Abstract
The protein deacetylase, sirtuin 1 (SIRT1), is a proposed master regulator of exercise-induced mitochondrial biogenesis in skeletal muscle, primarily via its ability to deacetylate and activate peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α). To investigate regulation of mitochondrial biogenesis by SIRT1 in vivo, we generated mice lacking SIRT1 deacetylase activity in skeletal muscle (mKO). We hypothesized that deacetylation of PGC-1α and mitochondrial biogenesis in sedentary mice and after endurance exercise would be impaired in mKO mice. Skeletal muscle contractile characteristics were determined in extensor digitorum longus muscle ex vivo. Mitochondrial biogenesis was assessed after 20 days of voluntary wheel running by measuring electron transport chain protein content, enzyme activity, and mitochondrial DNA expression. PGC-1α expression, nuclear localization, acetylation, and interacting protein association were determined following an acute bout of treadmill exercise (AEX) using co-immunoprecipitation and immunoblotting. Contrary to our hypothesis, skeletal muscle endurance, electron transport chain activity, and voluntary wheel running-induced mitochondrial biogenesis were not impaired in mKO versus wild-type (WT) mice. Moreover, PGC-1α expression, nuclear translocation, activity, and deacetylation after AEX were similar in mKO versus WT mice. Alternatively, we made the novel observation that deacetylation of PGC-1α after AEX occurs in parallel with reduced nuclear abundance of the acetyltransferase, general control of amino-acid synthesis 5 (GCN5), as well as reduced association between GCN5 and nuclear PGC-1α. These findings demonstrate that SIRT1 deacetylase activity is not required for exercise-induced deacetylation of PGC-1α or mitochondrial biogenesis in skeletal muscle and suggest that changes in GCN5 acetyltransferase activity may be an important regulator of PGC-1α activity after exercise.
Keywords: AMP-activated Kinase (AMPK), Mitochondria, Mitochondrial Metabolism, SIRT, Sirtuins, Skeletal Muscle
Introduction
Impaired mitochondrial function has been linked with physical inactivity, insulin resistance, and the pathogenesis of type 2 diabetes (1, 2). Importantly, aerobic exercise training increases mitochondrial biogenesis and oxidative capacity in skeletal muscle (3–7). The precise mechanisms, however, linking muscle contraction to mitochondrial plasticity are incompletely defined (8). Clearly, allosteric factors (e.g. Ca2+, AMP, NAD+, Pi) that are increased during contraction are important because they activate signaling pathways that ultimately converge at the transcriptional co-activator, peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α)2 (9–12). Once active, PGC-1α targets an array of transcription factors and nuclear receptors to coordinate gene expression of nuclear and mitochondrial-encoded genes (13, 14). However, how perturbations in cellular energy stress are sensed during exercise and subsequently how this signal is transduced to regulate mitochondrial biogenesis in a coordinated manner are still under intense investigation.
In recent years, lysine acetylation has emerged as a key regulatory process controlling enzyme activity and gene transcription (15, 16). To this end, the class III NAD+-dependent protein deacetylase, sirtuin 1 (SIRT1), has been proposed as a central regulator of exercise-induced mitochondrial biogenesis in skeletal muscle due to its sensitivity to perturbations in NAD+ and its ability to regulate PGC-1α activity (10, 17–20). Indeed, deacetylation of PGC-1α by SIRT1 is thought to be pivotal for regulating the activation of numerous genes involved in glucose and lipid metabolism, as well as oxidative capacity (19, 20) Consistent with this, SIRT1 gene expression and protein content and activity are increased in skeletal muscle by endurance exercise training in association with mitochondrial biogenesis (21–24). Moreover, overexpression of SIRT1 in PC12 cells leads to PGC-1α deacetylation and significantly increases oxygen consumption, which is indicative of increased mitochondrial biogenesis (19).
Whether SIRT1 regulates mitochondrial biogenesis in vivo is less clear. For example, mice with whole body knock-out of SIRT1 do not display decrements in muscle respiratory capacity (25), whereas transient overexpression of SIRT1 in rodent skeletal muscle reduces oxidative capacity and PGC-1α protein abundance (26). Moreover, total SIRT1 activity and protein content show a limited correlation with skeletal muscle oxidative capacity (27). Recently, it has been suggested that nuclear SIRT1 activity regulates mitochondrial biogenesis; although without loss of function studies this hypothesis has yet to be addressed directly (22). Thus, to better understand the precise role of SIRT1 deacetylase activity in regulating mitochondrial biogenesis in skeletal muscle, we studied mice with knock-out of SIRT1 deacetylase activity in skeletal muscle (referred to as mKO), in sedentary (SED) or exercise conditions. We hypothesized that skeletal muscle endurance and exercise-induced mitochondrial biogenesis would be impaired in mKO skeletal muscle, primarily due to lack of SIRT1-mediated deacetylation of PGC-1α.
EXPERIMENTAL PROCEDURES
Reagents and Materials
All reagents were from Sigma-Aldrich unless otherwise stated.
Generation of mKO Mouse
To generate the mKO mouse, floxed mice harboring loxP sites flanking exon 4 of the SIRT1 gene (28) (SIRT1FLX-exon4), which encodes the deacetylase domain of SIRT1, were crossed with mice expressing Cre recombinase under the control of the muscle creatine kinase promoter. In these mice, a truncated SIRT1 protein is present, but the protein lacks deacetylase activity (28). For simplicity, the SIRT1FLX-exon4 mice used to generate mKO are referred to as wild-type (WT) mice. Mice were housed on a 12:12-h light:dark cycle, and only male mice were studied. Mice were studied at 13 weeks of age. All experiments were approved by and were conducted in accordance with the Animal Care Program at the University of California, San Diego.
Voluntary Wheel Running (VWR)
10-week-old mice were given access to a running wheel for 20 days with time run per day recorded via a digital recorder. On day 22 (36 h following removal of the running wheel) mice were anesthetized, and the gastrocnemius muscle was removed, cleaned of visible fat, and frozen in liquid nitrogen for later analysis.
Acute Exercise (AEX) Protocol
13-week-old mice were habituated to a motorized treadmill (Columbus Instruments, Columbus, OH) on two occasions (10 min at 10 m/min and 10° grade) in the 2 days prior to the experimental exercise bout. After a 3-h fast mice completed 60 min of running exercise (10° grade) at repetitions of variable exercise intensity (5–25 m/min). Following exercise, mice were returned to their cages without food, but with access to water, until muscle collection. Muscle was collected either immediately (0h) or 3 h (3h) after AEX. A sedentary (SED) control group was used as a comparison. SED mice were fasted for the same period of time, and muscle was collected at the same time of day as the experimental group (i.e. after a 4-h or 7-h fast).
Tissue Homogenization
Tissue was prepared as described previously (29). Briefly, muscle was powdered under liquid nitrogen using a mortar and pestle and Polytron-homogenized in 10-fold mass of ice-cold sucrose lysis buffer (50 mm Tris, pH 7.5, 250 mm sucrose, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 50 mm NaF, 1 mm NaVO4 Na2(PO4)2, and 0.1% DTT). The homogenate was briefly vortexed and centrifuged at 4 °C for 10 min at 8000× g to remove insoluble material. Protein concentrations were determined using the DC protein assay (Bio-Rad).
Skeletal Muscle Nuclear Isolation
Nuclear and cytosolic fractions were isolated from SED and AEX gastrocnemius muscle using a commercially available kit (78835: NE-PER; Thermo Scientific) with the addition of the COMPLETE protease inhibitor mixture (11697498001; Roche Applied Science). Integrity of nuclear fractions was confirmed by immunoblotting for the cytosolic enzyme, lactate dehydrogenase (LDH, L7016; Sigma) and the nuclear protein, histone H2B (ab9408; AbCam).
Immunoprecipitation (IP)
Nuclear extracts from gastrocnemius muscle were used for IP experiments. For IP of acetylated lysine and acetylation measurements of p53 and PGC-1α, nuclear fractions were isolated with the addition of 1 μm trichostatin A and 10 mm nicotinamide. For IP of acetylated lysine, 150 μg of protein was precleared with 50 μl of protein A-agarose beads (16-156; Millipore), whereas for IP of p53, PGC-1α and general control of amino-acid synthesis 5 (GCN5), 150 μg of protein was precleared with 25 μl of protein A-agarose beads and 25 μl of protein G-agarose beads (16-266; Millipore) for 1 h at 4 °C. Protein was then rotated for 2 h (4 °C) with anti-acetylated lysine (9441; Cell Signaling), anti-PGC-1α (AB3242; Millipore), anti-p53 (sc-6243; Santa Cruz Biotechnology), or anti-GCN5 (607201; Biolegend), and then protein A-agarose (50 μl) or protein A/G beads (25 μl of each) were added and the samples and rotated overnight (4 °C). The following morning, agarose beads were washed three times with sucrose lysis buffer and three times in TAE before antigens were eluted from bead complexes with 1× Laemmli SDS buffer (30 μl). Samples were boiled for 10 min and centrifuged for 5 min at 8000× g (4 °C) prior to SDS-PAGE (20 μl) separation.
Immunoblotting
Equal amounts of protein (30 μg) were boiled for 5 min in 1 × Laemmli sample buffer and separated on 7.5–10% gels by SDS-PAGE as described previously (30). Antibody binding was detected using an enhanced chemiluminescence horseradish peroxidase (HRP) substrate detection kit (Millipore). Imaging and band quantification were carried out using a Chemi Genius Bioimaging Gel Doc System (Syngene).
Antibodies
All primary antibodies were used at a 1:1000 dilution in buffers according to the instructions of the manufacturer. Mitoprofile total OXPHOS antibody mixture (MS604) was from Mitosciences, Acetyl-lysine (9441), phospho-AMPKThr172 (2535), phospho-ACC-βSer79 (3661), HDAC5 (2082), phospho-p38 MAPKThr180/Tyr182 (9215), p38 MAPK (9212), phospho-ATF2Thr71 (9221), phospho-CaMKIIThr286 (3361), and CaMKII (3362) were from Cell Signaling Technology. PGC-1α (AB3242) was from Millipore. p53 (sc-6243) was from Santa Cruz Biotechnology. Phospho-HDAC5Ser259 (ab47283) was from AbCam. Phospho-CREBSer133 (600-401-270) and CREB (100-401-195) were from Rockland. AMPKα1, AMPKα2, and ACC-β antibodies were kindly provided by Professor Grahame Hardie, University of Dundee.
RNA Extraction and cDNA Synthesis
RNA was extracted from gastrocnemius muscle using the phenol/chloroform method, and cDNA was synthesized as described previously (30). Quantitative real-time PCR was performed using an Eppendorf Light Cycler PCR machine, SYBR Green PCR plus reagents (Sigma Aldrich), and custom-designed primers. 10-μl PCRs were assayed in triplicate on a 96-well heat-sealed PCR plate (Thermo Fisher Scientific). Each reaction contained 5 μl of SYBR Green Taq, 1 μl of forward and reverse primers, and 3 μl of cDNA (1:10 dilution). The target gene expression was calculated relative to values from glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed normalized to pretreatment values. Absolute CT for GAPDH was unchanged by any of the treatments (data not shown). Primers for quantitative PCR were designed to span exon-exon boundaries to avoid amplification of genomic DNA. The following primer sequences were used: SIRT1 forward, 5′-GGC TAC CGA GAC AAC CTC CTG-3′ and reverse 5′-AGT CCA GTC ACT AGA GCT GGC-3′; SIRT1-Exon3 forward, 5′-GAT GCT GTG AAG TTA CTG CAG GAG TG-3′ and reverse SIRT1-Exon5, 5′-AAT TTG TGA CAC AGA GAC GGC TGG-3′. PGC-1α forward, 5′-CCC TGC CAT TGT TAA GAC CG-3′ and reverse, 5′-TCT GCT GCT GTT CCT GTT TTC-3′; mitofusin-2 forward, 5′-AGA CAC CCA CAG GAA CAC AG-3′ and reverse, 5′-TCT TCC CAT TGC TCG TCC-3′; cytochrome c forward, 5′-GCA AGC ATA AGA CTG GAC CAA A-3′ and reverse, 5′-TTG TTG GCA TCT GTG TAA GAG AAT C-3′; pyruvate dehydrogenase kinase 4 forward, 5′-CCA AGA TGC CTT TGA GTG TG-3′ and reverse, 5′-ACA CAA TGT GGA TTG GTT GG-3′; GAPDH forward, 5′-TGG AAA GCT GTG GCG TGA T-3′ and reverse 5′-TGC TTC ACC ACC TTC TTG AT-3′.
Isolated Whole Muscle Stimulation Protocol
Mice were anesthetized (sodium pentobarbital 60 mg/kg), and the fifth toe muscle of the extensor digitorum longus and the tendon insertion were isolated and mounted in a specialized muscle chamber containing Ringer solution (137 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgSO4, 1 mm NaH2PO4, 24 mm NaHCO3, 11 mm glucose containing 10 mg/liter curare) at room temperature. The muscle origin was tied with 6-0 silk suture to a rigid post, and the insertion was secured to the arm of a dual-mode ergometer (model 300B; Aurora Scientific, ON, Canada) that could impose precise length changes. Muscle activation was provided by an electrical stimulator (model S88; Astro-Med, West Warwick, RI) via parallel platinum plate electrodes that extended the length of the muscle. Optimal muscle length was determined as the length at which supramaximal stimulation produced maximal isometric tetanic force. Isometric tetanic contractions were elicited with 300-ms train duration and 0.2-ms pulse duration at 70 Hz with variable rest intervals. Rest intervals were decreased every 1 min in a progressive manner (from a contraction every 8 to 4 to 3 to 2 to 1 s). The test was terminated when the developed force had fallen to ∼40% of the initial maximal developed force. Time to fatigue was measured at the time it took each muscle to reach 60% of the maximal developed force. Specific force was calculated by normalizing muscle force to muscle physiological cross-sectional area calculated using standard equations (31). The maximal isometric tetanic force was expressed as newtons/cm2.
Mitochondrial Enzyme Activity Assays
To isolate mitochondria, 40 mg of powdered gastrocnemius was homogenized in 2 ml of buffer A (210 mm mannitol, 70 mm sucrose, 1 mm EGTA, 0.5% BSA, 5 mm HEPES, 10 μg/ml subtilisin, pH 7.2). The homogenate was centrifuged at 1000× g for 10 min (4 °C), and the resulting supernatant was transferred to a separate microtube and recentrifuged at 10,000× g for 20 min (4 °C). Following aspiration, the remaining pellet was resuspended in 100 μl of buffer B (210 mm mannitol, 70 mm sucrose, 1 mm EGTA, 5 mm HEPES, pH 7.2) and frozen at −80 °C. Complex II–IV and citrate synthase (CS) enzyme activity assays were performed using standard methods as previously described (32) with each experiment repeated in triplicate.
Quantification of Mitochondrial DNA (mtDNA)
20 mg of powdered gastrocnemius muscle was suspended in 150 μl of digestion buffer (1 m Tris-HCl, pH 8.0, 0.5 mm EDTA, 5 m NaCl, 10% SDS, + 4 μl of proteinase K) and incubated overnight at 60 °C. The following day, samples were centrifuged at 14,000× g for 10 min, the pellet was washed twice with 500 μl of 70% ethanol and then allowed to air dry at room temperature for 20 min. The pellet was resuspended in 100 μl of TE (1 m Tris, pH 8.0, 0.5 m EDTA, ddH2O) and incubated at 60 °C for 2 h before being stored overnight at 4 °C. RT-PCR was performed against primers specific to a mitochondrial (cytochrome b forward, 5′-TTC GCA GTC ATA GCC ACA G-3′ and reverse 5′-AGA TGA AGT GGA AAG CGA AG-3′) and nuclear (β-globulin forward, 5′-TGC CAT CCC ATC ACA ACA AG-3′ and reverse, 5′-GCC AAT ACA CAG GTC ACA GAG-3′) encoded gene using an Eppendorf Light Cycler PCR machine.
Statistical Analysis
A Student's t test or two-way repeated measures analysis of variance (SigmaPlot® 11.2, Systat Software, Inc. San Jose CA) was used to determine differences between groups and experimental conditions, and Tukey post hoc test was used where appropriate. Values are displayed as mean ± S.E., with statistical significance set at p < 0.05.
RESULTS
Generation of mKO Mouse
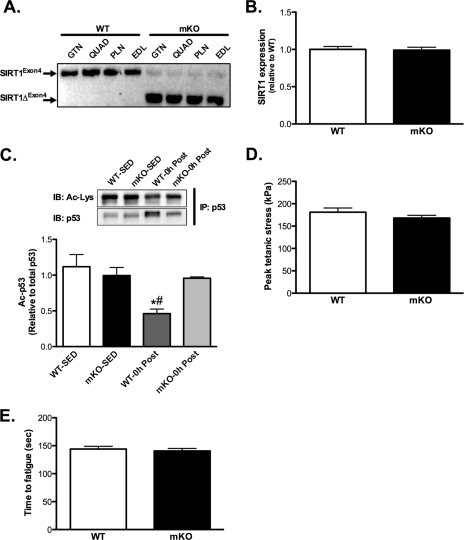
Exon4 was efficiently excised in various skeletal muscles in mKO versus WT (i.e. SIRT1FLX-exon4; Fig. 1A) mice, but this deletion did not occur in adipose tissue or liver (data not shown). Despite deletion of exon4, SIRT1 gene expression was similar in mKO versus WT mice (Fig. 1B). To verify that SIRT1 deacetylase activity was disrupted in mKO muscle, we measured the acetylation of the SIRT1 target, p53, in nuclear pellets from skeletal muscle collected immediately (0h) after AEX. The acetylation of p53 (Ac-p53) was significantly reduced in WT mice 0 h after AEX, but this reduction was completely abrogated in mKO mice (Fig. 1C), confirming that SIRT1 deacetylase activity was diminished in mKO skeletal muscle. Nuclear p53 abundance was significantly (p < 0.05) increased after AEX only in WT mice and not in mKO mice (WT-SED, 1.00 ± 0.09; KO-SED, 1.12 ± 0.15; WT-AEX, 2.41 ± 0.18; KO-AEX, 1.17 ± 0.31).
FIGURE 1.
Loss of SIRT1 activity does not affect skeletal muscle contractile characteristics. A, semiquantitative PCR analysis using primers directed across exon4 of the SIRT1 gene in skeletal muscle (gastrocnemius (GTN), quadriceps (QUAD), plantaris (PLN), and extensor digitorum longus (EDL)) of WT and mKO mice. Note the shorter PCR product in mKO mice (SIRT1ΔFLX-Exon4), indicating efficient deletion of exon4. B, deletion of exon4 does not affect SIRT1 gene expression in gastrocnemius muscle. C, deacetylation of p53 occurs after AEX (0 h after) in gastrocnemius muscle from WT but not mKO mice, compared with SED mice. D and E, maximal tetanic force (D) and time to fatigue (E) in isolated fifth toe of the extensor digitorum longus muscle of WT and mKO mice. All data are presented as mean ± S.E. (error bars). #, within 0h; *, within genotype p < 0.05.
Skeletal Muscle Contractile Performance
Maximal tetanic force and time to fatigue of isolated extensor digitorum longus muscles were not different between mKO versus WT mice (Fig. 1, D and E), demonstrating that loss of SIRT1 deacetylase activity does not impair skeletal muscle function or endurance capacity.
Mitochondrial Biogenesis in Response to Exercise Training Is Not Impaired in mKO Mice
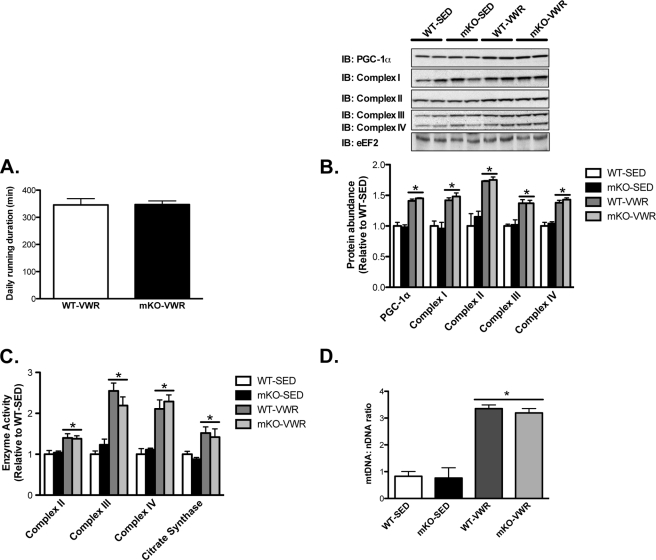
Next, we studied adaptations to exercise training using a 20-day VWR model. Despite loss of SIRT1 deacetylase activity, time spent running was similar between mKO and WT mice, suggesting no impairments in running ability (Fig. 2A). Training-induced increases in the skeletal muscle protein abundance of PGC-1α and the electron transport chain subunit complexes I–IV (∼40–80%; Fig. 2B), the activity of complexes II–IV and citrate synthase (∼40–125%, Fig. 2C), and the mitochondrial DNA to nuclear DNA ratio (∼3.5-fold; mtDNA:nDNA, Fig. 2D) were also comparable between mKO versus WT mice. Thus, loss of SIRT1 deacetylase activity in skeletal muscle did not impair basal muscle function, endurance, or oxidative capacity, nor did it compromise induction of mitochondrial biogenesis in response to VWR.
FIGURE 2.
Loss of SIRT1 activity does not impair mitochondrial biogenesis following 20-days of voluntary wheel running (VWR). A, average running time per day during 20 days of VWR in mKO and WT mice. B, immunoblotting and quantification of PGC-1α and electron transport chain (complexes I–IV) in SED and following 20 days of VWR, in mKO and WT mice. Elongation factor 2 (eEF2) was used as a loading control. C and D, enzyme activity of electron transport chain complexes II-IV and citrate synthase (C) and mitochondrial to nuclear DNA ratio (mtDNA:nDNA) (D) in SED and VWR, mKO, and WT mice. Data represent n = 6–8/group. All data are presented as mean ± S.E. (error bars). *, within genotype p < 0.05.
Activation of Mitochondrial Biogenesis and PGC-1α by AEX Are Not Impaired in mKO Mice
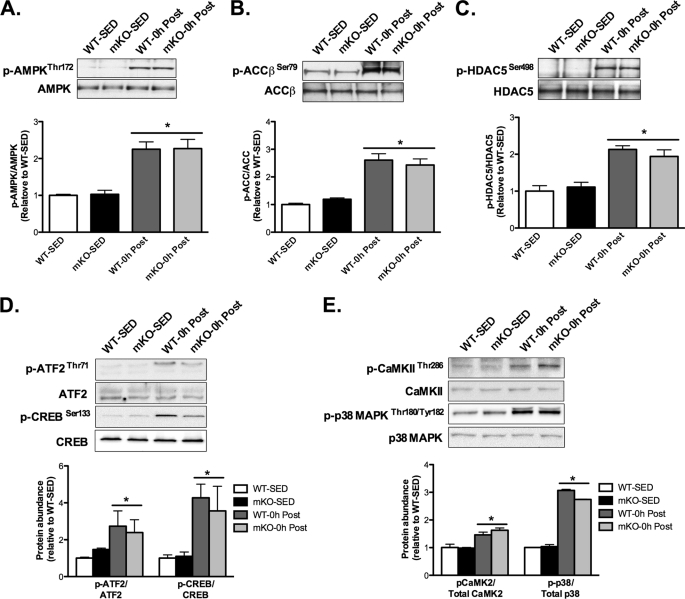
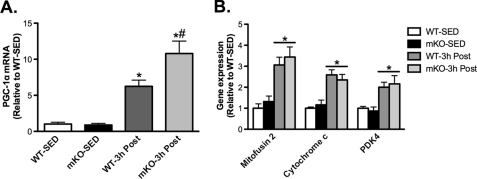
Considering that mitochondrial biogenesis and induction of PGC-1α were not impaired in mKO mice following VWR, we studied other signaling pathways that could potentially compensate for the loss of SIRT1 deacetylase activity in mKO mice and thus explain the normal adaptation to VWR. Immediately after exercise (i.e. 0h), phosphorylation of the AMP-activated protein kinase (AMPKThr172) and the AMPK substrates, acetyl-CoA carboxylase (ACC-βSer79) and histone deacetylase 5 (HDAC5Ser498) were similarly increased ∼2–2.5-fold above SED values in mKO and WT mice (Fig. 3, A–C). We also observed similar increases 0 h after exercise in the phosphorylation of activating transcription factor-2 (ATF2Thr71), cAMP response element-binding protein (CREBSer133; Fig. 3D), Ca2+/calmodulin-dependent protein kinase II (CaMKIIThr286) and p38 mitogen-activated protein kinase (p38 MAPKTyr180/182; Fig. 3E) in AEX mKO and WT mice compared with SED mice. Therefore, altered activation of these pathways does not appear to explain the ability of mKO to adapt to VWR and AEX. PGC-1α gene transcription is well known to be induced by exercise (7, 9, 33), and SIRT1 has been reported to be involved in regulating PGC-1α promoter activity (19, 20, 34). However, we found that skeletal muscle PGC-1α expression was significantly increased 3 h after AEX in both WT and mKO mice compared with SED mice, and in fact, the increase in mKO mice 3 h after AEX was ∼2-fold greater than that of WT mice (Fig. 4A). In parallel with increased PGC-1α expression 3 h after AEX, the activation of proposed PGC-1α target genes, mitofusin-2, cytochrome c, and pyruvate dehydrogenase kinase 4 were similarly increased by AEX in mKO and WT mice (Fig. 4B).
FIGURE 3.
mKO mice display normal activation of exercise-responsive proteins following AEX. Protein phosphorylation and total abundance were determined in gastrocnemius whole muscle lysates of sedentary (SED) mice or immediately after AEX (0 h post). A–C, phosphorylation of AMPKThr172 and its substrates (A), ACCβSer79 (B), and HDAC5Ser498 (C) were increased equally 0 h after AEX versus SED, in WT and mKO. D and E, phosphorylation of ATF2Thr71 and CREBSer133 (D) and CaMKIIThr286 and p38 MAPKTHR180/TYR182 (E) were increased equally 0 h after AEX versus SED in WT and mKO. Data represent n = 4–6/group. All data are presented as mean ± S.E. (error bars). *, within genotype p < 0.05.
FIGURE 4.
mKO mice display normal activation of PGC-1α and exercise-responsive genes following AEX. A, PGC-1α and, B, mitofusin-2, cytochrome c, and pyruvate dehydrogenase kinase 4 (PDK4) skeletal muscle gene expression on sedentary (SED) or 3 h post (3h Post) AEX, in WT and mKO mice. Data represent n = 4–6/group. All data are presented as mean ± S.E. (error bars). *, within genotype; #, within exercise p < 0.05.
Deacetylation of PGC-1α after Exercise Is Not Impaired in mKO Mice
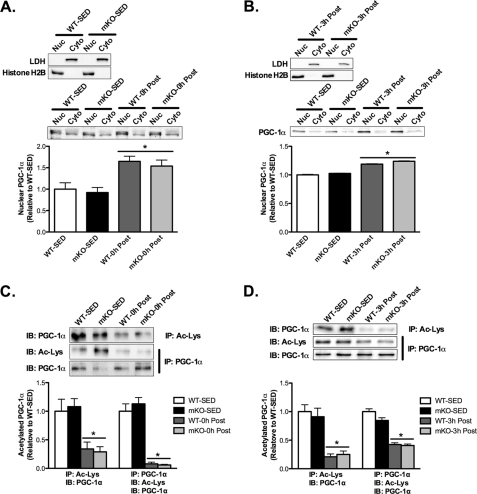
Because SIRT1 is the purported regulator of PGC-1α acetylation status (10, 19, 20) and therefore PGC-1α activity, we performed detailed analysis of PGC-1α nuclear localization and acetylation status at 0 and 3 h after AEX. We obtained isolated purified nuclear and cytosolic fractions in 0 and 3-h post-AEX samples, as evidenced by the lack of LDH in nuclear fractions and histone H2B in cytosolic fractions (Fig. 5, A and B, upper panels). In WT and mKO mice, at both 0 and 3 h after AEX, the nuclear abundance of PGC-1α was increased ∼60 and ∼20%, respectively (Fig. 5, A and B, respectively). In addition, the acetylation of nuclear PGC-1α at 0 h after AEX was reduced ∼70–90% (Fig. 5C) and at 3 h after AEX was reduced ∼60–80% compared with SED values (Fig. 5D), which is in line with previously published studies (10, 35). Contrary to our hypothesis, however, the deacetylation of PGC-1α after AEX was not impaired in mKO mice (Fig. 5, C and D).
FIGURE 5.
PGC-1α nuclear translocation and deacetylation following AEX are unaffected by loss of SIRT1 activity. A and B, integrity of nuclear and cytosolic fractions was determined via immunoblotting for lactate dehydrogenase (LDH) (cytosolic; Cyto) and histone H2B (nuclear; Nuc) abundance. PGC-1α nuclear abundance increases 0 h (A) and 3 h (B) after AEX (0h Post and 3h Post, respectively) compared with sedentary (SED) in both mKO and WT mice. C and D, to determine PGC-1α acetylation, we immunoprecipitated (IP) with an antibody to acetylated proteins (Ac-Lys) or PGC-1α and immunoblotted (IB) for Ac-PGC-1α or total PGC-1α. PGC-1α was deacetylated to the same extent 0 h (C) and 3 h (D) after AEX in mKO and WT mice compared with SED mice. Data represent n = 4–6/group. All data are presented as mean ± S.E. (error bars). *, within genotype p < 0.05.
The Association of PGC-1α and GCN5 Is Reduced after AEX
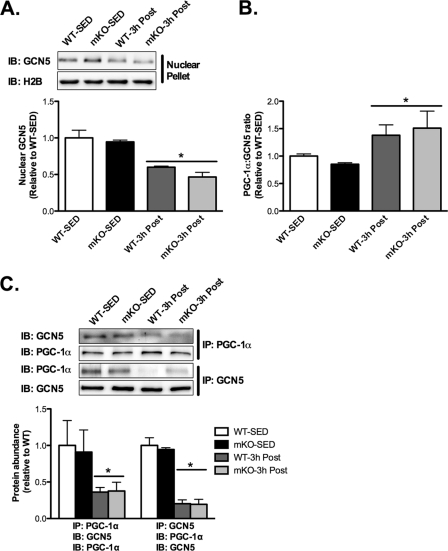
Because protein acetylation represents a balance between acetyltransferase and deacetylase activity, we examined whether AEX affected the association of PGC-1α with the acetyltransferase, GCN5, because GCN5 has been shown to acetylate PGC-1α in C2C12 myotubes (15) and fao hepatocytes (36). In line with reduced acetylation of nuclear PGC-1α after AEX, the nuclear abundance of GCN5 3 h after AEX was decreased ∼40–50% in both mKO and WT compared with SED groups (Fig. 6A), resulting in an increase in the nuclear PGC-1α:GCN5 ratio (Fig. 6B). Complementing these findings, we found that GCN5 and PGC-1α co-immunoprecipitate in skeletal muscle and interestingly, that the abundance of GCN5 in PGC-1α immunoprecipitates, and the abundance of PGC-1α in GCN5 immunoprecipitates, are significantly reduced ∼60–80% in 3-h post-AEX versus SED muscle, of both mKO and WT mice (Fig. 6C). Thus, these results suggest a novel paradigm for regulation of PGC-1α acetylation status in vivo after AEX, whereby reduced acetylation of PGC-1α after AEX results, not from increased SIRT1 deacetylase activity, but rather dissociation of GCN5 from PGC-1α.
FIGURE 6.
Nuclear abundance of GCN5 and the association of GCN5 with PGC-1α are decreased after AEX. A, immunoblotting (IB) of GCN5 and H2B in nuclear fractions of sedentary (SED) and 3 h after AEX (3h Post) skeletal muscle of WT and mKO mice. 3 h after AEX the nuclear abundance of GCN5 is decreased in both mKO and WT mice compared with SED mice. B, significant increase in the nuclear PGC-1α to GCN5 ratio in both WT and mKO mice 3 h after AEX. C, immunoprecipitation of GCN5 or PGC-1α and immunoblotting of GCN5 or PGC-1α demonstrate that GCN5 and PGC-1α interact in vivo and that this interaction is reduced 3 h after AEX in both mKO and WT mice compared with SED mice. Data represent n = 4–6/group. All data are presented as mean ± S.E. (error bars). *, within genotype p < 0.05.
DISCUSSION
Mitochondrial biogenesis is a fundamental adaptation observed in skeletal muscle following endurance exercise training (3–7). Importantly, aspects of this adaptive response appear to be altered or compromised during physical inactivity, insulin resistance, and type 2 diabetes (1, 2). Recently, SIRT1 has garnered attention as a potential integrator and effector of the adaptive response to exercise, due to its ability to alter the transcriptional activity of PGC-1α, a principal regulator of mitochondrial biogenesis (10, 17–20). In line with this, recent reports have demonstrated increased SIRT1 activity in rodents and humans in response to exercise (21, 22, 37). Further, exercise-mediated deacetylation of PGC-1α has been shown to be compromised in a rodent model of insulin resistance, linking reduced SIRT1 function to impaired activation of PGC-1α and reduced mitochondrial function (38). Thus, herein we hypothesized that skeletal muscle endurance, deacetylation of PGC-1α after endurance exercise, and endurance exercise-induced mitochondrial biogenesis would be impaired in mKO mice. Contrary to our hypothesis, however, neither skeletal muscle endurance nor skeletal muscle oxidative capacity was compromised in mKO mice. Further, exercise-mediated deacetylation of PGC-1α was not impaired in mKO mice, nor was induction of PGC-1α expression, PGC-1α signaling, or mitochondrial biogenesis. Alternatively, our results suggest that reduced acetylation of PGC-1α by the acetyltransferase, GCN5, and not increased deacetylation by SIRT1, could be the predominant regulator of PGC-1α acetylation status after exercise.
The acetylation status of PGC-1α has been implicated as a fundamental post-translational modification that regulates its transcriptional activity (17, 19, 39–41). Our observation that the acetylation of PGC-1α is reduced following AEX in mKO mice is novel and suggests, in contrast to previous in vitro work (20), that in vivo regulation of PGC-1α acetylation has substantial redundancy and likely occurs via SIRT1-independent mechanisms. To this end, the acetyltransferase GCN5 has previously been shown to localize with and alter the acetylation state of PGC-1α (19, 36). Complementing this, whole body deletion of SRC-3, an upstream activator of GCN5, results in decreased PGC-1α acetylation and increased mitochondrial biogenesis (40). Considering that GCN5 is the proposed principle acetyltransferase regulating PGC-1α (19, 36), we performed co-immunoprecipitation experiments to examine whether PGC-1α and GCN5 interact in vivo. We demonstrate, for the first time, that PGC-1α and GCN5 co-immunoprecipitate in skeletal muscle at rest and that this interaction is significantly reduced 3 h following AEX. Further, we demonstrate that the nuclear localization of GCN5 is reduced after AEX, whereas the nuclear localization of PGC-1α is increased, all of which would facilitate an increase in PGC-1α-activity. Based on this interesting finding, our working hypothesis is that endurance exercise reduces the interaction of GCN5 and PGC-1α, which leads to PGC-1α deacetylation and induction of mitochondrial biogenic pathways.
A key, currently unresolved question is whether lysine acetylation regulates the localization and function of PGC-1α in skeletal muscle in vivo. It is clear from in vitro studies that the acetylation of PGC-1α regulates its nuclear distribution (36) and subsequent activity toward target genes (19, 20, 39). Moreover, PGC-1α is highly acetylated, containing 13 lysine residues along the length of the protein (39, 41), and mutating these residues prevents nicotinamide (a SIRT1 inhibitor)-induced acetylation and inhibition of PGC-1α activity toward HNF4α by as much as ∼75% (39, 41). In the present study we found the nuclear abundance of PGC-1α to be increased at 0 and 3 h after AEX, in parallel with significant deacetylation of PGC-1α. However, whether deacetylation of PGC-1α is required for the translocation of PGC-1α to the nucleus remains to be determined, and future studies that utilize PGC-1α acetylation mutants in vivo in combination with exercise would greatly advance our knowledge in this respect.
Relatively little is known about the regulation of GCN5 in skeletal muscle. Overexpression of GCN5 in fao hepatoma cells increases PGC-1α association with the transcriptional co-repressor RIP140 and reduces PGC-1α transcriptional activity (36). It has been suggested that post-translational modifications may alter GCN5 acetyltransferase activity toward target proteins (42). Given that AEX increases the activity of numerous kinases, such as AMPK, p38 MAPK, and CaMKII, the GCN5-PGC-1α interaction may also be regulated by phosphorylation following AEX. Clearly, further investigation is required to understand fully how AEX modulates GCN5 function and cellular localization in skeletal muscle.
In addition to post-translational modification of PGC-1α, SIRT1 has also been reported to regulate PGC-1α transcriptional activity via interaction with myogenic regulatory factors at an autoregulatory region in the PGC-1α promoter (34). At 3 h after AEX, we observed a marked increase in PGC-1α gene expression in mKO compared with WT mice. This is in contrast to the hypothesis that SIRT1 deacetylase function is required for PGC-1α transactivation (10, 19, 20, 22, 34). In fact, our observation that exercise-induced PGC-1α gene expression was greater in mKO compared with WT suggests that SIRT1 may even play a restrictive role in the regulation of the PGC-1α promoter. Indeed, this observation agrees with in vivo studies that found a disconnect between SIRT1 abundance/activity and PGC-1α (26, 27) and a previous report suggesting that overexpression of SIRT1 can negatively regulate PGC-1α promoter activity (19). Our results also conflict with those of a recent paper that suggests that exercise-mediated mitochondrial biogenesis is regulated by nuclear SIRT1 activity (22), although whether SIRT1 is required for PGC-1α deacetylation or mitochondrial biogenesis was not measured directly by the authors.
As a sensor of cellular energy status, the function and action of SIRT1 has drawn parallels to AMPK (43). There has been some debate in the literature regarding how this reciprocal interaction is facilitated. Functional AMPK is required for activation of SIRT1 following energy depletion (17) and in response to exercise (10). In contrast, overexpression of SIRT1 in human embryonic kidney 293T cells leads to deacetylation, relocalization, and activation of LKB1, an upstream regulator of AMPK, and results in elevated AMPK activity (44). In parallel, knockdown of SIRT1 in vitro and starvation-refeeding in rodents suppress SIRT1 and AMPK activity. Collectively, these studies implicate SIRT1 as an upstream regulator of AMPK function via deacetylation of LKB1 (43). However, in the present study, we report that increases in AMPKThr172 phosphorylation in response to exercise are comparable in mKO versus WT mice, as is the phosphorylation of the AMPK substrates ACC-βSer79 and HDAC5Ser498. Our data therefore suggest that SIRT1 activity is not required for normal AMPK function in skeletal muscle, in vivo, in the basal state or in response to endurance exercise.
Differential protein acetylation has received attention recently as a means of regulating metabolic protein function and activity (15, 16). Accordingly, protein deacetylases capable of modulating acetylation status, such as SIRT1, have been proposed as key regulators of metabolism and mitochondrial biogenesis. The results from the present study, however, do not support a requirement for SIRT1 in regulating PGC-1α activity, acetylation, or mitochondrial biogenesis in skeletal muscle after exercise. Rather, our results suggest that modifications by acetyltransferases, such as GCN5, may play an equal or perhaps more important role in the regulation of PGC-1α acetylation and activity and, by extension, mitochondrial biogenesis in skeletal muscle after endurance exercise. Thus, four decades on from the seminal studies of Holloszy (3), important questions clearly remain regarding how exercise increases skeletal muscle mitochondrial biogenesis in vivo. Given the well described decline in mitochondrial metabolism following physical inactivity and in disease states such as obesity, insulin resistance and type 2 diabetes (1, 2), determining the molecular regulators of skeletal muscle mitochondrial biogenesis is important if targeted therapies are to be developed to maintain skeletal muscle function, metabolic potential, and health (45).
Acknowledgments
We thank Fred W. Alt (Harvard Medical School) and David S. Lombard (University of Michigan) for kindly sharing SIRT1 floxed mice and expertise. We thank Johan Auwerx and Carlés Canto (Ecole Polytechnique Fédérale, Lausanne) for advice regarding PGC-1α acetylation measurements and Jerrold M. Olefsky (University of California at San Diego) for input throughout this study.
This work was supported, in whole or in part, by National Institutes of Health Grants P30 AR058878-02 and K12 HD057022. This work was also supported by National Skeletal Muscle Research Center Grant R24 HD050837.
- PGC-1α
- peroxisome proliferator-activated receptor-γ coactivator 1-α
- ACC
- acetyl-CoA carboxylase
- AEX
- acute exercise
- AMPK
- AMP-activated protein kinase
- ATF 2
- activating transcription factor
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- CREB
- cAMP response element-binding protein
- GCN5
- general control of amino-acid synthesis 5
- HDAC
- histone deacetylase
- mKO
- knock-out of SIRT1 deacetylase activity in skeletal muscle
- SED
- sedentary
- SIRT1
- sirtuin 1
- VWR
- voluntary wheel running.
REFERENCES
- 1. Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Handschin C., Spiegelman B. M. (2008) Nature 454, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holloszy J. O. (1967) J. Biol. Chem. 242, 2278–2282 [PubMed] [Google Scholar]
- 4. Constable S. H., Favier R. J., McLane J. A., Fell R. D., Chen M., Holloszy J. O. (1987) Am. J. Physiol. 253, C316–322 [DOI] [PubMed] [Google Scholar]
- 5. Gollnick P. D., Armstrong R. B., Saltin B., Saubert C. W., 4th, Sembrowich W. L., Shepherd R. E. (1973) J. Appl. Physiol. 34, 107–111 [DOI] [PubMed] [Google Scholar]
- 6. Spina R. J., Chi M. M., Hopkins M. G., Nemeth P. M., Lowry O. H., Holloszy J. O. (1996) J. Appl. Physiol. 80, 2250–2254 [DOI] [PubMed] [Google Scholar]
- 7. Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) FASEB J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 8. Yan Z., Okutsu M., Akhtar Y. N., Lira V. A. (2011) J. Appl. Physiol. 110, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akimoto T., Pohnert S. C., Li P., Zhang M., Gumbs C., Rosenberg P. B., Williams R. S., Yan Z. (2005) J. Biol. Chem. 280, 19587–19593 [DOI] [PubMed] [Google Scholar]
- 10. Cantó C., Jiang L. Q., Deshmukh A. S., Mataki C., Coste A., Lagouge M., Zierath J. R., Auwerx J. (2010) Cell Metab. 11, 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wright D. C., Geiger P. C., Han D. H., Jones T. E., Holloszy J. O. (2007) J. Biol. Chem. 282, 18793–18799 [DOI] [PubMed] [Google Scholar]
- 12. Wende A. R., Schaeffer P. J., Parker G. J., Zechner C., Han D. H., Chen M. M., Hancock C. R., Lehman J. J., Huss J. M., McClain D. A., Holloszy J. O., Kelly D. P. (2007) J. Biol. Chem. 282, 36642–36651 [DOI] [PubMed] [Google Scholar]
- 13. Kelly D. P., Scarpulla R. C. (2004) Genes Dev. 18, 357–368 [DOI] [PubMed] [Google Scholar]
- 14. Scarpulla R. C. (2008) Physiol. Rev. 88, 611–638 [DOI] [PubMed] [Google Scholar]
- 15. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 19. Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 20. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gurd B. J., Perry C. G., Heigenhauser G. J., Spriet L. L., Bonen A. (2010) Appl. Physiol. Nutr. Metab. 35, 350–357 [DOI] [PubMed] [Google Scholar]
- 22. Gurd B. J., Yoshida Y., McFarlan J. T., Holloway G. P., Moyes C. D., Heigenhauser G. J., Spriet L., Bonen A. (2011) Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R67–75 [DOI] [PubMed] [Google Scholar]
- 23. Little J. P., Safdar A., Wilkin G. P., Tarnopolsky M. A., Gibala M. J. (2010) J. Physiol. 588, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suwa M., Nakano H., Radak Z., Kumagai S. (2008) Metabolism 57, 986–998 [DOI] [PubMed] [Google Scholar]
- 25. Boily G., Seifert E. L., Bevilacqua L., He X. H., Sabourin G., Estey C., Moffat C., Crawford S., Saliba S., Jardine K., Xuan J., Evans M., Harper M. E., McBurney M. W. (2008) PLoS One 3, e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gurd B. J., Yoshida Y., Lally J., Holloway G. P., Bonen A. (2009) J. Physiol. 587, 1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chabi B., Adhihetty P. J., O'Leary M. F., Menzies K. J., Hood D. A. (2009) J. Appl. Physiol. 107, 1730–1735 [DOI] [PubMed] [Google Scholar]
- 28. Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hamilton D. L., Philp A., MacKenzie M. G., Baar K. (2010) PLoS One 5, e11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philp A., Perez-Schindler J., Green C., Hamilton D. L., Baar K. (2010) Am. J. Physiol. 299, C240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sacks R. D., Roy R. R. (1982) J. Morphol. 173, 185–195 [DOI] [PubMed] [Google Scholar]
- 32. Trounce I. A., Kim Y. L., Jun A. S., Wallace D. C. (1996) Methods Enzymol. 264, 484–509 [DOI] [PubMed] [Google Scholar]
- 33. Pilegaard H., Saltin B., Neufer P. D. (2003) J. Physiol. 546, 851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amat R., Planavila A., Chen S. L., Iglesias R., Giralt M., Villarroya F. (2009) J. Biol. Chem. 284, 21872–21880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wright D. C., Han D. H., Garcia-Roves P. M., Geiger P. C., Jones T. E., Holloszy J. O. (2007) J. Biol. Chem. 282, 194–199 [DOI] [PubMed] [Google Scholar]
- 36. Lerin C., Rodgers J. T., Kalume D. E., Kim S. H., Pandey A., Puigserver P. (2006) Cell Metab. 3, 429–438 [DOI] [PubMed] [Google Scholar]
- 37. Little J. P., Safdar A., Cermak N., Tarnopolsky M. A., Gibala M. J. (2010) Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R912–917 [DOI] [PubMed] [Google Scholar]
- 38. Li L., Pan R., Li R., Niemann B., Aurich A. C., Chen Y., Rohrbach S. (2011) Diabetes 60, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 40. Coste A., Louet J. F., Lagouge M., Lerin C., Antal M. C., Meziane H., Schoonjans K., Puigserver P., O'Malley B. W., Auwerx J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dominy J. E., Jr., Lee Y., Gerhart-Hines Z., Puigserver P. (2010) Biochim. Biophys. Acta 1804, 1676–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poux A. N., Cebrat M., Kim C. M., Cole P. A., Marmorstein R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14065–14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ruderman N. B., Xu X. J., Nelson L., Cacicedo J. M., Saha A. K., Lan F., Ido Y. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lan F., Cacicedo J. M., Ruderman N., Ido Y. (2008) J. Biol. Chem. 283, 27628–27635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hawley J. A., Holloszy J. O. (2009) Nutr. Rev. 67, 172–178 [DOI] [PubMed] [Google Scholar]