Abstract
By providing contacts between hematopoietic cells and the bone marrow microenvironment, integrins are implicated in cell adhesion and thereby in control of cell fate of normal and leukemia cells. The ASB2 gene, initially identified as a retinoic acid responsive gene and a target of the promyelocytic leukemia retinoic acid receptor α oncoprotein in acute promyelocytic leukemia cells, encodes two isoforms, a hematopoietic-type (ASB2α) and a muscle-type (ASB2β) that are involved in hematopoietic and myogenic differentiation, respectively. ASB2α is the specificity subunit of an E3 ubiquitin ligase complex that targets filamins to proteasomal degradation. To examine the relationship of the ASB2α structure to E3 ubiquitin ligase function, functional assays and molecular modeling were performed. We show that ASB2α, through filamin A degradation, enhances adhesion of hematopoietic cells to fibronectin, the main ligand of β1 integrins. Furthermore, we demonstrate that a short N-terminal region specific to ASB2α, together with ankyrin repeats 1 to 10, is necessary for association of ASB2α with filamin A. Importantly, the ASB2α N-terminal region comprises a 9-residue segment with predicted structural homology to the filamin-binding motifs of migfilin and β integrins. Together, these data provide new insights into the molecular mechanisms of ASB2α binding to filamin.
Keywords: E3 Ubiquitin Ligase, Hematopoiesis, Leukemia, Proteasome, Protein Degradation, ASB, Filamin
Introduction
Characterization of hematopoietic stem cell and leukemia stem cell properties and understanding the molecular mechanisms that control the early steps of hematopoietic differentiation, which are deregulated in leukemia cells, are major challenges. These issues are relevant not only for the development of therapeutic approaches to target leukemia stem cells in vivo, but also for engraftment of normal hematopoietic stem cells following transplantation. Hematopoietic stem cells reside predominantly in a complex bone marrow microenvironment, the stem cell niche (1, 2). Hematopoietic stem cell fate decisions are governed by the integrated effects of niche-independent intrinsic and niche-dependent extrinsic signals. Recent studies indicate that changes in the hematopoietic stem cell niche may have a role in hematopoietic malignancies (3–5). Available data show that during development and following transplantation, integrin adhesion molecules play a major role in anchoring stem cells in the hematopoietic niche (6–8). Although adhesion of CD34 positive cells to fibronectin inhibits cell proliferation, adhesion of acute myeloid leukemia (AML)5 cells to fibronectin stimulates proliferation (9). In addition, AML cells showed increased survival as a result of the interaction of β1 integrins (α4β1/VLA-4 and α5β1/VLA-5) on leukemia cells with fibronectin leading to their reduced chemosensitivity (10, 11). Accordingly, antibodies directed against VLA-4 prolong survival of mice in a bone marrow minimal residual disease model (10). Moreover, the FNIII14 peptide of fibronectin that impairs VLA-4- and VLA-5-mediated adhesion to fibronectin overcomes cell adhesion-mediated drug resistance (12). In this context, proteins controlling integrin-dependent adhesion of hematopoietic cells may represent novel therapeutic targets in AML.
Our previous work identified ASB2 as a retinoic acid response gene and a target gene for the oncogenic promyelocytic leukemia retinoic acid receptor α (PML-RARα) fusion protein in acute promyelocytic leukemia cells (13, 14). Expression of PML-RARα has been shown to induce the myeloid differentiation arrest observed in acute promyelocytic leukemia (15–18). At the molecular level, PML-RARα acts as a transcriptional repressor that interferes with gene expression programs normally leading to full myeloid differentiation. Recently, PML-RARα was shown to be bound to the ASB2 promoter in acute promyelocytic leukemia cells in the absence of retinoic acid leading to hypoacetylation of histone H3 (19). Moreover, following retinoic acid treatment of acute promyelocytic leukemia cells, hyperacetylation and recruitment of RNA polymerase II to the ASB2 promoter were observed (19). Furthermore, ASB2 is also a target of another oncoprotein that acts as a transcriptional repressor, the AML1-ETO fusion protein,6 indicating that ASB2 mis-expression is associated with AML. However, ASB2 is specifically expressed in normal immature hematopoietic cells (13, 14) and so is likely to be relevant during early hematopoiesis. Importantly, Notch activation stimulated ASB2 expression (20). ASB2 encodes two isoforms, a hematopoietic-type (ASB2α) and a muscle-type (ASB2β) that are involved in hematopoietic and myogenic differentiation, respectively (21, 22). ASB2 proteins belong to the family of ASB proteins that harbor a variable number of ankyrin repeats (ANK) followed by a suppressor of cytokine signaling box located at the C-terminal end of the protein (23). These proteins are the specificity subunits of E3 ubiquitin ligase complexes (21, 22). Indeed, suppressor of cytokine signaling box-mediated interactions with the Elongin B-Elongin C (EloB-EloC) complex and the Cul5/Rbx2 module allow ASB2 proteins to assemble a multimeric E3 ubiquitin ligase complex, and so regulate the turnover of specific proteins involved in cell differentiation. We have recently shown that ASB2α ubiquitin ligase activity drives proteasome-mediated degradation of actin-binding proteins filamin A (FLNa), FLNb, and FLNc (24, 25). In addition to their role as actin cross-linkers, FLNs bind many adaptor and transmembrane proteins (26–28). In this way, FLNs can regulate cell shape and cell motility. We have demonstrated that ASB2α-mediated degradation of FLNs can regulate integrin-mediated spreading of adherent cells and initiation of migration of both HT1080 and Jurkat cells (24, 25, 29). FLNs are composed of an N-terminal actin-binding domain followed by 24 immunoglobulin-like domains (IgFLN(1–24)) (30). The CD face of Ig-like repeats of FLNa (IgFLNa), the major nonmuscle isoform of FLNs, represents a common interface for FLN-ligand interaction (31–33). Interestingly, it was recently demonstrated that FLN ligands can associate with several IgFLNa domains belonging to the same subgroup (34). Among group A, which contains seven IgFLNa repeats, IgFLNa21 binds GPIbα, β7 integrin, and migfilin with the highest affinity (31, 32, 34). Here, molecular modeling, site-directed mutagenesis, and cell biological studies were used to obtain structural and functional insights into the ASB2α E3 ubiquitin ligase complex.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture Conditions
Myeloblastic PLB985 cells stably transfected with ZnSO4-inducible vectors expressing ASB2αwt, ASB2αLA, ASB2ΔN, and ASB2αY9F were used as described (24). FLNa knockdown PLB985 cells were obtained by transfecting PLB985 cells with short hairpin RNA (shRNA) against human FLNa in pSM2c vector (Open Biosystems). After 2 days, transfected cells were selected using 0.5 μg/ml of puromycin. PLB985 cells expressing an shRNA targeting luciferase were used as controls. HeLa and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter of glucose (Invitrogen), 10% fetal bovine serum (PAA Laboratories), and penicillin-streptomycin (Invitrogen).
Plasmid Constructs
The pcDNA3-FLNa-GFP, pEGFP-C3-ASB2α, and pEGFP-C3-ASB2αLA expression constructs have been used previously (22, 24). The pGEX-IgFLNa21 and pGEX-IgFLNa21AA/DK plasmids were described previously (32). IgFLNb21 was generated by PCR and subcloned into a derivative of pGEX-2T vector (GE Healthcare). Deletion of the amino-terminal region of ASB2α (amino acids 1 to 20) was generated by PCR amplification. Constructions of the ASB2αY9F, ASB2αS11D, and ASB2αF13E mutated plasmids were achieved using the QuikChange site-directed mutagenesis kit. For this, forward mutated oligonucleotide sequences were (mutated bases in bold face): 5′-CTCCTATGCAGAGTTCTTTTCCCTCTTTCAC-3′, 5′-CCTATGCAGAGTACTTTGACCTCTTTCACTCCTGCTCTGC-3′, and 5′-CAGAGTACTTTTCCCTCGAGCACTCCTGCTCTGCACCC-3′, respectively. The ASB2ΔN and ASB2αY9F fragments were subcloned into the pEGFP-C3 expression vector (Clontech), the pBacPAK8 plasmid (Clontech), and the pMTCB6+-derived expression vectors to direct the expression of ASB2 proteins fused to the FLAG epitope at their amino terminus under control of the zinc-inducible sheep metallothionein promoter. ASB2αS11D and ASB2αF13E fragments were subcloned into the pEGFP-C3 expression vector. The pEGFP-C3-ASB2α(1–20), pEGFP-C3-ASB2αANK(1–3), pEGFP-C3-ASB2αANK(1–5), pEGFP-C3-ASB2αANK(1–7), pEGFP-C3-ASB2αANK(1–9), pEGFP-C3-ASB2αANK(1–10), pEGFP-C3-ASB2αANK(1–12), and pEGFP-C3-ASB2αΔNANK(1–12) plasmids were constructed as follows: (i) cloning sites were introduced using the QuikChange site-directed mutagenesis kit (Stratagene) at the 3′ end of the DNA fragments of interest, (ii) the 3′ parts of the ASB2 open reading frame were deleted following digestion and ligation of the vectors. The ASB2αANK(1–10) fragment was cloned into a derivative of pET21a (Novagen), in-frame with His6 codons located downstream from the ASB2α sequence. All constructs were verified by DNA sequencing.
Cell Adhesion to Fibronectin
Fibronectin (BD Biosciences) was immobilized overnight at 4 °C in 96-well plates (50 μg/ml) in PBS. Wells were then saturated with 5% BSA in PBS for 1 h at room temperature and washed three times with PBS. PLB985 cells stably transfected with zinc-inducible vectors encoding ASB2 proteins or the corresponding empty vector were cultured with or without ZnSO4 for 16 h, loaded with 0.5 μm calcein AM in HBSS without Ca2+ and Mg2+ (HBSS−) containing 0.5% BSA, and washed once in HBSS−, 1 mm EDTA. Adhesion to fibronectin was assayed using 200,000 cells/well in HBSS−, 0.5% BSA wells in the absence or presence of 1 mm MnCl2 to activate integrins for 10 min. Nonadherent cells were removed with 1 to 3 gentle washes with PBS containing 1 mm MnCl2 (to maintain integrin activation) or PBS alone (to suppress integrin activation). Fluorescence intensity was quantified using a microplate fluorescence reader FLx-800 (Bio-TEK). The percentage of adherent cells was calculated as follows: (fluorescence intensity of adherent cells/fluorescence intensity of cells plated) × 100. Each assay was performed in triplicate and at least four independent experiments were done. Statistical analyses were performed using Prism software. All p values were calculated using the Mann-Whitney t test.
Cell Spreading
Cell spreading assays were carried out as described (24).
Immunofluorescence Microscopy
Immunofluorescence analyses were performed essentially as described (24). To better visualize ASB2α and FLNa localization on stress fibers 8 h after transfection of HeLa cells, immunofluorescence analyses were performed in cytoskeleton buffer containing 10 mm MES, pH 6.9, 5 mm glucose, 150 mm NaCl, 5 mm EGTA, 5 mm MgCl2 and the cells were pretreated with the same buffer containing 0.1% Triton X-100 for 30 s before paraformaldehyde fixation. Secondary antibodies used were Cy3-coupled goat anti-mouse (Jackson Laboratories). F-actin was visualized with Alexa 633-phalloidin (Fisher) diluted 1:500. Slides were viewed with a Zeiss Axio Imager M2 using a ×63/1.3 oil DIC Plan Apochromat objective (Zeiss). Images were acquired and processed using AxioVision software and AxioCam MRm camera (Zeiss). Pearson correlation coefficients of the ASB2 protein co-localization with FLNa are mean ± S.E. from 5 random regions from 5 ASB2α-expressing cells.
In Vivo Expression and Protein Extracts
HeLa and NIH3T3 cells were transfected using the Jet PEI reagent (Polyplus transfection) as recommended by the manufacturer. PLB985 cells were transfected using the nucleofector T solution and the O17 program (Amaxa). Cells were washed twice in PBS and, when indicated, treated with 0.1% Triton X-100 in cytoskeleton buffer for 30 s before lysis in whole cell extract buffer containing 50 mm Tris-HCl, pH 7.9, 150 mm NaCl, 1 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, or RIPA buffer containing 10 mm NaPi buffer, pH 7.2, 0.59 m NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 10% glycerol supplemented with 1 mm dithiothreitol, 1 mm Na3VO4, 50 mm NaF, 25 mm β-glycerophosphate, 2 mm sodium pyrophosphate, and 1% protease inhibitor mixture (P8340; Sigma). After three freeze-thaw cycles in liquid nitrogen, the resulting cell lysates were cleared by a 10-min 20,000 × g centrifugation at 4 °C.
Antibodies, Immunoblotting, and Immunoprecipitation
The serum raised against ASB2 (1PNA) has been described previously (14). All monoclonal antibodies were purchased from commercial suppliers as follows: anti-polyubiquitinylated proteins (clone FK1; Biomol International), anti-GFP (Rockland), anti-GST (Euromedex), anti-FLNa (clone PM6/317; Millipore), and anti-α-actinin 1 (Clone AT6.172; Millipore). Immunoblot analyses were performed essentially as described (24).
2 μg of anti-FLNa antibodies or control IgG and 30 μl of protein A-Sepharose suspension (GE Healthcare) were added to 880 μg of whole cell extracts of demembranated cells. After 3 h in ice, beads were washed twice in XT buffer (50 mm Pipes, 50 mm NaCl, 150 mm sucrose, 40 mm Na4P2O7·10H2O, 0.05% Triton X-100) supplemented with 1 mm Na3VO4, 50 mm NaF, 25 mm β-glycerophosphate, 2 mm sodium pyrophosphate, and 1% protease inhibitor mixture. After 5 min of boiling in Laemmli buffer, samples were resolved by SDS-PAGE and analyzed by immunoblotting.
Recombinant Protein Production
GST, GST-IgFLNa21, GST-IgFLNa21AA/DK, and GST-IgFLNb21 proteins were produced as described (35). The recombinant ASB2αANK(1–10)-His6, which was produced as inclusion bodies, was solubilized in 8 m urea, 10 mm Tris-HCl, 100 mm NaH2PO4, pH 8, and purified by chromatography using a nickel-nitrilotriacetic acid-agarose column and eluted with 100 mm imidazole in 8 m urea, 10 mm Tris-HCl, 100 mm NaH2PO4, pH 4.5. The purified ASB2αANK(1–10)-His6 protein was then re-natured by dialysis into 50 mm Hepes, pH 7.9, 150 mm NaCl, 5% sucrose, 1 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, 50 mm glycine.
In Vitro Binding Assays
7 μl of 1PNA serum or 15 μl of its preimmune counterpart and 30 μl of protein A-Sepharose suspension was added to 500 μg of whole cell extracts of demembranated cells. After 3 h in ice, beads were washed twice in XT buffer supplemented with 1 mm Na3VO4, 50 mm NaF, 25 mm β-glycerophosphate, 2 mm sodium pyrophosphate, and 1% protease inhibitor mixture. The immobilized GFP-ASB2αANK(1–10) protein was further incubated with 10 μg of Escherichia coli extracts expressing GST or GST-IgFLNa21 in whole cell extract buffer for 3 h in ice. Beads were washed twice as described above. After 5 min of boiling in Laemmli buffer, samples were resolved by SDS-PAGE and analyzed by immunoblotting. Alternatively, pulldown assays were performed using 5 μg of purified GST-IgFLNa21, GST-IgFLNa21AA/DK, or GST-IgFLNb21 bound to glutathione-agarose beads (Macherey-Nagel) and 0.4 μg of purified ASB2αANK(1–10)-His6. After two washes in dialysis buffer, bound proteins were fractionated by SDS-PAGE and analyzed by protein staining and Western blotting.
Ubiquitylation Assays
Recombinant baculoviruses encoding ASB2ΔN were generated with the BacPAK baculovirus expression system (Clontech). In vitro ubiquitylation assays were carried out as described (21, 24).
Molecular Modeling
Modeling was performed using the Accelrys modules Homology, Discover, Docking, and Biopolymer, run within InsightII (2005 version) on a Silicon Graphics Fuel work station.
Docking of the ASB2α N-terminal Peptide into the IgFLNa21 Domain
The FLNa-binding motif within the 20-residue ASB2α N-terminal peptide was identified and modeled based on structural homology to the integrin β2 (PDB code 2JF1_T), integrin β7 (PDB code 2BRQ_C), and migfilin (PDB code 2W0P_C) motifs, using the Align 123 and Homology programs. The residues located at the N- and C-terminals of the ASB2α β-strand motif were initially built in an extended conformation. Energy was minimized using the Discover consistent valence force field, a forcing constant of 100 kcal/mol, and the steepest descent and conjugate gradients protocols. The common β-strand served to preposition the ASB2α N-terminal peptide into the IgFLNa21 CD groove in the same way as migfilin. The resulting preliminary structure was then submitted to the Affinity program, an automatic flexible docking refinement procedure applied to predefined residues from the binding interface. The final interaction energy was −90 and −42 kcal for the van der Waals and electrostatic components, respectively. The criteria used for identifying hydrogen bonds were a donor-acceptor distance ≤3 Å and a minimum donor-proton-acceptor angle of 120°. The criterion used for identifying an interaction between hydrophobic groups was a distance ≤5 Å.
RESULTS
ASB2α-induced FLNa Degradation Regulates Adhesion of Hematopoietic Cells to Fibronectin
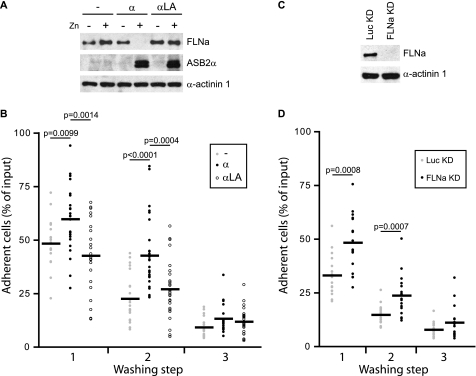
We previously found that ASB2α expression prevented cell spreading and inhibited initiation of migration, and that these effects were recapitulated by knocking down FLNa and FLNb (24, 29). To assess the role of ASB2α-induced FLN degradation in hematopoietic cell adhesion, myeloblastic PLB985 cells stably transfected with zinc inducible vectors encoding ASB2α, E3 ubiquitin ligase defective mutant ASB2αLA, or the corresponding empty vector control were cultured with or without ZnSO4, labeled with calcein AM, and allowed to adhere on fibronectin-coated wells in the absence or presence of Mn2+ to activate integrins. Loss of FLNa was observed only in cells expressing ASB2α (Fig. 1A). As expected, when adhesion was performed in the absence of Mn2+, only a low level of adhesion of cells was observed (supplemental Fig. S1A). However, cell adhesion was greatly enhanced with Mn2+ (Fig. 1B). Intriguingly, upon integrin activation by cultivating the cell in the presence of Mn2+, percentages of adherent cells expressing ASB2α after the first and second washing steps in PBS alone were significantly increased (Fig. 1B). However, no significant differences were observed following washes in PBS containing Mn2+ (supplemental Fig. S1B). To extend and further validate the finding that ASB2α controls integrin-mediated adhesion of hematopoietic cells to fibronectin, we established PLB985 cells lacking FLNa expression by transfecting cells with a vector encoding an shRNA against FLNa (Fig. 1C). As was observed for ASB2α-expressing cells, adhesion of PLB985 FLNaKD cells was significantly higher than adhesion of PLB985 LucKD control cells after washing steps in PBS alone (Fig. 1D). Taken together, our results suggest that integrin-dependent adhesion of hematopoietic cells is sustained in the absence of FLNa and establish a role for ASB2α in the regulation of hematopoietic cell adhesion through FLNa degradation.
FIGURE 1.
ASB2α-induced FLNa degradation enhances adhesion of hematopoietic cells to fibronectin. PLB985/MT-FLAG (−), PLB985/MT-ASB2α (α), and PLB985/MT-ASB2αLA (α LA) cells were cultured for 16 h without or with 60 μm ZnSO4 (A and B), loaded with calcein AM, serum arrested for 30 min, and plated on fibronectin-coated wells for 10 min in the presence of 1 mm Mn2+ (B). A, ASB2α and FLNa expression in cells cultured in the absence (−) or presence (+) of ZnSO4. Protein extracts (10 μg) were separated by SDS-PAGE and immunoblotted for FLNa, ASB2α, and α-actinin 1 as loading control. B, adhesion of PLB985/MT-FLAG, PLB985/MT-ASB2α, and PLB985/MT-ASB2αLA cells to fibronectin was assayed following the first, second, and the third washing steps with PBS. Dot plots show the overall distribution, the line shows the median value. p values were calculated using the Mann-Whitney t test. (Sample size: PLB985/MT-FLAG = 21, PLB985/MT-ASB2α = 27, and PLB985/MT-ASB2αLA = 27; from 7, 9, and 9 independent experiments, respectively.) C, FLNa expression in PLB985 FLNaKD and LucKD cells. Protein extracts (10 μg) were separated by SDS-PAGE and immunoblotted for FLNa and α-actinin 1. D, adhesion of PLB985 FLNaKD and PLB985 LucKD cells. Adhesion assays were performed as described above (sample size: PLB985 FLNaKD = 18 and PLB985 LucKD = 18; from 6 independent experiments).
The N-terminal Region of ASB2α Is Required to Induce Degradation of FLNa
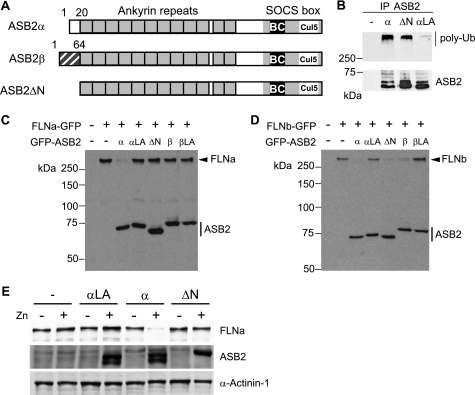
We previously demonstrated that although ASB2α drives ubiquitin-mediated degradation of both FLNa and FLNb (24), ASB2β induces degradation of FLNb but not FLNa (22). To better understand the molecular basis of this specificity, a deletion mutant lacking the 20-amino acid N-terminal region specific to ASB2α (ASB2ΔN) was constructed (Fig. 2A). When immunopurified from Sf21 cells, the recombinant ASB2ΔN-EloB-EloC-Cul5-Rbx2 complex stimulated formation of ubiquitin conjugates by UbcH5a, as observed with the wild-type ASB2α complex (Fig. 2B). This indicated that the ASB2ΔN-EloB-EloC-Cul5-Rbx2 complex harbors intrinsic E3 ubiquitin ligase activity. NIH3T3 cells were then co-transfected with vectors expressing GFP, GFP-ASB2α, GFP-ASB2β, or GFP-ASB2ΔN together with a FLNa-GFP or FLNb-GFP expression vectors. Twenty-four hours post-transfection, Western blotting revealed that GFP-ASB2α expression resulted in a loss of FLNa-GFP, whereas FLNa-GFP levels were not altered in cells expressing GFP-ASB2ΔN or GFP-ASB2β (Fig. 2C). In contrast, loss of FLNb was observed in cells expressing GFP-ASB2α, GFP-ASB2ΔN, or GFP-ASB2β (Fig. 2D). Furthermore, whereas equivalent amounts of ASB2α, ASB2ΔN, and ASB2αLA proteins were expressed in PLB985 hematopoietic cells, only wild-type ASB2α induced loss of endogenous FLNa (Fig. 2E). Collectively, our results demonstrated that ASB2α residues 1–20 are dispensable for ASB2α intrinsic E3 ubiquitin ligase activity but are required for ASB2α-induced FLNa degradation.
FIGURE 2.
The N-terminal region of ASB2α is required for ASB2α to target FLNa. A, schematic representation of the domains of ASB2α, ASB2β, and ASB2ΔN. B, ASB2ΔN activates formation of polyubiquitin chains by the E2 enzyme. EloB, EloC, Cul5, and Rbx2 together with wild-type ASB2α, ASB2αLA (LA), and ASB2ΔN were expressed in Sf21 cells. ASB2α complexes were immunoprecipitated (IP) with anti-ASB2 antibodies, and incubated with Uba1, Ubc5a, ubiquitin, and ATP. Their ability to stimulate polyubiquitylation was assessed by Western blotting with antibodies to polyubiquitylated proteins. ASB2 immunoprecipitation was assessed by blotting with anti-ASB2 antibodies. C, ASB2α-induced FLNa-GFP degradation is dependent on the N-terminal region of ASB2α. NIH3T3 cells were transfected for 24 h with FLNa-GFP together with GFP (−), GFP-ASB2α, GFP-ASB2αLA, GFP-ASB2ΔN, GFP-ASB2β, and GFP-ASB2βLA expression vectors, as indicated. 20-μg aliquots of whole cell extracts were immunoblotted with antibodies to GFP. D, the N-terminal region of ASB2α is dispensable for ASB2α-induced FLNb-GFP degradation. NIH3T3 cells were transfected for 24 h with FLNb-GFP together with GFP (−), GFP-ASB2α, GFP-ASB2αLA, GFP-ASB2ΔN, GFP-ASB2β, and GFP-ASB2βLA expression vectors, as indicated. 20-μg aliquots of whole cell extracts were immunoblotted with antibodies to GFP. E, ASB2α-induced FLNa degradation is dependent on the N-terminal region of ASB2α in hematopoietic cells. PLB985/MT-FLAG, PLB985/MT-ASB2α, PLB985/MT-ASB2αLA, and PLB985/MT-ASB2ΔN cells were cultured without (−) or with 95, 70, 85, and 95 μm ZnSO4, respectively, for 8 h to induce the expression of equivalent amounts of ASB2α proteins. Protein extracts (10 μg) were separated by SDS-PAGE and immunoblotted for FLNa, ASB2α, and α-actinin 1.
The N-terminal Region and ANK 1 to 10 of ASB2α Are Necessary to Target FLNa
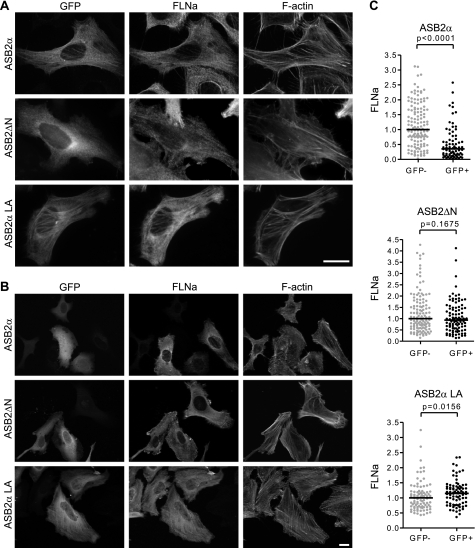
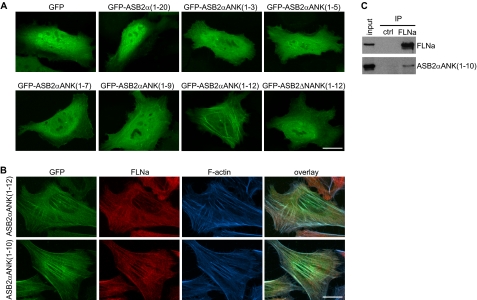
As previously observed in transfected HeLa cells (24), (i) ASB2α was transiently colocalized with F-actin (Fig. 3A); (ii) ASB2α is diffuse throughout the cytoplasm when FLNa is degraded (Fig. 3B); and (iii) E3 ubiquitin ligase defective mutants of ASB2α do not degrade FLNa and accumulate on stress fibers (Fig. 3, A and B). These results suggest that colocalization of ASB2α with F-actin may be the result of ASB2α association with FLNa. We therefore examined the subcellular localization of the ASB2ΔN N-terminal deletion mutant in transfected HeLa cells. ASB2ΔN did not accumulate on stress fibers (Fig. 3, A and B). Accordingly, deletion of the N-terminal region in ASB2α abrogated its ability to induce degradation of endogenous FLNa in transfected HeLa cells (Fig. 3, B and C). Collectively, our data indicated that ASB2α residues 1–20 encompass a major determinant for ASB2α colocalization with FLNa and subsequent polyubiquitylation and degradation by the proteasome. We next questioned whether ASB2α residues 1–20 were sufficient for colocalization with FLNa onto stress fibers. This domain was therefore fused to GFP and expressed in HeLa cells. The resulting GFP-ASB2α (residues 1–20) protein was diffuse throughout the cytoplasm and the nucleus (Fig. 4A) indicating that the N-terminal region specific to ASB2α is not sufficient for ASB2α recruitment to stress fibers. To further delineate the ASB2α domain required for the targeting of FLNa, several C-terminal deletion mutants of ASB2α were constructed and their subcellular localization assessed in HeLa cells. In contrast to GFP-ASB2αANK(1–3), GFP-ASB2αANK(1–5), GFP-ASB2αANK(1–7), and GFP-ASB2αANK(1–9), GFP-ASB2αANK(1–12) accumulates onto stress fibers (Fig. 4A). As expected, deletion of amino acids 1–20 (GFP-ASB2ΔNANK(1–12)) abrogated stress fiber localization of this deletion mutant (Fig. 4A). To better visualize ASB2α and FLNa localization on stress fibers, cells were permeabilized with Triton X-100 before fixation. This treatment removes the cell membrane and membrane-associated proteins but leaves behind the cytoskeleton and cytoskeletally associated proteins. Eight hours post-transfection, GFP-ASB2αANK(1–12) and GFP-ASB2αANK(1–10) colocalized with FLNa and F-actin in stress fibers (Fig. 4B). These results indicate that ANK 1 to 10, together with the N-terminal region specific to ASB2α, is required for colocalization of ASB2α with FLNa on stress fibers.
FIGURE 3.
ASB2α residues 1–20 encompass a major determinant for ASB2α colocalization with FLNa and subsequent degradation. A and B, HeLa cells were transfected with GFP-ASB2α, GFP-ASB2αLA, or GFP-ASB2ΔN expression vectors and analyzed 8 (A) and 48 h (B and C) after transfection using an antibody against FLNa. C, dot plots show relative FLNa fluorescence intensities of GFP negative (gray) and GFP positive (black) cells measured using the Axiovision software. p values were calculated with the Mann-Whitney t test. Scale bar represents 20 μm.
FIGURE 4.
Identification of the minimal region of ASB2α necessary for association with FLNa. A, HeLa cells were transfected with GFP, GFP-ASB2α(1–20), GFP-ASB2αANK(1–3), GFP-ASB2αANK(1–5), GFP-ASB2αANK(1–7), GFP-ASB2αANK (1–9), GFP-ASB2αANK(1–12), or GFP-ASB2ΔN/ANK(1–12) expression vectors and analyzed 8 h after transfection. B, HeLa cells were transfected with GFP-ASB2αANK(1–12) or GFP-ASB2αANK(1–10) expression vector. Eight hours after transfection, cells were pretreated with Triton X-100 before fixation and stained with anti-FLNa and phalloidin. C, ASB2αANK(1–10) associates with FLNa. Protein extracts from demembranated cells expressing ASB2αANK(1–10) were subjected to immunoprecipitation (IP) with anti-FLNa or control antibodies and then assessed for ASB2α or FLNa. Scale bar represents 20 μm.
To determine whether ASB2αANK(1–10) can associate with FLNa we performed co-immunoprecipitation assays. HeLa cells were transfected with a vector expressing GFP-ASB2αANK(1–10), 8 h later cells were demembranated by permeabilization with Triton X-100 and lysed. ASB2αANK(1–10) was detected in FLNa but not in control immunoprecipitates (Fig. 4C). Together with the co-localization assays, our results indicate that ASB2αANK(1–10) is the minimal ASB2α construct that associates with endogenous FLNa.
Identification of an IgFLNa21-binding Motif Encompassing ASB2α Residues 8–16
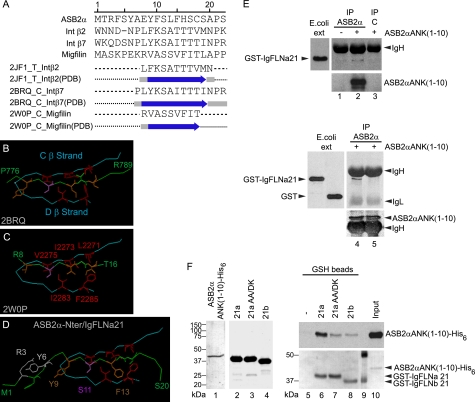
We next questioned whether ASB2α could target FLNa for degradation through an interaction between its N-terminal region and FLNa (Fig. 5A). A prototypic structural FLNa-binding motif has recently been characterized through a number of x-ray and NMR-based studies of complexes between the IgFLNa21 and peptides derived from either the tails of integrin β2 or β7, or the N-terminal region of migfilin (31, 32, 34, 36). This 8–10-residue long β-strand motif forms an anti-parallel β-sheet with the IgFLNa21 C β-strand and anchors into the CD groove through hydrophobic contacts, especially with Ile-2283 and Phe-2285 residues from the D β-strand (Fig. 5B,C). Alignment of the ASB2α N-terminal peptide with filamin-binding peptides derived from either the tails of integrin β2 or β7, or the N-terminal region of migfilin predicts the presence of a 9-residue hydrophobic β-strand in the ASB2α N-terminal peptide (residues 8–16) (Fig. 5A), structurally homologous to those in migfilin and β integrins. Interestingly, the most stringent element of sequence conservation detected is Ser-11, which was shown to play a critical role in anchoring the motif to the D strand via hydrogen bonding. These results suggest that this newly identified structural ASB2α motif might dock into the cavity delineated by the IgFLNa21 CD hairpin similarly to the migfilin and integrin β FLNa-binding motifs. Indeed, the molecular modeling of the complex between the ASB2α N-terminal peptide and the IgFLNa21 domain, based on the x-ray structure of the migfilin motif-IgFLNa21 complex, revealed that the ASB2α motif could fit into the hydrophobic IgFLNa21 CD groove without steric clashes (Fig. 5D). Binding is first mediated by hydrogen bonding between the ASB2α and IgFLNa21 C β-strand backbones and by a side chain/main chain hydrogen-bond between ASB2α Ser-11 and IgFLNa21 Ala-2281. The interaction is then anchored by a dense network of stacking interactions between ASB2α Phe-13 and IgFLNa21 Ile-2273 (C strand) and Ile-2283 and Phe-2285 (D strand). The complex is further stabilized by interactions involving residues specific to ASB2α, such as Leu-12 and His-14, both stacked with IgFLNa21 Ala-2272.
FIGURE 5.
Identification of a FLNa-binding motif within the N-terminal peptide of ASB2α. A, combined sequence and secondary structure alignment of ASB2α peptide (residues 1–20) against integrin β2 (residues 747–765), integrin β7 (residues 770–789), migfilin (residues 1–20) peptides, and their respective IgFLNa21-binding motifs solved by crystallography (PDB codes 2JF1_T, 2BRQ_C, and 2W0P_C). Predicted and x-ray-derived (indicated by arrows) locations of β-strands were in good agreement, validating the method of the secondary structure prediction used by Align 123. B and C, main structural determinants of the interaction between IgFLNa21 and either the integrin β7 (B) or migfilin (C) motifs, as derived from crystallographic structures (respective PDB codes 2BRQ and 2W0P). Interacting residues color code: red for IgFLNa21 (Leu-2271, Ile-2273, Val-2275, Ile-2283, and Phe-2285), brown for integrin (Tyr-778, Ile-782, and Thr-784) or migfilin (Val-9, Val-13, and Ile-15), and pink for Ser-780 or Ser-11. D, modeled structure resulting from docking ASB2α N-terminal peptide into the IgFLNa21 CD groove. The display code used was described for B and C. E, ASB2αANK(1–10) associates with IgFLNa21. Protein extracts from HeLa cells mock transfected (lane 1) or transfected with the GFP-ASB2αANK(1–10) vector (lanes 2–5) were immunoprecipitated (IP) with anti-ASB2 serum (lanes 1, 2, 4, and 5) or with control ASB2 preimmune serum (lane 3). Immobilized proteins were further incubated with extracts from E. coli expressing GST-IgFLNa21 (lanes 1–4) or GST alone (lane 5). The interacting complexes, as well as aliquots of E. coli extracts (E. coli ext.), were resolved by SDS-PAGE and immunoblotted with anti-GST and/or anti-ASB2 antibodies. The heavy (IgH) and light (IgL) chains of immunoglobulins are indicated. F, ASB2αANK(1–10) binds IgFLNa directly. ASB2αANK(1–10)-His6 and GST-IgFLN proteins were purified by affinity chromatography. 0.2 μg of ASB2αANK(1–10)-His6 (lane 1), 5 μg of GST-IgFLNa21 (lane 2), 5 μg of GST-IgFLNa21AA/DK (lane 3), and 5 μg of GST-IgFLNb21 (lane 4) were run on a SDS-PAGE. The gel was stained with Coomassie Blue. Direct pulldown assays were performed using purified ASB2αANK(1–10)-His6 and glutathione beads that were uncoated (lane 5) or coated with purified GST-IgFLNa21 (lane 6), GST-IgFLNa21AA/DK (lane 7), and GST-IgFLNb21 (lane 8). Bound proteins were detected by Ponceau staining (GST fusion proteins) and immunoblotting with anti-ASB2 antibodies (ASB2αANK(1–10)-His6). Lane 9, molecular mass marker.
To explore whether ASB2α can associate with the IgFLNa21 domain, affinity immobilized GFP-ASB2αANK(1–10) from demembranated cell extracts was incubated with GST or GST-IgFLNa21 proteins expressed in E. coli. As shown in Fig. 5E, GST-IgFLNa21 was detected in ASB2α but not in control pulldowns. Furthermore, GFP-ASB2αANK(1–10) associates with GST-IgFLNa21 but not with GST alone (Fig. 5E) suggesting that IgFLNa21 is an ASB2α binding domain. To further demonstrate an interaction between ASB2αANK(1–10) and IgFLNa21, recombinant proteins were produced in E. coli, purified by affinity chromatography (Fig. 5F), and their direct binding tested in pulldown assays. Indeed, ASB2αANK(1–10) bound strongly to IgFLNa21, whereas no binding was seen for glutathione beads (Fig. 5F). Importantly, binding of ASB2αANK(1–10) to the IgFLNa21AA/DK mutant was greatly reduced. Furthermore, despite the high degree of similarity between β strands C and D of IgFLNa21 and IgFLNb21, the interaction between ASB2αANK(1–10) and IgFLNb21 is reduced compared with IgFLNa21 (Fig. 5F).
Tyr-9, Ser-11, and Phe-13 of ASB2α Are Required to Target FLNa to Degradation
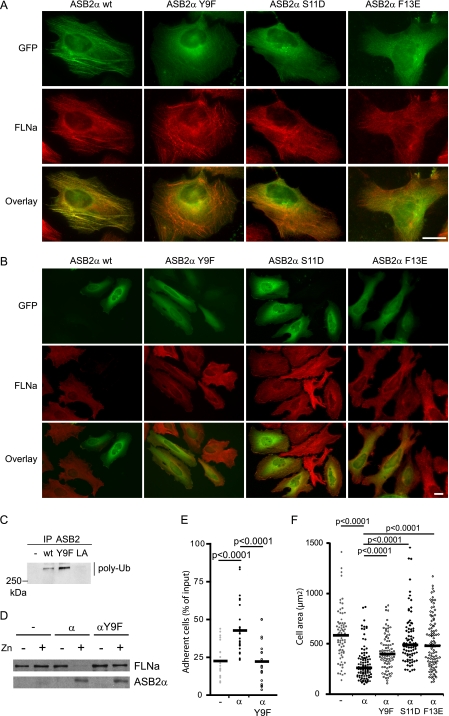
Because we previously showed that colocalization of ASB2α with F-actin may be the result of ASB2 association with FLNs, we examined subcellular localization of ASB2α proteins mutated within the putative IgFLNa-binding site in transfected HeLa cells. In contrast to GFP-ASB2α, GFP-ASB2αY9F, GFP-ASB2αS11D, and GFP-ASB2αF13E did not co-localize with FLNa to stress fibers (Fig. 6A). To better quantify these observations, we measured the co-localization correlation coefficient between ASB2α and FLNa staining. The Pearson coefficients for GFP-ASB2α expressing cells was 0.539 ± 0.117, for GFP-ASB2αY9F expressing cells, 0.006 ± 0.085; for GFP-ASB2αS11D expressing cells, −0.003 ± 0.107; and for GFP-ASB2αF13E expressing cells, −0.050 ± 0.129. To understand the structural determinants of these observations, ASB2αS11D- and F13E-mutated peptides were modeled and docked into the IgFLNa21 hydrophobic CD groove as described for the wild-type ASB2α N-terminal peptide (supplemental Fig. S2, A and B). In both cases, there was hydrogen pairing between the ASB2α-mutated motifs (residues 8–16) and the C β-strand backbones, but the respective complexes presented different degrees of stability relatively to the wild-type complex, as estimated by an energy score. ASB2αF13E mutation was the most deleterious, inducing a marked decrease in the van der Waals energy absolute value, well in line with the loss of the critical hydrophobic contacts between ASB2α Phe-13 and IgFLNa21 Ile-2273, Ile-2283, and Phe-2285. Surprisingly, the ASB2αS11D mutation caused a slight increase in the van der Waals energy absolute value, partially compensating for the decrease in the electrostatic energy absolute value, in correlation with a subtle reorganization and stabilization of the hydrophobic cluster. In addition, there was still a possibility of a side chain/main chain hydrogen bond between Asp-11 and IgFLNa21 Ala-2281. Remarkably, it was impossible to obtain a low energy conformation for the ASB2αY9F-IgFLNa21 complex, probably due to the observed perturbation in the initial orientations of the mutated motif side chains. These results further suggest that ASB2α residues 8–16 comprise a determinant for ASB2α binding to FLNa. Accordingly, mutation of Tyr-9, Ser-11, or Phe-13 in ASB2α abrogated its ability to induce degradation of endogenous FLNa in transfected HeLa cells (Fig. 6B). Although the ASB2αY9F protein has intrinsic E3 ubiquitin ligase activity (Fig. 6C), it does not induce degradation of endogenous FLNa in PLB985/MT-ASB2αY9F cells induced to express ASB2αY9F with zinc (Fig. 6D). We then assessed whether mutation of one of these key residues, Tyr-9, disrupted the ability of ASB2α to stabilize adhesion of hematopoietic cells. Indeed, in contrast to cells expressing wild-type ASB2α, adhesion of ASB2αY9F-expressing cells was not sustained following integrin activation by Mn2+ and washes with PBS alone (Fig. 6E). Furthermore, expression of ASB2αY9F but also of ASB2αS11D and ASB2αF13E did not affect the spreading of transfected NIH3T3 cells on fibronectin-coated slides (Fig. 6F) as previously observed for ASB2α E3 ligase defective mutants (24). Collectively, our data indicated that ASB2α residues 8–16 encompass a major determinant for FLNa binding, subsequent polyubiquitylation and degradation by the proteasome, and regulate cell spreading and cell adhesion.
FIGURE 6.
ASB2α Tyr-9, Ser-11, and Phe-13 are required for ASB2α to target FLNa. HeLa cells were transfected with GFP-ASB2α, GFP-ASB2αY9F, GFP-ASB2αS11D, and GFP-ASB2αF13E expression vectors and analyzed 8 (A) and 48 h (B) after transfection using an antibody against FLNa. A, ASB2α Tyr-9, Ser-11, and Phe-13 are required for co-localization of ASB2α and FLNa onto stress fibers. B, ASB2α-induced FLNa degradation in HeLa cells is dependent on Tyr-9, Ser-11, and Phe-13 of ASB2α. Scale bar represents 20 μm. C, ASB2αY9F activates formation of polyubiquitin chains by the E2 enzyme. EloB, EloC, Cul5, and Rbx2 together with wild-type ASB2α, ASB2αLA, and ASB2αY9F were expressed in Sf21 cells. ASB2α complexes were immunoprecipitated (IP) with anti-ASB2 antibodies, and incubated with Uba1, Ubc5a, ubiquitin, and ATP. Their ability to stimulate polyubiquitylation was assessed by Western blotting with antibodies to polyubiquitylated proteins. D and E, effects of ASB2αY9F on adhesion of hematopoietic cells to fibronectin. PLB985/MT-FLAG (−), PLB985/MT-ASB2α (α), and PLB985/MT-ASB2αY9F (αY9F) cells were cultured for 16 h without or with 60 μm ZnSO4 (D and E), loaded with calcein AM, serum arrested for 30 min, and plated on fibronectin-coated wells for 10 min in the presence of 1 mm Mn2+. D, 10-μg aliquots of protein extracts of cells cultured in the absence (−) or presence (+) of ZnSO4 were immunoblotted with antibodies to ASB2α and FLNa. E, adhesion of PLB985/MT-FLAG, PLB985/MT-ASB2α, and PLB985/MT-ASB2αY9F cells to fibronectin was assayed following the second washing step with PBS. Dot plots show the overall distribution, the line shows the median value. (Sample size: PLB985/MT-FLAG = 21, PLB985/MT-ASB2α = 27, and PLB985/MT-ASB2αY9F = 21; from 7, 9, and 7 independent experiments, respectively.) F, effects of ASB2αY9F, ASB2αS11D, and ASB2αF13E on cell spreading on fibronectin of NIH3T3 cells. NIH3T3 cells were transfected with GFP (−), GFP-ASB2α (α), GFP-ASB2αY9F (αY9F), GFP-ASB2αS11D (αS11D), or GFP-ASB2αF13E (αF13E) expression vectors for 24 h, trypsinized, and serum arrested for 1 h in suspension. Cells were plated on fibronectin-coated coverslips and fixed after 45 min. Cell areas of at least 100 cells from three independent experiments were measured. Dot plots show the overall distribution, lines show the median values. p values were calculated using the Mann-Whitney t test.
DISCUSSION
Physical contact between leukemia cells and the bone marrow microenvironment provides a refuge for minimal residual disease. Of importance, interaction of β1 integrins with fibronectin is involved in acquired chemoresistance of AML cells (10, 11, 37). In this context, deciphering the molecular mechanisms controlling integrin-dependent adhesion of normal hematopoietic and leukemia cells may ultimately lead to new treatment strategies that specifically target leukemia cells. Although players that activate integrins have been described, few players that inhibit integrins have been identified so far (38, 39). Among them, FLNa has been proposed to compete with talin for binding to the cytoplasmic tail of integrin β subunits (32). We recently demonstrated that ASB2α regulates FLNa functions via proteasomal degradation of FLNa (24), suggesting that ASB2 may contribute to integrin activation. We therefore assessed whether ASB2α, through FLNa degradation, plays a role in the regulation of integrin-dependent functions in hematopoietic cells. We found that expression of ASB2α significantly sustained integrin-dependent adhesion of hematopoietic cells to fibronectin. Of importance, FLNa knockdown recapitulated ASB2α effects on hematopoietic cell adhesion and concomitant FLNb knockdown was not required. This is in contrast to the spreading or migration defects previously observed in adherent cells following ASB2α expression or FLNa and FLNb double knockdown (24, 29). However, the levels of FLNb and FLNc are low in PLB985 cells (25) and so may be insufficient to compensate for the loss of FLNa in these cells. Our results are in agreement with previous work in HT1080 fibrosarcoma cells and Jurkat T lymphoblasts showing that loss of FLNs increased the percentage of non-motile cells plated on fibronectin (29).
The role of ubiquitin-mediated proteasomal degradation in the control of hematopoiesis has recently been highlighted by the fact that c-Myc stability is controlled by the SCFFbw7 E3 ubiquitin ligase in hematopoietic stem cells (40). In fact, ubiquitin-mediated degradation is one of the major pathways for controlled proteolysis in eukaryotes. In this pathway, E3 ubiquitin ligases that determine the specificity of protein substrates represent a class of potential drug targets for pharmaceutical intervention. Although proteasome inhibition has proved to be of therapeutic utility, the strategy of modulating the activity of E3 ubiquitin ligases is more specific. In this regard, characterization of the various E3 ubiquitin ligases and their respective substrates and understanding the signals that regulate specific ubiquitin ligation events should contribute to the development of new therapies that target the ubiquitin system. Our data provide evidence that the N-terminal region specific to the hematopoietic isoform of ASB2 plays roles in the targeting of FLNa to proteasomal degradation. Indeed, deletion of this domain or mutation of Tyr-9, Ser-11, or Phe-13 abolished the recruitment of ASB2α to actin stress fibers and completely abrogated the ability of ASB2α to induce FLNa degradation. Of interest, mutation of Tyr-9 of ASB2α abolished ASB2α effects on adherent cell spreading and adhesion of hematopoietic cells. The most striking feature of this N-terminal region is that it encompasses residues (8 to 16) that share structural homology with the binding domains of several IgFLNa21 ligands (31–34). However, our results indicate that, in addition to this region, ankyrin repeats 1 to 10 of ASB2α are necessary for in vivo colocalization with FLNa. Interestingly, we further showed that the ASB2αANK(1–10) protein, which contains the N-terminal region, can bind directly to the IgFLNa21 domain. As expected, mutation of Ala-2272 and Ala-2274 in strand C of IgFLNa21 strongly inhibited ASB2αANK(1–10) binding as previously observed for β7 integrin or migfilin (31, 32, 34), suggesting that ASB2α, integrin β7, and migfilin may bind to a similar site on IgFLNa21. However, because the whole group A of IgFLNa repeats can bind a set of ligands including β integrins and migfilin (34), we do not exclude that other regions of FLNa may also contribute to the ASB2α-FLNa interaction.
Our results reinforce the view that the CD face of IgFLNa is a common binding interface for FLN partners and suggest that ASB2α may compete with FLN partners such as β integrins and migfilin for FLNa binding. We therefore cannot exclude the possibility that ASB2α affects integrin-dependent functions through the dissociation of FLNa partners from FLNa. In this context, it should be mentioned that as observed in FLN-depleted cells following ASB2α expression or FLN knockdown, cells depleted in migfilin exhibit less cell spreading (41). Moreover, it is worth noting that binding of migfilin to FLNa may promote integrin activation by dissociating FLNa from integrins (42). Furthermore, displacement of FLNa from integrin tails or from migfilin by ASB2α may allow FLNa polyubiquitylation and subsequently, acute proteasomal degradation of all FLNa molecules. We have previously demonstrated that ASB2α induces degradation of all three filamins (29), whereas ASB2β induces degradation of FLNb but not FLNa (21, 22). Our findings that the N-terminal region specific to the ASB2α isoform is required for FLNa degradation, and that ASB2α-ANK(1–10) preferentially binds IgFLNa21, may help to explain the specificity of ASB2 proteins toward FLNa and FLNb. It will nonetheless be important to identify residues involved in the molecular interactions between ASB2 proteins and FLNa and FLNb to further our understanding of the specificity of ASB2 proteins toward FLNs.
In conclusion, our structural and cell biology studies have revealed a region of ASB2α that is involved in the recruitment of its substrate, FLNa. By inducing FLNa degradation by the proteasome, ASB2α may regulate integrin-dependent functions and thus hematopoietic stem cell fate within the niche.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health (Grant GM068600 to D. A. C.), and by grants from the Centre National de la Recherche Scientifique, the Université Paul Sabatier, the Comité Leucémie de la Fondation de France and the Association pour la Recherche sur le Cancer (to P. G. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
J. H. A. Martens and H. G. Stunnenberg, personal communication.
- AML
- acute myeloid leukemia
- ANK
- ankyrin repeats
- EloB
- Elongin B
- EloC
- Elongin C
- FLN
- filamin
- PML-RARα
- promyelocytic leukemia retinoic acid receptor α.
REFERENCES
- 1. Li Z., Li L. (2006) Trends Biochem. Sci. 31, 589–595 [DOI] [PubMed] [Google Scholar]
- 2. Kiel M. J., Morrison S. J. (2008) Nat. Rev. Immunol. 8, 290–301 [DOI] [PubMed] [Google Scholar]
- 3. Walkley C. R., Olsen G. H., Dworkin S., Fabb S. A., Swann J., McArthur G. A., Westmoreland S. V., Chambon P., Scadden D. T., Purton L. E. (2007) Cell 129, 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walkley C. R., Shea J. M., Sims N. A., Purton L. E., Orkin S. H. (2007) Cell 129, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raaijmakers M. H., Mukherjee S., Guo S., Zhang S., Kobayashi T., Schoonmaker J. A., Ebert B. L., Al-Shahrour F., Hasserjian R. P., Scadden E. O., Aung Z., Matza M., Merkenschlager M., Lin C., Rommens J. M., Scadden D. T. (2010) Nature 464, 852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Teixidó J., Hemler M. E., Greenberger J. S., Anklesaria P. (1992) J. Clin. Invest. 90, 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirsch E., Iglesias A., Potocnik A. J., Hartmann U., Fässler R. (1996) Nature 380, 171–175 [DOI] [PubMed] [Google Scholar]
- 8. Potocnik A. J., Brakebusch C., Fässler R. (2000) Immunity 12, 653–663 [DOI] [PubMed] [Google Scholar]
- 9. Fernandez-Vidal A., Ysebaert L., Didier C., Betous R., De Toni F., Prade-Houdellier N., Demur C., Contour-Galcéra M. O., Prévost G. P., Ducommun B., Payrastre B., Racaud-Sultan C., Manenti S. (2006) Cancer Res. 66, 7128–7135 [DOI] [PubMed] [Google Scholar]
- 10. Matsunaga T., Takemoto N., Sato T., Takimoto R., Tanaka I., Fujimi A., Akiyama T., Kuroda H., Kawano Y., Kobune M., Kato J., Hirayama Y., Sakamaki S., Kohda K., Miyake K., Niitsu Y. (2003) Nat. Med. 9, 1158–1165 [DOI] [PubMed] [Google Scholar]
- 11. Becker P. S., Kopecky K. J., Wilks A. N., Chien S., Harlan J. M., Willman C. L., Petersdorf S. H., Stirewalt D. L., Papayannopoulou T., Appelbaum F. R. (2009) Blood 113, 866–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsunaga T., Fukai F., Miura S., Nakane Y., Owaki T., Kodama H., Tanaka M., Nagaya T., Takimoto R., Takayama T., Niitsu Y. (2008) Leukemia 22, 353–360 [DOI] [PubMed] [Google Scholar]
- 13. Moog-Lutz C., Cavé-Riant F., Guibal F. C., Breau M. A., Di Gioia Y., Couraud P. O., Cayre Y. E., Bourdoulous S., Lutz P. G. (2003) Blood 102, 3371–3378 [DOI] [PubMed] [Google Scholar]
- 14. Guibal F. C., Moog-Lutz C., Smolewski P., Di Gioia Y., Darzynkiewicz Z., Lutz P. G., Cayre Y. E. (2002) J. Biol. Chem. 277, 218–224 [DOI] [PubMed] [Google Scholar]
- 15. Brown D., Kogan S., Lagasse E., Weissman I., Alcalay M., Pelicci P. G., Atwater S., Bishop J. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2551–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grisolano J. L., Wesselschmidt R. L., Pelicci P. G., Ley T. J. (1997) Blood 89, 376–387 [PubMed] [Google Scholar]
- 17. He L. Z., Tribioli C., Rivi R., Peruzzi D., Pelicci P. G., Soares V., Cattoretti G., Pandolfi P. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guibal F. C., Alberich-Jorda M., Hirai H., Ebralidze A., Levantini E., Di Ruscio A., Zhang P., Santana-Lemos B. A., Neuberg D., Wagers A. J., Rego E. M., Tenen D. G. (2009) Blood 114, 5415–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martens J. H., Brinkman A. B., Simmer F., Francoijs K. J., Nebbioso A., Ferrara F., Altucci L., Stunnenberg H. G. (2010) Cancer Cell 17, 173–185 [DOI] [PubMed] [Google Scholar]
- 20. Nie L., Zhao Y., Wu W., Yang Y. Z., Wang H. C., Sun X. H. (2011) Cell Res. 21, 754–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heuzé M. L., Guibal F. C., Banks C. A., Conaway J. W., Conaway R. C., Cayre Y. E., Benecke A., Lutz P. G. (2005) J. Biol. Chem. 280, 5468–5474 [DOI] [PubMed] [Google Scholar]
- 22. Bello N. F., Lamsoul I., Heuzé M. L., Métais A., Moreaux G., Calderwood D. A., Duprez D., Moog-Lutz C., Lutz P. G. (2009) Cell Death Differ. 16, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hilton D. J., Richardson R. T., Alexander W. S., Viney E. M., Willson T. A., Sprigg N. S., Starr R., Nicholson S. E., Metcalf D., Nicola N. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heuzé M. L., Lamsoul I., Baldassarre M., Lad Y., Lévêque S., Razinia Z., Moog-Lutz C., Calderwood D. A., Lutz P. G. (2008) Blood 112, 5130–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burande C. F., Heuzé M. L., Lamsoul I., Monsarrat B., Uttenweiler-Joseph S., Lutz P. G. (2009) Mol. Cell Proteomics 8, 1719–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., Shapiro S. S. (2001) Nat. Rev. Mol. Cell Biol. 2, 138–145 [DOI] [PubMed] [Google Scholar]
- 27. Popowicz G. M., Schleicher M., Noegel A. A., Holak T. A. (2006) Trends Biochem. Sci. 31, 411–419 [DOI] [PubMed] [Google Scholar]
- 28. Calderwood D. A., Shattil S. J., Ginsberg M. H. (2000) J. Biol. Chem. 275, 22607–22610 [DOI] [PubMed] [Google Scholar]
- 29. Baldassarre M., Razinia Z., Burande C. F., Lamsoul I., Lutz P. G., Calderwood D. A. (2009) PLoS One 4, e7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van der Flier A., Sonnenberg A. (2001) Biochim. Biophys. Acta 1538, 99–117 [DOI] [PubMed] [Google Scholar]
- 31. Lad Y., Jiang P., Ruskamo S., Harburger D. S., Ylänne J., Campbell I. D., Calderwood D. A. (2008) J. Biol. Chem. 283, 35154–35163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kiema T., Lad Y., Jiang P., Oxley C. L., Baldassarre M., Wegener K. L., Campbell I. D., Ylänne J., Calderwood D. A. (2006) Mol. Cell 21, 337–347 [DOI] [PubMed] [Google Scholar]
- 33. Nakamura F., Pudas R., Heikkinen O., Permi P., Kilpeläinen I., Munday A. D., Hartwig J. H., Stossel T. P., Ylänne J. (2006) Blood 107, 1925–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ithychanda S. S., Hsu D., Li H., Yan L., Liu D. D., Liu D., Das M., Plow E. F., Qin J. (2009) J. Biol. Chem. 284, 35113–35121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Denis F. M., Benecke A., Di Gioia Y., Touw I. P., Cayre Y. E., Lutz P. G. (2005) J. Biol. Chem. 280, 9043–9048 [DOI] [PubMed] [Google Scholar]
- 36. Takala H., Nurminen E., Nurmi S. M., Aatonen M., Strandin T., Takatalo M., Kiema T., Gahmberg C. G., Ylänne J., Fagerholm S. C. (2008) Blood 112, 1853–1862 [DOI] [PubMed] [Google Scholar]
- 37. Bendall L. J., Makrynikola V., Hutchinson A., Bianchi A. C., Bradstock K. F., Gottlieb D. J. (1998) Leukemia 12, 1375–1382 [DOI] [PubMed] [Google Scholar]
- 38. Harburger D. S., Calderwood D. A. (2009) J. Cell Sci. 122, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Legate K. R., Fässler R. (2009) J. Cell Sci. 122, 187–198 [DOI] [PubMed] [Google Scholar]
- 40. Reavie L., Della Gatta G., Crusio K., Aranda-Orgilles B., Buckley S. M., Thompson B., Lee E., Gao J., Bredemeyer A. L., Helmink B. A., Zavadil J., Sleckman B. P., Palomero T., Ferrando A., Aifantis I. (2010) Nat. Immunol. 11, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tu Y., Wu S., Shi X., Chen K., Wu C. (2003) Cell 113, 37–47 [DOI] [PubMed] [Google Scholar]
- 42. Ithychanda S. S., Das M., Ma Y. Q., Ding K., Wang X., Gupta S., Wu C., Plow E. F., Qin J. (2009) J. Biol. Chem. 284, 4713–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.