2-AM in Krebs buffer containing 0.001% pluronic acid for 30 min and rinsed with Krebs buffer, and the Ca2+ response to S1P, FTY720P, or FTY720PN was measured using a Flex Station II fluorescence plate reader (Molecular Devices, Sunnyvale, CA). The ratio of peak emissions at 510 nm 2 min after ligand addition was determined for excitation wavelengths of 340/380 nm. All samples were run in triplicate, and assays were performed at least three times with a different transfection for each receptor construct. The response elicited by the agonists was measured, and maximal activation (Emax; efficacy) and potency (EC50) were determined. Activation of the FLAG-tagged chimeric S1P1/S1P2 receptors constructs was examined in transiently transfected HTC4 cells, which show no endogenous Ca2+ response to S1P. The assays were performed similarly to the assays that utilized RH7777 cells except that the HTC4 cells were replated into poly-l-lysine-coated 96-well microplates 16 h after transfection at a density of 40,000 cells/well and cultured for 24 h before performing the Ca2+ mobilization assays.
Receptor Internalization
HEK293 cells stably transfected with S1P receptor-GFP constructs were plated on poly-l-lysine-coated glass coverslips in 24-well plates (30,000–60,000 cells/well). Forty hours later, the complete medium was replaced with serum-free medium. After 3 h, cells were treated with either vehicle (100 nm charcoal-stripped BSA in PBS) or agonist for 30 min. The cells were either washed in ice-cold PBS and fixed for 15 min in 100% methanol at −20 °C or in some cases rinsed twice with serum-free medium and incubated for 2 h in serum-free medium that contained 15 μg/ml cycloheximide before fixation. Microscopy was performed using a Zeiss LSM 510 Pascal laser-scanning confocal microscope.
RESULTS
Mutagenesis Strategy
To validate and refine the previously published S1P2 computational model (31), 22 residues in TMs 3, 4, 5, 6, and 7 predicted to line the ligand-binding pocket (Fig. 1) were mutated to alter their charge, size, and/or steric properties to create 37 point mutants. The receptor constructs were transiently expressed in RH7777 cells, and the cell surface expression was measured by anti-FLAG epitope flow cytometric analysis. The constructs showed variable expression at the cell surface, ranging from 5 to 41% FLAG-positive cells (supplemental Table 1).
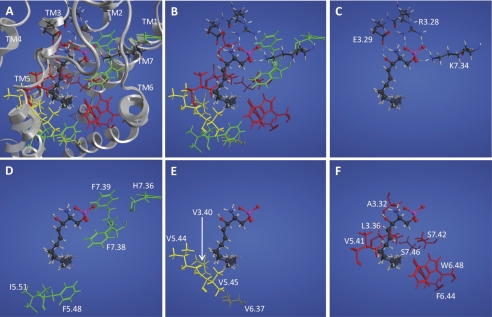
FIGURE 1.
Model S1P2 complex with S1P. All panels show the same view of the complex with S1P (ball and stick) and different subsets of S1P2 residues (stick) shown. A, complex showing all mutation sites with the ribbon (gray) with transmembrane domains labeled at the extracellular end. B, complex showing all mutation sites. C, complex showing mutated residues interacting with the polar head group. Hydrogen bonding/ionic interactions are shown as dotted lines. D, mutation sites in green at which all mutations showed WT activation by S1P. E, mutation sites in yellow at which a mutant showed only moderately diminished activation by S1P. F, mutation sites in red at which a mutant showed impaired or abolished activation by S1P.
WT S1P2 plasmid was diluted with different amounts of empty vector plasmid and transfected into RH7777 cells to determine the effect of cell surface expression on EC50 and Emax values in cellular Ca2+ mobilization assays. Transfection with receptor plasmid that was diluted 6-fold resulted in only 8% FLAG-positive cell staining compared with 20% for cells transfected with undiluted receptor plasmid; however, EC50 and Emax values were similar to those obtained for cells transfected with the undiluted receptor (supplemental Table 2). Seven of the S1P2 point mutant constructs, F3.33A, F3.33S, L3.43E, W4.64A, K5.38A, K5.38N, and K5.38Q, showed less than 8% cell surface expression and were excluded from further analysis. For the receptors that did show at least 8% surface expression, we assessed the effect of each mutation on receptor activation. The receptor constructs were transiently coexpressed in RH7777 cells with Gαq protein to facilitate coupling of the receptors to cellular calcium mobilization, and the EC50 and Emax of the Ca2+ responses elicited by S1P and FTY720P were measured (Table 1).
TABLE 1.
Properties of S1P2 receptors with point mutations
EC50 values (mean ± S.D., n = 3) and Emax values were determined in RH7777 cells transiently expressing S1P2 receptor constructs. 100% represents the maximal S1P-elicited Ca2+ mobilization response of the WT S1P2 receptor. na, not activated.
| Receptor construct | S1P |
FTY720P |
||
|---|---|---|---|---|
| EC50 ± S.D. | Emax | EC50 ± S.D. | Emax | |
| nm | % | nm | % | |
| S1P2 WT | 10 ± 3 | 100 | 359 ± 69 | 56 |
| Mutations of residues interacting with the polar head group | ||||
| R3.28A | na | na | na | na |
| R3.28K | 191 ± 98 | 96 | na | na |
| E3.29A | >1000 | 106 | na | na |
| E3.29Q | na | na | na | na |
| K7.34A | 123 ± 75 | 58 | na | na |
| K7.34R | 28 ± 6 | 97 | 383 ± 63 | 52 |
| Mutations in the hydrophobic binding pocket that retained wild-type-like activation by S1P (EC50 < 30 nm and Emax > 80% of WT S1P2) | ||||
| V3.40T | 22 ± 5 | 101 | 892 ± 419 | 40 |
| V5.44C | 18 ± 12 | 87 | 210 ± 208 | 52 |
| V5.45T | 14 ± 7 | 94 | 384 ± 345 | 49 |
| F5.48A | 8 ± 3 | 97 | 509 ± 142 | 56 |
| I5.51A | 8 ± 3 | 83 | 77 ± 33 | 56 |
| I5.51L | 10 ± 1 | 111 | 473 ± 524 | 43 |
| H7.36E | 22 ± 8 | 107 | 379 ± 259 | 45 |
| F7.38A | 10 ± 4 | 96 | 292 ± 92 | 56 |
| F7.39L | 9 ± 3 | 92 | 132 ± 74 | 46 |
| S7.42A | 23 ± 8 | 100 | 518 ± 99 | 51 |
| Mutations in the hydrophobic binding pocket with moderately diminished activation by S1P (EC50 > 30 nm or Emax < 80% of WT S1P2) | ||||
| L3.36A | 50 ± 23 | 76 | na | na |
| V3.40A | 32 ± 18 | 79 | na | na |
| V5.44A | 48 ± 16 | 88 | na | na |
| V5.45A | 16 ± 6 | 76 | na | na |
| V6.37T | 37 ± 9 | 102 | na | na |
| W6.48E | 30 ± 9 | 102 | na | na |
| S7.46H | 40 ± 17 | 112 | na | na |
| Mutations in the hydrophobic binding pocket with impaired or abolished activation by S1P (EC50 > 50 nm or Emax < 70% of WT S1P2) | ||||
| A3.32K | na | na | na | na |
| A3.32M | 93 ± 27 | 118 | 420 ± 132 | 59 |
| L3.36Q | na | na | na | na |
| V5.41A | 25 ± 14 | 59 | na | na |
| F6.44A | 56 ± 28 | 62 | na | na |
| W6.48A | na | na | na | na |
| S7.42L | 15 ± 11 | 67 | na | na |
| S7.42H | 81 ± 28 | 54 | na | na |
| S7.46L | na | na | na | na |
| S1P2 receptor constructs with multiple point mutations | ||||
| CVVTI5.43-47FCTTV | 7 ± 2 | 89 | na | na |
| CVVTI5.43-47FCTTV+F7.30L | 14 ± 6 | 84 | na | na |
| SII5.49-51TLL | 7 ± 3 | 89 | na | na |
| FAVST7.39-43LVLAV | 9 ± 6 | 51 | na | na |
Effects of Mutations of Conserved Residues Interacting with Polar Head Group of S1P
In previous studies of the S1P1 and S1P4 ligand-binding pocket, we concluded that the amino acid residues Arg3.28 and Glu3.29 form obligate ion pair interactions with the phosphate and hydroxyl groups of S1P and that the Arg7.34 residue of S1P1 also forms an ionic interaction with the phosphate group of S1P that is required for ligand binding and activation (22, 25). The model complex of S1P with S1P2 (Fig. 1, A–C) suggests similar roles for residues at these positions in S1P2. Experimentally, the R3.28A mutant of S1P2 was not activated in our assays, whereas the R3.28K mutant was activated by S1P albeit with a 19-fold reduced potency (EC50 = 191 ± 98 versus 10 ± 3 nm for WT S1P2), indicating that Arg3.28 is also required for S1P2-S1P head group interactions, but could be partially compensated for by a positively charged lysine residue. The E3.29A showed a 100-fold reduced potency for S1P (EC50 > 1000 nm), and the E3.29Q mutant was completely inactive. The K7.34A mutant showed decreased activation (EC50 = 123 ± 75 nm, Emax = 58% of WT S1P2), indicating the key role of this residue in S1P2-S1P head group interactions. The charge-conservative substitution of Lys7.34 with arginine found at this position in S1P1, K7.34R, yielded a receptor with nearly WT S1P2-like activity albeit slightly decreased potency for S1P (EC50 = 28 ± 6 versus 10 nm ± 3 nm for WT S1P2). Thus, our data indicate that the Arg3.28, Glu3.29, and Lys7.34 residues form a critical tripartite interaction with the polar head group of S1P (Fig. 1C) similar to the Arg3.28/Glu3.29/Arg7.34 triad of S1P1 (25, 26).
Effects of Mutations of Residues Lining Predicted Hydrophobic Binding Pocket of S1P2
Previously, we identified residues in the hydrophobic binding pocket of S1P1 that interact with the aliphatic chain of S1P; some of these residues are essential for ligand activation (26). We mutated the corresponding residues in the predicted hydrophobic binding pocket of S1P2 to examine their impact on activation by S1P. Based on the responses measured in our Ca2+ mobilization assays, we categorized the effects of the mutations into three categories: mutants with WT-like S1P activation (EC50 < 30 nm and Emax > 80% of WT S1P2), mutants with diminished S1P activation (EC50 > 30 nm or Emax < 80%), and mutants with impaired or abolished S1P activation (EC50 > 50 nm or Emax < 70%) (Table 1). Mutations of five residues, F5.48A, I5.51A, H7.36E, F7.38A, and F7.39L, produced receptors that retained WT-like activation by S1P consistent with limited contact between these sites and S1P in the model (Fig. 1D). Mutations of five other residues, V3.40A, V5.44A, V5.45A, V6.37T, and W6.48E, showed moderately diminished activation by S1P. Four of these residues show moderate interactions with the S1P hydrophobic tail in our homology model (Fig. 1E). Seven other residues predicted by our model to interact with the S1P hydrophobic tail (Fig. 1F), Ala3.32, Leu3.36, Val5.41, Phe6.44, Trp6.48, Ser7.42, and Ser7.46, when mutated impaired or abolished activation by S1P in our cellular Ca2+ mobilization assays (Fig. 1F and Table 1).
Effects of Mutations on S1P2 Receptor Activation by FTY720P
FTY720P is a high affinity agonist of all the S1P receptors except S1P2, although high concentrations of FTY720P activate S1P2-mediated Ca2+ mobilization in some experimental settings (21). In RH7777 cells co-transfected with Gαq, FTY720P elicited a weak but measurable calcium efflux response from the WT S1P2 receptor (EC50 = 359 ± 69 nm, Emax = 56% of the maximal S1P response; Table 1). Prior modeling studies did not identify substantial differences in the interactions of S1P and FTY720P with S1P2 (31). To investigate the molecular basis of the negative selectivity of S1P2 for FTY720P, we tested the S1P2 ligand-binding pocket mutants for Ca2+ mobilization in response to FTY720P. Most of the receptor constructs with WT-like S1P activation retained weak activation by high concentrations of FTY720P similar to WT S1P2, whereas most mutants with moderately diminished, impaired, or abolished activation by S1P lacked any detectable activation by FTY720P (Table 1).
As part of our strategy to uncover the basis for ligand selectivity of the S1P1 and S1P2 receptors, we generated mutants in which we replaced one or more amino acids of S1P2 with the corresponding S1P1 residue. This strategy had only limited success. The S1P1/S1P2 swap mutation A3.32M impaired potency by S1P (93 ± 27 versus 10 ± 3 nm for WT S1P2) but retained WT S1P2-like activation by FTY720P (EC50 = 430 ± 132 nm, Emax = 59%), and the alanine mutant rather than the swap leucine mutant of Ile5.51 showed enhanced potency for FTY720P (77 ± 33 versus 359 ± 69 nm for WT S1P2). The other S1P1/S1P2 swap mutants, V5.44C, V5.45T, F7.39L, S7.42A, and H7.36E, retained activation by S1P but still showed only weak activation by FTY720P.
To further investigate the negative selectivity of S1P2 for FTY720P, we created receptor constructs with multiple S1P1/S1P2 swap mutations of residues in TM5 and TM7 that line the predicted ligand-binding pocket. Although these receptor constructs could all be activated by S1P to some degree, none of them were activated by FTY720P (Table 1). Although the point mutants we generated revealed many similarities between the S1P2 model and our previously validated S1P1 and S1P4 models, we were unable to identify single or multiple residue replacements that could bestow FTY720P responsiveness to S1P2. Therefore, we undertook a different experimental strategy.
Chimeric S1P1/S1P2 Receptors
Because none of the S1P2 receptors we made with single or multiple S1P1 swap mutations in the ligand-binding pocket gave a robust response to FTY720P in our assays, we considered that the FTY720P selectivity determinants might reside in other regions of the receptors. FTY720P functions as a biased ligand for the S1P3 receptor, selectively stimulating S1P3-coupled Gαi activation but inhibiting S1P3-coupled Gαq activation (34). This suggested that G protein-coupling and receptor conformation may coordinately impact ligand specificity. We hypothesized that the FTY720P ligand specificity determinants of the S1P1 and S1P2 receptors might partly reside in the regions that determine G-protein coupling selectivity, the ICs and C-terminal tail (CT) of the receptors. We constructed a panel of chimeric S1P1/S1P2 receptors in which we swapped these domains between the two receptors to examine their effect on receptor activation and ligand selectivity. To test the activation of the chimeric receptors by S1P or FTY720P, the receptors were transiently expressed in HTC4 cells, which lack any endogenous Ca2+ response to S1P. Gα16 was coexpressed with the chimeric receptors because it allowed both S1P1 and S1P2 to couple to Ca2+ mobilization in our cellular assays.
Characterization of Single Splice Site Chimeric Receptors
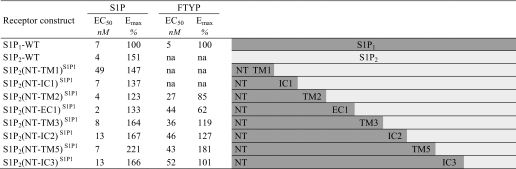
In constructing the S1P1/S1P2 domain swap chimeras, we first generated a panel of receptors in which the N-terminal sequence of either S1P1 or S1P2 was fused to the C-terminal part of the other receptor. The single splice junction chimeric receptors were characterized for their cellular surface expression by anti-FLAG antibody flow cytometry analysis (supplemental Table 3). Cells transfected with vector showed 2% background staining, whereas the WT S1P1-transfected cells exhibited 13% positive staining, and the WT S1P2-transfected cells showed 18% positive staining. The chimeric S1P2 receptors with replacements of the N-terminal domains (NT) with S1P1 sequence, S1P2(NT-TM1)S1P1, S1P2(NT-IC1)S1P1, S1P2(NT-TM2)S1P1, S1P2(NT-EC1)S1P1, S1P2(NT-TM3)S1P1, S1P2(NT-IC2)S1P1, S1P2(NT-TM5)S1P1, and S1P2(NT-IC3)S1P1, all resulted in receptors that were expressed on the cell surface at comparable levels, ranging from 12 to 19% (supplemental Table 3). The chimeric receptors all retained nearly WT S1P2-like activation by S1P except for the S1P2(NT-TM1)S1P1 receptor, which had moderately reduced potency for S1P (EC50 = 49 versus 4 nm for WT S1P2) (Table 2). The receptors were also tested for activation by FTY720P. Similar to WT S1P2, the S1P2(NT-TM1)S1P1 and S1P2(NT-IC1)S1P1 receptors were unresponsive to FTY720P. However, S1P2 chimeric receptors further substituted with S1P1 N-terminal proximal domains, S1P2(NT-TM2)S1P1 and S1P2(NT-EC1)S1P1, were each activated by FTY720P although with decreased potency and efficacy compared with WT S1P1 (EC50 = 27 or 44 nm versus 5 nm for WT S1P1 and Emax = 85 or 62% of the WT S1P1 response). The S1P2 chimeric receptors that were even more substituted from the N terminus with S1P1 sequence, S1P2(NT-TM3)S1P1, S1P2(NT-IC2)S1P1, S1P2(NT-TM5)S1P1, and S1P2(NT-IC3)S1P1, were also activated by FTY720P with a similar decreased potency (EC50 ranging from 36 to 52 nm) but with an increase in efficacy for FTY720P (Emax ranging from 101 to 181% of the WT S1P1 response) (Table 2). The chimeric receptors we generated containing C-terminal S1P1 sequence, S1P2(TM6-CT)S1P1, S1P2(IC3-CT)S1P1, S1P2(TM4-CT)S1P1, S1P2(IC2-CT)S1P1, S1P2(TM3-CT)S1P1, S1P2(EC1-CT)S1P1, and S1P2(TM2-CT)S1P1, all showed dramatically reduced or no surface expression (supplemental Table 3) and were unresponsive to both S1P and FTY720P in Ca2+ mobilization assays (data not shown).
TABLE 2.
Properties of S1P1 and S1P2 chimeric receptors
EC50 (mean, n = 3) and Emax values were determined in HTC4 cells transiently expressing WT S1P1, WT S1P2, and S1P1/S1P2 chimeric receptors. 100% represents the maximal response for Ca2+ mobilization of the WT S1P1 receptor for S1P or FTY720P. na, not activated.
Characterization of S1P2 Chimeric Receptors with S1P1 Domain Insertions
In the S1P2(NT-TM3)S1P1 chimera, the collective replacement of the NT, TM1, IC1, TM2, EC1, and TM3 domains with S1P1 sequence bestowed responsiveness to FTY720P (Table 2). To assign the FTY720P selectivity determinants to individual domains, we created a panel of S1P1 and S1P2 chimeric receptors with each of these domains individually replaced with the corresponding region of the other receptor. We also created constructs in which we swapped the CT, IC2, or IC3 domains, which are known determinants of G protein selectivity (35). These chimeric receptors were tested for surface expression and Ca2+ mobilization in response to S1P and FTY720P (Table 3).
TABLE 3.
Properties of S1P1 and S1P2 domain replacement chimeric receptors
EC50 values (mean, n = 3) and Emax values were determined in HTC4 cells transiently expressing WT S1P1, WT S1P2, and S1P1/S1P2 chimeric receptors. 100% represents the maximal response for calcium mobilization of the wild-type S1P2 receptor (for S1P) or S1P1 (for FTY720P). na, not activated.
| S1P |
FTY720P |
FLAG | |||
|---|---|---|---|---|---|
| EC50 | Emax | EC50 | Emax | ||
| nm | % | nm | % | % | |
| S1P2 receptor constructs | |||||
| S1P2-WT | 4 | 151 | na | na | 22 |
| S1P2(NT)S1P1 | 10 | 142 | na | na | 13 |
| S1P2(TM1)S1P1 | 9 | 148 | na | na | 23 |
| S1P2(IC1)S1P1 | 1 | 155 | 112 | 78 | 11 |
| S1P2(IC1-TM2)S1P1 | 1 | 150 | 35 | 100 | 14 |
| S1P2(TM2)S1P1 | 5 | 173 | 112 | 97 | 19 |
| S1P2(EC1)S1P1 | 4 | 160 | na | na | 23 |
| S1P2(TM3)S1P1 | 9 | 155 | na | na | 24 |
| S1P2(IC2)S1P1 | 3 | 162 | na | na | 27 |
| S1P2(IC3)S1P1 | 2 | 166 | na | na | 19 |
| S1P2(CT)S1P1 | na | na | na | na | 10 |
| S1P1 receptor constructs | |||||
| S1P1-WT | 7 | 100 | 5 | 100 | 13 |
| S1P1(NT)S1P2 | 9 | 142 | 33 | 127 | 22 |
| S1P1(TM1)S1P2 | 10 | 70 | 35 | 71 | 10 |
| S1P1(IC1)S1P2 | 15 | 60 | na | na | 7 |
| S1P1(TM2)S1P2 | 5 | 105 | 20 | 113 | 12 |
| S1P1(EC1)S1P2 | 7 | 90 | 27 | 99 | 15 |
| S1P1(TM3)S1P2 | 3 | 157 | 21 | 91 | 9 |
| S1P1(IC2)S1P2 | 3 | 75 | 4 | 101 | 9 |
| S1P1(IC3)S1P2 | 9 | 102 | 11 | 88 | 11 |
| S1P1(CT)S1P2 | na | na | na | na | 4 |
S1P2 chimeric receptors with individual internal S1P1 domain insertions all showed surface expression ranging from 11 to 27% FLAG-positive stained cells. The receptors maintained nearly WT S1P2-like activation by S1P with EC50 values ranging from 1 to 10 nm (versus 4 nm for WT S1P2) and efficacies that ranged from 142 to 173% of the S1P1 Emax for S1P (versus 151% for WT S1P2). The S1P2(CT)S1P1 construct showed diminished surface expression (10% FLAG-positive) compared with the other receptors and showed no activation by S1P.
Replacement of either the IC1 or TM2 domain in S1P2 with the corresponding sequence of S1P1 enabled the chimera to be partially activated by FTY720P although to a lesser extent than WT S1P1. The S1P2(IC1)S1P1 was activated by FTY720P with an EC50 of 60 nm and Emax that was 78% of WT S1P1. The S1P2(TM2)S1P1 receptor showed a similar partial responsiveness to FTY720P with lower potency (EC50 = 112 nm) but WT S1P1-like efficacy (Emax = 97% of WT S1P1). None of the other single domain-swapped S1P2 chimeras showed any response to FTY720P. Because the IC1 or TM2 S1P1 domain alone could impart partial activation by FTY720P, we tested whether the receptor with both domains replaced with the corresponding S1P1 sequence, S1P2(IC1-TM2)S1P1, would be even more responsive to FTY720P. The S1P2(IC1-TM2)S1P1 receptor retained full WT S1P2-like activation by S1P and was activated by FTY720P with an efficacy comparable with WT S1P1 but with a diminished potency (EC50 = 35 versus 5 nm for WT S1P1; Table 3).
Characterization of S1P1 Chimeric Receptors with S1P2 Domain Insertions
Replacement of any of the NT, TM1, IC1, TM2, EC1, TM3, IC2, or IC3 of S1P1 with the corresponding domain of S1P2 all resulted in receptors that were expressed at the cell surface with the percentage of FLAG-positive stained cells ranging from 7 to 22% (Table 3). The S1P1(NT)S1P2 and S1P1(EC1)S1P2 constructs showed relatively high expression (22 and 15%, respectively). In contrast, replacement of the S1P1 CT with that of S1P2 had a detrimental effect on cell surface expression (4% FLAG-positive stained cells), and the receptor construct was not activated in our cellular assays. The S1P1(IC1)S1P2 and S1P1(IC2)S1P2 constructs both showed somewhat decreased expression and activation in response to S1P. The S1P1(IC1)S1P2 receptor was not activated by FTY720P, whereas the S1P1(IC2)S1P2 receptor was, supporting the role of the IC1 domain as a determinant of FTY720P selectivity.
Radioligand Binding Experiments
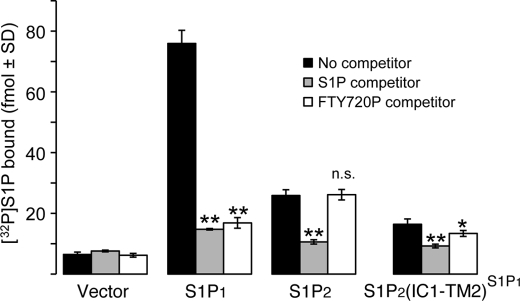
To obtain additional evidence for the specific interaction of S1P2(IC1-TM2)S1P1 chimera with S1P and FTY720P, we examined the ability of S1P and FTY720P to compete with [32P]S1P for binding to WT S1P1, WT S1P2, or S1P2(IC1-TM2)S1P1 receptors (Fig. 2). Expression of WT S1P1, WT S1P2, or S1P2(IC1-TM2)S1P1 in HEK293T cells enabled the specific binding of [32P]S1P. [32P]S1P radioligand binding was competed from all three receptors with 3 μm excess cold S1P. However, competition took place with 3 μm cold FTY720P only in cells transfected with WT S1P1 or S1P2(IC1-TM2)S1P1 but not WT S1P2. These results provide added support to the data obtained from functional assays for the specific interaction of FTY720P with the S1P2(IC1-TM2)S1P1 receptor chimera.
FIGURE 2.
Ability of S1P and FTY720P to compete with [32>P]S1P for binding to S1P1, S1P2, and S1P1/S1P2 chimeric receptors. HEK293T cells were transfected with WT S1P1, WT S1P2, or S1P2(IC1-TM2)S1P1 receptors. [32P]S1P radioligand binding was determined using 3 nm [32P]S1P. Cells were incubated with 3 μm unlabeled S1P or FTY720P as the competitor. The means ± S.D. were plotted. *, p < 0.05; **, p < 0.01 relative to no competitor. n.s., not significant.
Identification of Motifs in IC1-TM2 Affecting Activation by FTY720P
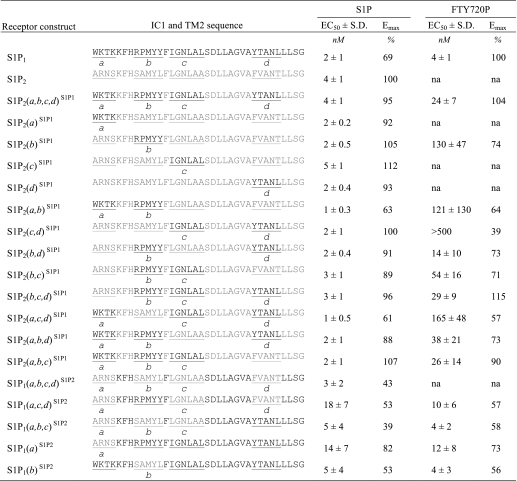
The S1P2(IC1-TM2)S1P1 chimera has two domain replacements from S1P1 consisting of 36 amino acid residues. However, because of the high homology in the endothelial differentiation gene family S1P receptors, only 12 of the 36 residues actually differ between S1P1 and S1P2. The 12 residues that differed could be clustered into four motifs, a–d (Table 4). To determine which S1P1 motif was necessary for activation by FTY720P, each motif was separately mutated back to the S1P2 sequence, and the resultant receptors were tested for activation by S1P and FTY720P. Individual removal of any of the motifs b, c, or d but not a diminished receptor activation by FTY720P with motif b having the greatest negative impact (Table 4).
TABLE 4.
Effects of swapping IC1 and TM2 motifs between S1P1 and S1P2 on receptor activation
Calcium mobilization in response to S1P and FTY720P was measured in WT S1P1, WT S1P2, and chimeric receptors in which different combinations of motifs a–d in IC1 and TM2 were replaced with the corresponding sequence from the other receptor subtype. EC50 values (mean ± S.D., n = 3) and Emax values were determined in HTC4 cells transiently expressing the receptor constructs. 100% represents the maximal response measured in the WT S1P1 or S1P2 receptors for S1P or the maximal S1P1 response for FTY720P. na, not activated.
Insertion of motif a in IC loop 1 alone into S1P2 in the S1P2(a)S1P1 chimera did not impart responsiveness to FTY720P, although the receptor remained fully responsive to S1P. Conversely, the S1P2(b,c,d)S1P1 receptor with motif a swapped back to the S1P2 sequence retained full activation by FTY720P, indicating that differences in motif a alone are not required for FTY720P activation.
Motif b, when inserted alone into S1P2, bestowed FTY720P responsiveness (EC50 = 130 ± 47 nm, Emax = 74% of WT S1P1). In contrast, insertion of either motif c or d in S1P2(c)S1P1 or S1P2(d)S1P1 did not introduce FTY720P responsiveness. However, the combination of the two motifs in S1P2(c,d)S1P1 did impart a weak response to FTY720P. When both of these motifs were combined with b, the resulting chimera, S1P2(b,c,d)S1P1, was activated as much as S1P2(a,b,c,d)S1P1 by FTY720P in terms of both efficacy and potency. Thus, the eight residues unique to S1P1 in motifs b, c, and d contribute to the FTY720P responsiveness of the S1P2(IC1-TM2)S1P1 chimeric receptor.
Reverse swaps of motifs a, b, c, and d together from S1P2 inserted into S1P1 but not individually abolished the response to FTY720P (Table 4). Insertion of sequences into S1P1 limited to motifs a, b, and c; a, c, and d; a; or b of S1P2 did not abolish the activation of the chimeras by FTY720P. These results reinforce the hypothesis that the distant allosteric constraint exerted on the ligand-binding pocket required for activation by FTY720P resides in a combination of multiple residues that differ between S1P1 and S1P2.
Structural Influence of IC1-TM2 Motifs on S1P and FTY720P Complexes
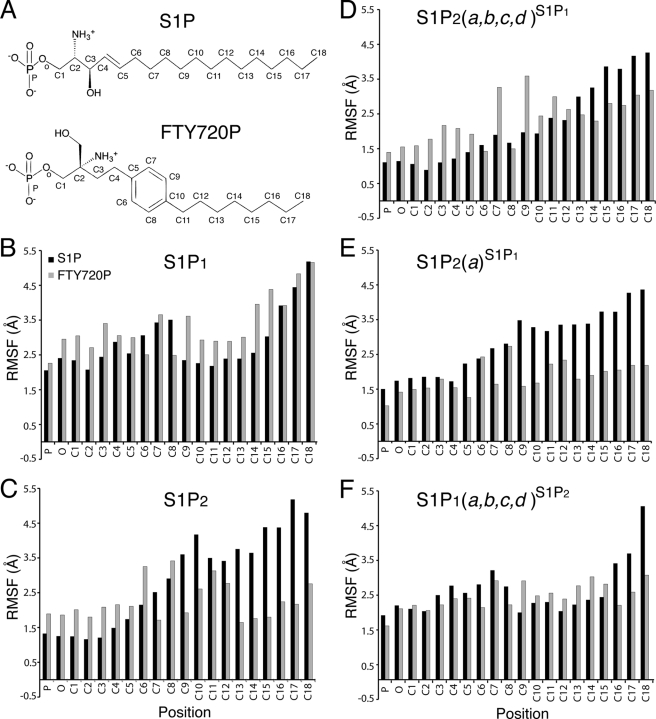
We utilized molecular dynamics simulations to investigate how the IC1-TM2 motifs, which are not located in close proximity to the ligand-binding site, influence binding preferences. Multiple simulations were performed for WT S1P1, WT S1P2, the S1P2(a)S1P1 chimera, the S1P2(a,b,c,d)S1P1 chimera, and the S1P1(a,b,c,d)S1P2 chimera. Fig. 3 shows the RMSFs of the ligand atoms during the molecular dynamics simulations. All complexes are characterized by relatively low mobility of the tightly ion-paired phosphate groups as reflected by RMSF values less than 2.5 Å for the phosphorus and oxygen of the phosphate ester bond. All S1P complexes show substantially higher mobility of the hydrophobic tail (RMSF values in excess of 4 Å), and some S1P complexes show higher mobility (RMSF values in excess of 3.5 Å) near the center of the chain. In contrast, higher mobility as reflected by RSMF values in excess of 3.5 Å was observed for FTY720P complexes only for WT S1P1 and S1P2(a,b,c,d)S1P1. Molecular dynamics simulations with the reverse S1P1(a,b,c,d)S1P2 chimera as well as the S1P2(a)S1P1 chimera showed low RMSF values consistent with the experimentally validated lack of activation of these receptors by FTY720P (Table 4 and Fig. 3). These results suggest that the combination of unique aspects of the S1P2 IC1-TM2 sequence and the planar para-substituted aromatic ring of FTY720P produce an unfavorable entropic penalty due to the loss of conformational degrees of freedom upon FTY720P binding that confers the negative selectivity of S1P2 for FTY720P.
FIGURE 3.
RMSF by ligand atom position. RMSF values in Å are reported as averages from three to four simulations of each construct in complex with either S1P (black bars; A, top structure) or FTY720P (gray bars; A, bottom structure) using the position labels shown on the structures. Note that the gray bars corresponding to the RMSFs of FTY720P are considerably smaller in S1P2 (C) and S1P2(a)S1P1 (E) than S1P1 (B), indicating that this ligand has restricted mobility in the ligand-binding pocket. In contrast, the RMSF values of FTY720P carbons in chimera S1P2(a,b,c,d)S1P1 (C) are substantially larger than in S1P2 or S1P2(a)S1P1 (C and E). The reverse chimera S1P1(a,b,c,d)S1P2 (F) shows S1P2-like (C) RMSF values for FTY720P carbons.
Activation of S1P2(IC1-TM2)S1P1 Chimera by Other S1P1-specific Ligands
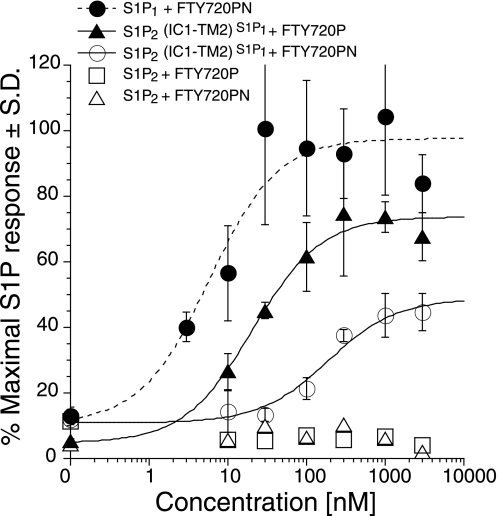
We investigated whether the insertion of the IC1-TM2 domains into S1P2 will allow activation by other FTY720-like S1P1-specific agonists. Previously, we characterized FTY720PN against the S1P1–5 receptors expressed in HTC4 stable cell lines and found that the FTY720PN was a strong activator of S1P1 but did not activate S1P2 (21). We tested activation of S1P1, S1P2, and the S1P2(IC1-TM2)S1P1 receptors for activation by FTY720PN in transiently transfected HTC4 cells (Fig. 4). The S1P1 receptor was activated by FTY720PN although with a decreased potency compared with FTY720P (EC50 = 6 ± 3 versus 1 nm for FTY720P) but similar efficacy (Emax = 98% of maximal S1P response). The chimeric receptor S1P2(IC1-TM2)S1P1 was activated by FTY720PN but to a lesser extent than by FTY720P in terms of potency and efficacy (EC50 = 188 ± 69 nm for FTY720PN versus 21 ± 5 nm for FTY720P and Emax = 49% for FTY720PN versus 74% for FTY720P).
FIGURE 4.
FTY720PN activates S1P1 and S1P2(IC1-TM2)S1P1 chimeric receptor but not S1P2. HTC4 cells were transfected with WT S1P1, WT S1P2, or S1P2(IC1-TM2)S1P1 receptors, and intracellular Ca2+ transients were measured in response to FTY720PN. S1P2 and S1P2(IC1-TM2)S1P1 responses to FTY720P are shown as a reference. Samples were run in triplicate and normalized to the maximal S1P response for each receptor, and the means ± S.D. were plotted.
Receptor Trafficking of S1P2(IC1-TM2)S1P1
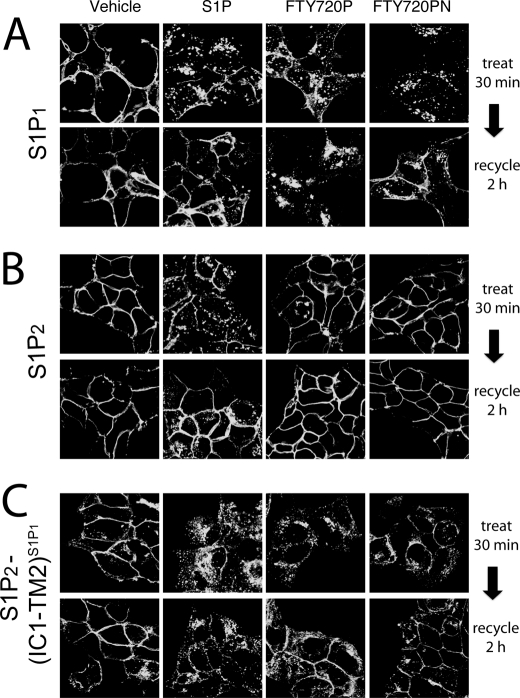
Upon activation by S1P, S1P1 and S1P2 are rapidly internalized and within 2 h recycle back to the plasma membrane. In contrast, activation of S1P1 by FTY720P leads to C-terminal polyubiquitination of the receptor, selectively targeting it for proteasomal degradation (18, 19). We examined the effects of S1P or FTY720P on the internalization of S1P1, S1P2, or S1P2(IC1-TM2)S1P1 receptor constructs C-terminally fused to GFP. These receptor constructs were stably expressed in HEK293 cells, and receptor localization and trafficking in response to ligands were examined by confocal fluorescence microscopy (Fig. 5). Exposure to S1P caused internalization of all three receptor constructs as evidenced by disappearance of plasma membrane-localized signal and appearance of punctate intracellular fluorescence. Consistent with the activation profiles in our functional assays, treatment with FTY720P caused internalization of the S1P1 and S1P2(IC1-TM2)S1P1 receptors, whereas S1P2 remained localized at the plasma membrane and did not internalize (Fig. 5, A–C, top rows).
FIGURE 5.
Receptor internalization of S1P1-, S1P2-, and S1P2(IC1-TM2)S1P1-GFP. Stably transfected HEK293 cells expressing WT S1P1 (A), WT S1P2 (B), or S1P2(IC1-TM2)S1P1 (C) GFP-fusion proteins were exposed to a 100 nm concentration of either vehicle (BSA), S1P, FTY720P, or FTY720PN for 30 min. Cells were rinsed and incubated for 2 h in the presence of cycloheximide to allow receptor recycling in the absence of new protein synthesis. The cells were fixed, and receptor localization was examined by laser-scanning confocal fluorescence microscopy.
After a 2-h washout, the majority of S1P-stimulated S1P1-GFP receptors recycled back to the plasma membrane, whereas after treatment with FTY720P, the receptor remained intracellularly localized. The SIP1-GFP receptor responded similarly to FTY720PN as to S1P with the majority of the receptor recycling back to the plasma membrane (Fig. 5A, bottom row). The S1P2-GFP receptor recycled back to the cell membrane 2 h after washout of S1P and retained its plasma membrane localization following exposure to FTY720P or FTY720PN (Fig. 5B, bottom row). The S1P2(IC1-TM2)S1P1-GFP receptor recycled back to the plasma membrane after activation with either S1P, FTY720P, or FTY720PN, although some intracellular staining was still evident (Fig. 5C, bottom row). These results indicate that the S1P2(IC1-TM2)S1P1-GFP receptor chimera recycles back to the plasma membrane after exposure to FTY720P; this is a key difference from the behavior of WT S1P1, which is retained intracellularly.
DISCUSSION
Our objective in the present study was to identify the structural basis of the negative specificity of FTY720P as an agonist of S1P2. The five S1P receptors share a high degree of amino acid identity (5), which supports the use of homology modeling strategies for the identification of structural differences that account for ligand selectivity. However, prior homology modeling of the S1P2 receptor (31) based on validated homology models of the S1P1 and S1P4 receptors (22, 23, 25, 26) failed to identify a structural basis for the negative selectivity of S1P2 for FTY720P. The S1P1 model has identified basic Arg3.28 and Arg7.34 and acidic Glu3.29 residues that form salt bridges with the phosphate and amino groups of S1P and are essential for ligand binding. The S1P4 model reiterated the importance of Arg3.28 and Glu3.29 and additionally revealed that Trp4.64 and Lys5.38 but not Arg7.33 (homologous to Arg7.34 in S1P1) are essential for ligand binding and activation. The S1P2 model suggested similar importance for the corresponding sites, but this prediction had not been experimentally validated.
Experimental validation of the S1P2 residues confirmed the predictions of the model as mutations to Arg2.28, Glu3.29, and Lys7.34 all produced a negative impact on S1P activation of S1P2 (Table 1). Mutation of these residues to alanine completely abolished the marginal activation by FTY720P seen in WT S1P2. This observation suggests some degree of interaction between FTY720P and the head group-interacting residues and suggests that this ligand also may engage these polar residues in the ligand-binding pocket albeit more weakly than S1P.
We have also identified 11 residues lining the putative ligand-binding pocket of S1P2 that are predicted to interact with the non-polar segments of S1P. Mutation of any of these residues diminished the responsiveness to S1P; the negative impact on receptor activation varied according to the nature of the amino acid substitution. However, all of these mutations completely abolished the weak activation of WT S1P2 by FTY720P. These observations support the hypothesis that S1P and FTY720P appear to engage similar residues in the putative ligand-binding pocket of S1P2.
We also swapped individual and multiple residues in S1P2 with residues that are found in identical positions in the S1P1 sequence (Table 1). These replacements, although still responsive to S1P, failed to improve activation by FTY720P, suggesting that residues outside the putative ligand-binding pocket play an important role in activation by this synthetic ligand.
FTY720P also functions in a unique manner on the S1P3 receptor, which couples to Gαi and Gαq when activated by S1P. FTY720P selectively activates S1P3-mediated Gαi signaling but inhibits S1P3-mediated Gαq signaling (34). An emerging concept in G protein-coupled receptor signaling is that of biased ligand signaling, where receptors are able to adopt different activation states in response to different ligands, leading to different downstream signaling outcomes (36). The unique actions of FTY720P as a biased ligand for at least two S1P receptors led us to examine whether the FTY720P specificity determinants might in part reside outside the ligand-binding pocket. This strategy yielded an unexpected result in that the chimera S1P2(NT-TM2)S1P1 containing just the NT to TM2 sequence of S1P1 became activated by FTY720P. Because the S1P2(NT-IC1)S1P1 chimera showed no activation by FTY720P, we concluded that motifs present in TM2 might be responsible for activation by FTY720P. These observations also support our findings with the mutagenesis of the ligand-binding pocket that suggested that FTY720P can make interactions necessary for activation with residues found in TMs 3, 5, 6, and 7 of S1P2.
Next, we dissected further the motifs responsible for FTY720P activation of the receptor by swapping individual domains between the S1P1 and S1P2 receptors. Replacement of either IC1 or TM2 of S1P2 with the corresponding S1P1 sequence imparted activation by FTY720P, and replacement of both domains enhanced the activation (Table 3). Furthermore, a reverse strategy in which the same IC and TM domains in S1P1 were replaced with S1P2 sequence showed that swapping just the IC1 loop was sufficient to abolish the activation by FTY720P.
Sequence alignment of IC1 and TM2 of S1P1 and S1P2 shows that the region necessary to confer activation by FTY720P encompasses 32 amino acids and differs at only 12 residues in motifs a, b, c, and d (Table 4). Four of the 12 residues represent differences that involve property-conserving amino acid residues. The motifs b, c, and d were sufficient to bestow FTY720P activation on S1P2; these residues are all within TM2 and the adjacent N-terminal two amino acids (Arg78 and Pro79) in IC1 just before the start of the TM2 α-helix. Swapping these motifs between the two receptors revealed that the sequence motif 78RP(MY)Y (where (MY) are identical between the two receptors) could alone impart activation by FTY720P. Thus, residues 78RP and Tyr82 are sufficient to exert the structural constraint to the rest of the S1P2 structure that allows the recognition of FTY720P. Nonetheless, the five differing residues in motifs c and d of TM2 when present with the 78RPMYY motif further enhance the potency and efficacy of FTY720P of the respective chimeras (Table 4). Conversely, motifs a, b, c, and d from S1P2 together, but not individually, inserted into S1P1 abolished the response to FTY720P.
Modeling studies using two FTY720P-responsive and three FTY720P-nonresponsive constructs demonstrate that the mobility of the aliphatic chain is the key difference correlating with receptor activation. This finding, coupled with the common recognition points identified using single point mutations, indicates that the negative selectivity of S1P2 for FTY720P is due to an unfavorable entropy of binding rather than an unfavorable enthalpy of binding. Previous modeling studies (31) correctly noted that very similar types of receptor-ligand interactions were possible for both S1P and FTY720P in S1P2 because these interactions only contribute to the enthalpy of binding. The activation properties of our point mutants, together with the finding that the S1P2(IC1-TM2)S1P1 chimera without any change in the putative ligand-binding pocket was responsive to FTY720P, support this explanation.
Given the importance of the four differing sequence motifs between S1P1 and S1P2, we considered whether motifs b, c, and d are conserved among the other S1P receptors. Sequence alignment of the IC1-TM2 regions of S1P1–5 shows that in S1P3 and S1P4 at least two of the three motifs are conserved such that the individual amino acid differences represent property-conserving amino acid replacements (supplemental Table 4).
The S1P1 receptor plays a critical role in lymphocyte trafficking because its presence on activated T cells allows the cells to egress from the lymphoid tissue in response to an S1P gradient (20). The clinical efficacy of FTY720 in multiple sclerosis depends on its ability to induce long term down-regulation S1P1 receptor signaling. FTY720P-induced internalization of the S1P2(IC1-TM2)S1P1 chimera showed receptor recycling back to the plasma membrane. This result suggests that the unique FTY720P-elicited trafficking of S1P1 receptor is linked to determinant(s) other than those included in IC1 and TM2. In summary, we discovered that structural motifs unique to S1P2 in IC1 and TM2 exert a distant allosteric constraint on the degree of motion of the bound FTY720P in the ligand-binding pocket that in turn prevents receptor activation by this ligand.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA-092160 (to G. T.) and HL-083187 (to R. B.). This work was also supported by the Van Vleet Oncology Research Endowment and National Science Foundation Grant CHE-0851880 (to A. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4.
- S1P
- sphingosine 1-phosphate
- S1P1–5
- S1P receptor subtypes 1–5
- EC
- extracellular loop
- IC
- intracellular loop
- TM
- transmembrane domain
- CT
- C-terminal domain
- NT
- N-terminal domain
- RMSF
- root mean square fluctuation
- FTY720
- 2-amino-2-[2-(4-octylphenyl)ethyl]propane-1,3-diol
- FTY720PN
- (R)-FTY720 phosphonate
- FTY720P
- FTY720 phosphate.
REFERENCES
- 1. Gräler M. H. (2010) Cell. Physiol. Biochem. 26, 79–86 [DOI] [PubMed] [Google Scholar]
- 2. Hla T., Brinkmann V. (2011) Neurology 76, Suppl. 3, S3–S8 [DOI] [PubMed] [Google Scholar]
- 3. Choi J. W., Gardell S. E., Herr D. R., Rivera R., Lee C. W., Noguchi K., Teo S. T., Yung Y. C., Lu M., Kennedy G., Chun J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strub G. M., Maceyka M., Hait N. C., Milstien S., Spiegel S. (2010) Adv. Exp. Med. Biol. 688, 141–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parrill A. L., Sardar V. M., Yuan H. (2004) Semin. Cell Dev. Biol. 15, 467–476 [DOI] [PubMed] [Google Scholar]
- 6. Takuwa Y., Okamoto Y., Yoshioka K., Takuwa N. (2008) Biochim. Biophys. Acta 1781, 483–488 [DOI] [PubMed] [Google Scholar]
- 7. Takuwa Y. (2002) Biochim. Biophys. Acta 1582, 112–120 [DOI] [PubMed] [Google Scholar]
- 8. Hohlfeld R., Barkhof F., Polman C. (2011) Neurology 76, S28–S37 [DOI] [PubMed] [Google Scholar]
- 9. Mehling M., Johnson T. A., Antel J., Kappos L., Bar-Or A. (2011) Neurology 76, Suppl. 3, S20–S27 [DOI] [PubMed] [Google Scholar]
- 10. Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G. J., Card D., Keohane C., Rosenbach M., Hale J., Lynch C. L., Rupprecht K., Parsons W., Rosen H. (2002) Science 296, 346–349 [DOI] [PubMed] [Google Scholar]
- 11. Brinkmann V., Davis M. D., Heise C. E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., Foster C. A., Zollinger M., Lynch K. R. (2002) J. Biol. Chem. 277, 21453–21457 [DOI] [PubMed] [Google Scholar]
- 12. Sanchez T., Estrada-Hernandez T., Paik J. H., Wu M. T., Venkataraman K., Brinkmann V., Claffey K., Hla T. (2003) J. Biol. Chem. 278, 47281–47290 [DOI] [PubMed] [Google Scholar]
- 13. Skoura A., Hla T. (2009) Cardiovasc. Res. 82, 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skoura A., Sanchez T., Claffey K., Mandala S. M., Proia R. L., Hla T. (2007) J. Clin. Investig. 117, 2506–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J. F., Gordon S., Estrada R., Wang L., Siow D. L., Wattenberg B. W., Lominadze D., Lee M. J. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H33–H42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohmori T., Yatomi Y., Osada M., Kazama F., Takafuta T., Ikeda H., Ozaki Y. (2003) Cardiovasc. Res. 58, 170–177 [DOI] [PubMed] [Google Scholar]
- 17. Peter B. F., Lidington D., Harada A., Bolz H. J., Vogel L., Heximer S., Spiegel S., Pohl U., Bolz S. S. (2008) Circ. Res. 103, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oo M. L., Chang S. H., Thangada S., Wu M. T., Rezaul K., Blaho V., Hwang S. I., Han D. K., Hla T. (2011) J. Clin. Investig. 121, 2290–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oo M. L., Thangada S., Wu M. T., Liu C. H., Macdonald T. L., Lynch K. R., Lin C. Y., Hla T. (2007) J. Biol. Chem. 282, 9082–9089 [DOI] [PubMed] [Google Scholar]
- 20. Pappu R., Schwab S. R., Cornelissen I., Pereira J. P., Regard J. B., Xu Y., Camerer E., Zheng Y. W., Huang Y., Cyster J. G., Coughlin S. R. (2007) Science 316, 295–298 [DOI] [PubMed] [Google Scholar]
- 21. Valentine W. J., Kiss G. N., Liu J., E S., Gotoh M., Murakami-Murofushi K., Pham T. C., Baker D. L., Parrill A. L., Lu X., Sun C., Bittman R., Pyne N. J., Tigyi G. (2010) Cell. Signal. 22, 1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parrill A. L., Wang D., Bautista D. L., Van Brocklyn J. R., Lorincz Z., Fischer D. J., Baker D. L., Liliom K., Spiegel S., Tigyi G. (2000) J. Biol. Chem. 275, 39379–39384 [DOI] [PubMed] [Google Scholar]
- 23. Wang D. A., Lorincz Z., Bautista D. L., Liliom K., Tigyi G., Parrill A. L. (2001) J. Biol. Chem. 276, 49213–49220 [DOI] [PubMed] [Google Scholar]
- 24. Fujiwara Y., Sardar V., Tokumura A., Baker D., Murakami-Murofushi K., Parrill A., Tigyi G. (2005) J. Biol. Chem. 280, 35038–35050 [DOI] [PubMed] [Google Scholar]
- 25. Inagaki Y., Pham T. T., Fujiwara Y., Kohno T., Osborne D. A., Igarashi Y., Tigyi G., Parrill A. L. (2005) Biochem. J. 389, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fujiwara Y., Osborne D. A., Walker M. D., Wang D. A., Bautista D. A., Liliom K., Van Brocklyn J. R., Parrill A. L., Tigyi G. (2007) J. Biol. Chem. 282, 2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valentine W. J., Fells J. I., Perygin D. H., Mujahid S., Yokoyama K., Fujiwara Y., Tsukahara R., Van Brocklyn J. R., Parrill A. L., Tigyi G. (2008) J. Biol. Chem. 283, 12175–12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu X., Sun C., Valentine W. J., Shuyu E., Liu J., Tigyi G., Bittman R. (2009) J. Org. Chem. 74, 3192–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olivera A., Rosenthal J., Spiegel S. (1994) Anal. Biochem. 223, 306–312 [DOI] [PubMed] [Google Scholar]
- 30. Ballesteros J. A., Weinstein H. (1995) in Methods in Neurosciences (Conn P. M., Sealfon S. C. eds) pp. 366–428, Academic Press, San Diego, CA [Google Scholar]
- 31. Pham T. C., Fells J. I., Sr., Osborne D. A., North E. J., Naor M. M., Parrill A. L. (2008) J. Mol. Graph. Model. 26, 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Halgren T. A. (1996) J. Comput. Chem. 17, 490–519 [Google Scholar]
- 33. Heckman K. L., Pease L. R. (2007) Nat. Protoc. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 34. Sensken S. C., Stäubert C., Keul P., Levkau B., Schöneberg T., Gräler M. H. (2008) Cell. Signal. 20, 1125–1133 [DOI] [PubMed] [Google Scholar]
- 35. Ratnala V. R., Kobilka B. (2009) Methods Mol. Biol. 552, 67–77 [DOI] [PubMed] [Google Scholar]
- 36. Whalen E. J., Rajagopal S., Lefkowitz R. J. (2011) Trends Mol. Med. 17, 126–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.