Abstract
APOBEC3G (A3G) is a cytidine deaminase that catalyzes deamination of deoxycytidine (dC) on single-stranded DNA (ssDNA). The oligomeric state of A3G required to support deaminase activity remains unknown. We show under defined in vitro conditions that full-length and native A3G formed complexes with ssDNA in an A3G concentration-dependent but temperature-independent manner. Complexes assembled and maintained at 4 °C did not have significant deaminase activity, but their enzymatic function could be restored by subsequent incubation at 37 °C. This approach enabled complexes of a defined size range to be isolated and subsequently evaluated for their contribution to enzymatic activity. The composition of A3G bound to ssDNA was determined by protein-protein chemical cross-linking. A3G-ssDNA complexes of 16 S were necessary for deaminase activity and consisted of cross-linked A3G homotetramers and homodimers. At lower concentrations, A3G only formed 5.8 S homodimers on ssDNA with low deaminase activity. Monomeric A3G was not identified in 5.8 S or 16 S complexes. We propose that deaminase-dependent antiviral activity of A3G in vivo may require a critical concentration of A3G in viral particles that will promote oligomerization on ssDNA during reverse transcription.
Keywords: DNA-binding Protein, DNA Enzymes, HIV, Host Defense, Protein DNA-Interaction, APOBEC, Deaminase, Protein-Protein Interaction
Introduction
APOBEC3G is a cytidine deaminase whose expression has been implicated in host defense and conditionally in viral resistance (1). The APOBEC4 protein family includes APOBEC1 (after which the family is named), activation-induced deaminase, APOBEC2, APOBEC3A-H, and APOBEC4 (2–5). The catalytic activity of these enzymes involves hydrolytic deamination of cytidine to form uridine or deoxycytidine to form deoxyuridine in single-stranded RNA or single-stranded DNA. There is considerable interest in this class of proteins because they are capable of mutagenic activity and epigenetic regulation of protein variant expression that can influence tissue functions and disease progression (4, 6).
APOBEC proteins are characterized by a helix-strand-helix supersecondary structure containing conserved residues (His/Cys)-Xaa-Glu-Xaa25–30Pro-Cys-Xaa-Xaa-Cys that are integral to the function of the zinc-dependent deaminase domain or ZDD (5). Several of the APOBEC proteins, such as APOBEC1 and activation-induced deaminase, contain a single ZDD that was responsible for catalytic activity. Other family members, such as APOBEC3B and APOBEC3G, contain multiple ZDDs within a single molecule where both domains or a single domain were catalytically active (2, 3).
The N-terminal and C-terminal ZDD in A3G were required for nucleic acid-dependent higher-order oligomerization as well as many other interactions necessary for antiviral activity and wild type levels of deaminase activity (7–11). In isolation, the N-terminal ZDD could not support RNA or ssDNA binding (12), but the C-terminal half of A3G supported a modest level of deaminase activity (12–16).
The crystal structure of full-length APOBEC2 and lower resolution small angle x-ray scattering structure of native, full-length A3G revealed that APOBEC proteins formed elongated, end-to-end homomultimers with extensive surface exposure to solvent (17, 18). A2 formed a homodimer through protein-protein interactions within the N terminus, and an elongated homotetramer was suggested by crystal contacts involving the C termini (17). An elongated homodimer of A3G involving a dimer interface in the C termini was predicted by small angle x-ray scattering and corroborated through immunoprecipitation studies of A3G deletion constructs (18, 19). Analytical ultracentrifugation of native, full-length A3G also showed that in the absence of nucleic acids, A3G was predominantly a homodimer (20). At low A3G concentrations, less than 5% of the protein was monomeric. At high A3G concentrations, homodimers of A3G predominated with a small proportion of homotetramers. The most recent crystal structure of only the C-terminal half of A3G also suggested subunit contacts and the potential for interactions between ZDDs within adjacent subunits (13).
In the presence of ssDNA or RNA, a number of experimental approaches including enzyme turnover studies (21), gel shift analysis (22), and atomic force microscopy (7, 11) showed that A3G subunits formed heterogeneous homomultimeric complexes. However, these studies could not rule out a functional role for A3G monomers in nucleic acid binding. Consequently, current models incorporate the possibility that monomers may be integral to translocation of A3G along the DNA strand or in mediating transfer of A3G between DNA strands (7, 11, 15, 16, 21, 23).
In support of a catalytically active monomer, A3G N-terminal point mutants that impaired homomultimerization were shown to retain ssDNA binding and deoxycytidine deaminase activity (7, 11). Although these data affirmed the assertion that homomultimerization of A3G was not necessary for ssDNA binding and deaminase activity, what remains is that native A3G forms stable dimers (20) and that monomers are rare biological isolates.
The interpretation of prior studies has been limited by the fact that defined size classes of A3G-ssDNA complexes were identified but not individually analyzed for deaminase activity. In this study, we used a novel approach that has enabled the isolation and enzymatic analysis of A3G-ssDNA complexes of defined sizes and A3G compositions. We show that full-length, native A3G binds to ssDNA as a homodimer to form 5.8 S complexes. These complexes had low or no ability to deaminate ssDNA. Deaminase activity required the formation of 16 S complexes that consisted of A3G homotetramers and homodimers. Given that A3G homodimers alone were catalytically inactive, our data suggested that catalytically active holoenzyme complexes are higher-order oligomers assembled from A3G homodimers.
EXPERIMENTAL PROCEDURES
Substrates and Primer
Binding and deaminase assays used either a 41-nt ssDNA substrate 5′-ATTATTATTATTATTATTATTCCCAAGGATTTATTTATTTA-3′ (Sigma-Aldrich) or a 30-nt ssDNA substrate 5′-TATTATTATTCCCAAGGATTTATTTATTTA-3′ (Integrated DNA Technologies) with a 5′ Uni-LinkTM amino modifier for labeling with Alexa Fluor® 647 and primer for poisoned primer extension (5′-TAAATAAATAAATCC-3′) (Sigma-Aldrich).
5′ Radiolabeling of 41-nt Substrate and Primer
ssDNA substrate or primer (250 pmol of either) were 5′ end-radiolabeled with [γ-32P]ATP (6,000 Ci/mmol) using T4 polynucleotide kinase (Roche Applied Science) and purified using a 15% denaturing PAGE.
5′ Alexa Fluor 647 Labeling of DNA Substrates
The 30-nt substrate was incubated for 12 h at room temperature with Alexa Fluor 647 succinimidyl ester in 0.1 m sodium tetraborate buffer, pH 8.5, and purified using a 15% denaturing PAGE.
EMSA with Labeled DNA Substrates
The radio- or Alexa Fluor-labeled substrates were incubated at varying molar ratios of A3G to ssDNA in deaminase buffer (40 mm Tris, pH 7.2, 50 mm NaCl, 10 mm MgCl2, 1 mm DTT, 0.1% Triton X-100, 2% glycerol) for the indicated time at 37 °C. The resulting complexes were resolved on a 5% native gel and visualized using a TyphoonTM PhosphorImager by excitation at 633 nm and measuring fluorescence at 670 nm. Radiolabeled samples were exposed to PhosphorImager screens and quantified.
Deaminase Assay
The ssDNA substrate at a concentration of 0.06 μm was incubated with or without purified 0.1–1.75 μm A3G for 60 min in deaminase buffer at the indicated temperature. Deaminase activity on the ssDNA substrate was detected by a poisoned primer extension assay described previously (14) and quantified by PhosphorImager scanning densitometry (supplemental Experimental Procedures). The percentage of deamination was calculated by visualizing and quantifying the primer extension products by PhosphorImager densitometry, and percentages were calculated by dividing the volume of the deaminated substrate (dU) by the total of deaminated and unmodified substrates (dU+dC) times 100.
Glycerol Gradient Velocity Sedimentation Analysis
Complexes formed when A3G bound to ssDNA were assembled at concentrations of 0.26 (A3G) and 1.8 μm (ssDNA) in deaminase buffer at 4 °C. Reactions were applied to 11-ml 10–50% glycerol gradients in deaminase buffer and centrifuged at 200,000 × g for 10 h at 4 °C (supplemental Experimental Procedures).
Three-dimensional Native Gel/Denaturing Gel/Western Blotting
First dimension native gels were run as described above using the 5′ end-labeled substrates. The indicated lanes were cut out of the gel and incubated with 30 mm dimethyl 3,3′-dithiobispropionimidate (DTBP) in 0.2 m triethanolamine or buffer alone for 30 min (30). The first dimension gels were treated with SDS-PAGE buffer minus reducing agent (10 min). layered horizontally onto the stacking gel of a 10.5% denaturing SDS-PAGE, and electrophoresed. A3G, polymerized in a native gel fragment, was treated with SDS-PAGE buffer and run adjacent to the native PAGE as a standard for A3G migration in the second dimension along with molecular mass standards. Products were detected by Western blotting using anti-A3G polyclonal antibody 9906 (AIDS Research and Reference Reagent Program). The molecular masses of the cross-linked complexes were calculated based on the migration of A3G as compared with that of molecular weight standards run on each gel.
RESULTS
A3G Assembly and ssDNA Deaminase Activity
To characterize A3G interactions with nucleic acid, we first evaluated A3G complex formation relative to deaminase activity. Consistent with earlier studies (11, 23), A3G bound to ssDNA very rapidly when incubated at an A3G-ssDNA molar ratio of either 1:4 or 4:1 (supplemental Fig. 1, A and B). These ratios were chosen to drive the formation of specific A3G-ssDNA complexes referred to henceforth as C1 and C3 (Fig. 1). The electrophoretic mobility of complexes and the amount of substrate shifted into complexes were a function of the amount of A3G in the reaction relative to ssDNA. Once complexes formed, their abundance remained relatively constant over the course of the incubation (supplemental Fig. 2, A and B). Despite the rapid binding of A3G to ssDNA, the amount of ssDNA deamination increased with longer incubation (supplemental Fig. 1, C and D). The maximum amount of substrate deaminated in the time course of the assay was dependent on the A3G:ssDNA molar ratio in the reaction. This correlation suggested that reaction conditions that favor assembly of complexes with lower electrophoretic mobility such as C3 are better able to carry out deamination.
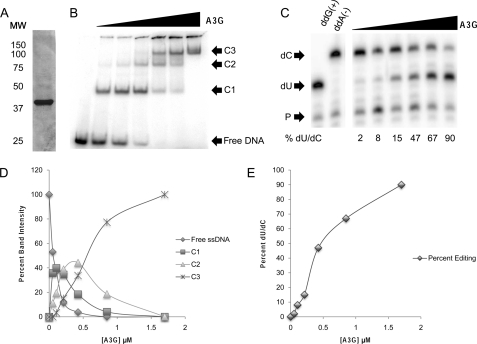
FIGURE 1.
A3G assembles multiple complexes on ssDNA and deaminates deoxycytidine. A, Coomassie Brilliant Blue stain of 5 μg of Sf9 cell-derived purified full-length human A3G. MW, molecular weight markers. B, EMSA using 5′-[γ-32P]ATP 5′ end-labeled 41-nt substrate ssDNA. The concentration of ssDNA in each reaction was 0.6 μm, whereas the concentration of A3G in each lane was 0, 0.05, 0.11, 0.22, 0.44, 0.86, and 1.75 μm (lanes 1–7, respectively). C1, C2, and C3 refer to A3G-ssDNA complexes 1, 2, and 3, respectively. Complexes were visualized by PhosphorImager scanning densitometry by virtue of their labeled ssDNA content. The gel shown is representative of four independent experiments. C, deaminase activity was assayed by primer extension on 41-nt ssDNA that was incubated with A3G. The concentration of ssDNA was 0.6 μm per reaction, and the concentration of A3G in each lane was 0.05, 0.11, 0.22, 0.44, 0.86, and 1.75 μm (lanes 1–6, respectively). The lowest arrow indicates free primer (P), the middle arrow indicates the primer extension product for deaminated ssDNA (dU), and the upper arrow indicates the primer extension product for unmodified ssDNA (dC). The percentage of deaminated ssDNA (%dU/dC) due to each reaction condition was determined by PhosphorImager scanning densitometry and calculated as dU divided by (dU+dC) times 100. The gel shown is representative of four independent experiments. D, graphic representation of the quantification of individual bands in B relative to input A3G in each reaction. E, graphic representation of deaminase activity in C demonstrating the positive correlation of deaminase activity with A3G concentration in the reaction and the appearance of complex C3 (as shown in B).
To examine this correlation, increasing amounts of purified A3G (Fig. 1A) were incubated with 5′ end-labeled ssDNA, and the complexes formed were resolved by EMSA and visualized via their content of labeled ssDNA (Fig. 1B). With increasing A3G input, more ssDNA was shifted into complexes, and eventually, free ssDNA was consumed in the binding reaction (fifth lane). Concomitantly, the faster migrating complexes that formed at lower A3G:ssDNA molar ratios (C1) became less apparent as slower migrating complexes (C2 and C3) increased in abundance. The data suggested that C1 complexes were fully shifted into C3 complexes in an A3G concentration-dependent manner. Quantification of ssDNA recovered within each EMSA band relative to A3G input corroborated this relationship (Fig. 1D).
We next evaluated the relationship between higher-order complex formation and deaminase activity. Reactions set up in parallel with those in Fig. 1B were assayed for A3G deaminase activity on the single “hot spot” deaminase site in each substrate using poisoned primer extension as described under “Experimental Procedures.” The data demonstrated low to background levels of deaminase activity in reactions containing A3G input only sufficient to assemble C1 complexes (Fig. 1C). Deaminase activity increased with additional input of A3G in parallel with the shift of C1 complexes into C2 and C3 complexes (Fig. 1D). Maximal deaminase activity was directly proportional to the formation of C3 complexes (Fig. 1E).
Glycerol Gradient Velocity Sedimentation Analysis
We next size-fractionated A3G-ssDNA complexes to evaluate the dependence of deaminase activity on the oligomeric state of A3G. The presence of sharply resolved EMSA bands in the foregoing experiment suggested that a finite number of A3G molecules assembled on ssDNA to form each complex. In the course of pursuing these experiments, we discovered that complex assembly proceeded in an A3G-dependent manner at both 4 °C and 37 °C, but deaminase activity was greatly reduced at 4 °C as compared with 37 °C (supplemental Fig. 2). This finding suggested an experimental approach by which A3G-ssDNA complexes of different size classes could be assembled at 4 °C, size-fractionated, and then evaluated for the ability of an individual size class of complexes to support deaminase activity at 37 °C using the substrates already contained in each complex.
We began our studies using glycerol gradient velocity sedimentation as we have used this method to separate size classes of APOBEC1-containing complexes for functional analyses (24). A3G was assembled with Alexa Fluor647 5′ end-labeled ssDNA at 4 °C at a molar ratio of ∼1:6 (sufficient to induce assembly of predominantly but not exclusively C1 complexes) (Fig. 2A). As anticipated, assembly reactions maintained at 4 °C did not efficiently deaminate the ssDNA substrate they bound to until they were incubated at 37 °C (Fig. 2B).
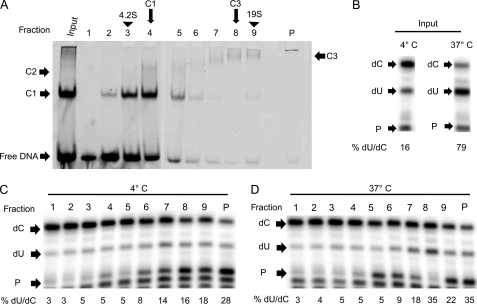
FIGURE 2.
Glycerol Gradient velocity sedimentation analysis of A3G-ssDNA complexes. A, A3G-ssDNA complexes were assembled using 0.26 μm A3G and 1.8 μm Alexa Fluor 647-labeled ssDNA at 4 °C and sedimented through a 10–50% glycerol gradient at 200,000 × g for 10 h at 4 °C. Gradients were calibrated using protein standards with known sedimentation values (supplemental Fig. 4). Fractions (fractions 1–9 and the pellet, P) were collected from the top of the gradients, and 10-μl aliquots were analyzed directly by EMSA for the complexes that had been assembled in the reaction that was loaded. Complexes were visualized by PhosphorImager scanning densitometry by virtue of their labeled ssDNA content. B, as controls for the effect of temperature on deaminase activity, separate reactions were incubated at either 4 °C or 37 °C for 2 h and assayed for deaminase activity. The lowest arrow indicates free primer (P), the middle arrow indicates the primer extension product for deaminated ssDNA (dU), and the upper arrow indicates the primer extension product for unmodified ssDNA (dC). The percentage of deaminated ssDNA (%dU/dC) due to each reaction condition was determined by PhosphorImager scanning densitometry and calculated as dU divided by (dU+dC) times 100. C and D, aliquots (100 μl) of each fraction were incubated at 4 °C for 2 h (C) or incubated at 37 °C for 2 h (D) and assayed for deaminase activity by primer extension assays. All panels shown are representative of three independent experiments.
A parallel reaction maintained continuously at 4 °C was sedimented through a 10–50% glycerol gradient, and after centrifugation, fractions were collected and aliquots were resolved directly by EMSA to visualize the complexes present in each gradient fraction as detected by the content of fluorescently labeled ssDNA in each complex. Free ssDNA in the reaction predominantly sedimented at the top of the gradient. Based on sedimentation of proteins with known S values in a parallel gradient (supplemental Fig. 3), the sedimentation values of C1 and C3 complexes were estimated to be 5.8 S and 16 S, respectively (Fig. 2A, 4th and 8th fractions, respectively).
Gradient fractions incubated for an additional 2 h at 4 °C did not demonstrate induction of deaminase activity, although the highest levels of deaminated substrate were observed in fractions 7, 8, and 9 and the pellet (Fig. 2C). In contrast, incubation of the gradient fractions for 2 h at 37 °C stimulated deaminase activity nonuniformly across the gradient fractions (Fig. 2D). A peak of activity was evident in fraction 8 corresponding to the sedimentation of the 16 S C3 complexes as well as higher molecular mass aggregates that accumulated in the gradient pellet. Despite the abundance of A3G in C1 complexes in fractions 3 and 4, elevated temperatures could not induce deaminase activity. The data suggested that although A3G could bind to ssDNA to form the 5.8 S C1 complexes, binding to ssDNA alone was not sufficient for deaminase activity. Given that additional A3G was required to produce the 16 S C3 complexes, the data suggested that higher-order A3G oligomers were required for deaminase activity.
The role of 16 S complexes as the oligomeric state necessary for deaminase activity was further supported by the finding that C3 complexes that were assembled and isolated at 4 °C appeared to be relatively stable following incubation at 37 °C for 2 h, with very little dissociation to C1 complexes (supplemental Fig. 4). Similarly, very little dissociation of free ssDNA from C1 complexes or on the other hand aggregation of C1 complexes to form C3 complexes occurred during the 2-h incubation at 37 °C.
Evaluation of A3G Composition of 5.8 S and 16 S Complexes
Previous analytical ultracentrifugation showed that nucleic acid-depleted A3G protein-protein dimer had 5.6 S (20). Given that the ssDNA molecular mass was 9.2 kDa, the C1 complex was likely to be a dimer of A3G plus one or two molecules of ssDNA. Nucleic acid-depleted homotetramers of A3G were shown to have 8.2 S (20), suggesting that the 16 S C3 complex in the present study was a higher-order oligomer of A3G and ssDNA.
To address these possibilities, a three-dimensional system was designed to evaluate the number of A3G subunits in the C1 and C3 complexes. A3G-ssDNA complexes were separated by EMSA and imaged, and the entire gel lane was excised and incubated with DTBP. This cross-linking reagent stabilized interacting A3G subunits through a 12 Å long linker that coupled appropriately spaced free amines on the subunit surfaces through imidoester reactive groups on either end of the DTBP (25). The gel lane was subsequently equilibrated with SDS-PAGE sample buffer to quench the cross-linking reagent and denature A3G with SDS, thereby liberating ssDNA from A3G. The treated gel was layered onto, and electrophoresed into, a 10.5% denaturing SDS-PAGE gel and Western blotted with A3G specific antibodies. Uncross-linked A3G that had been directly polymerized in a native gel and molecular mass standard proteins were run into the second dimension gel alongside the EMSA gel to calibrate the second dimension denaturing SDS-PAGE. The two-dimensional gel was transferred to nitrocellulose membrane and blotted with A3G-specific polyclonal antibody as the third dimension of analysis. The A3G used in this study has a His4 tag and was predicted to have a molecular mass of 46 kDa. However, under denaturing SDS-PAGE conditions, it migrated with an apparent molecular mass of 38 kDa (± 1.0 kDa S.E., n = 4) (Fig. 1A).
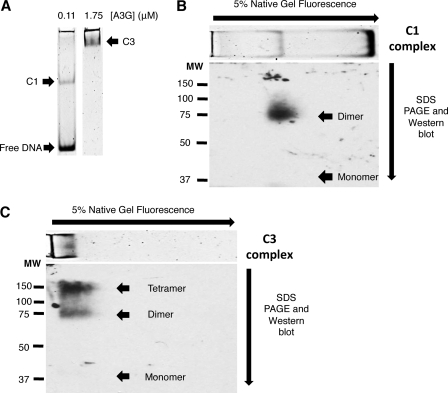
EMSA native gel lanes corresponding to A3G:ssDNA input ratios that yielded predominantly C1 complexes or C3 complexes (Fig. 3A) were excised, cross-linked in the gel, and processed through to the three-dimensional analysis. Western blotting revealed that cross-linked A3G migrated in the second dimension gel according to the composite molecular mass of A3G subunits in each complex. Cross-linked C1 complexes migrated under denaturing conditions with an apparent molecular mass of 75 kDa (± 5 kDa S.E., n = 4) (Fig. 3B). No A3G was evident at the molecular mass of the uncross-linked monomer or higher molecular mass multimers. Without cross-linking the first dimension EMSA, A3G in C1 complexes migrated as denatured monomers (supplemental Fig. 5). Densitometric scanning with a PhosphorImager revealed that fluorescent ssDNA was no longer detectable in the cross-linked and denatured 75-kDa complex (data not shown). This suggested that the 5.8 S glycerol gradient fraction and the C1 EMSA complex was composed of A3G homodimers bound to ssDNA.
FIGURE 3.
Protein subunit composition of A3G-ssDNA complexes. A, EMSA showing complexes that resulted from assembly reactions containing 0.6 μm Alexa Fluor 647-labeled 30-nt substrate ssDNA and either 0.11 μm (left) or 1.75 μm (right) A3G. These EMSA gel lanes were excised in their entirety and incubated with DTBP. EMSA complexes were visualized by PhosphorImager scanning densitometry by virtue of their labeled ssDNA content prior to cross-linking. Cross-linked complexes were resolved in the second dimension by denaturing SDS-PAGE and Western blotted with A3G-specific antibody to reveal A3G and its composite molecular mass in cross-linked complexes. The image of the EMSA, based on fluorescently tagged ssDNA, has been graphically inserted at the top of the two-dimensional gel Western image for reference and alignment of the EMSA with A3G detected by Western blotting. The arrows on the top and side of the image show the directions of electrophoresis of the EMSA and SDS-PAGE, respectively. C1 and C3 refer to A3G-ssDNA complexes 1 and 3, respectively. B, Western blot of two-dimensional native/denaturing gel of cross-linked C1. MW, molecular weight markers. C, Western blot of two-dimensional native/denaturing gel of cross-linked C3. Arrowheads in B and C point to the migration of monomeric A3G or cross-linked multimers of A3G. All panels shown are representative of four independent experiments. The electrophoretic migration in the second dimension denaturing PAGE was calibrated using purified A3G and molecular mass standard proteins run on the same gel.
The cross-linked C3 complexes migrated with an apparent molecular mass of 150 kDa (± 3.5 kDa S.E., n = 4) as well as some 75-kDa complexes (Fig. 3C). Densitometric scanning with a PhosphorImager revealed that fluorescent ssDNA was no longer detectable in the cross-linked and denatured 150-kDa complexes (data not shown). These findings suggested that the C3 complexes were composed of A3G homotetramers and homodimers. The recovery of A3G monomers from C3 complexes was below detection limits of our assay. We cannot rule out the possibility that 16 S C3 complexes may have consisted of higher-order A3G oligomers (such as homohexamers or homo-octamers) that may not have been detected because they did not cross-link efficiently, did not enter the second dimension gel, or did not transfer efficiently to nitrocellulose.
DISCUSSION
Using a defined in vitro system, we have shown that full-length and native A3G binds ssDNA to form a 5.8 S homodimer and that further oligomerization of A3G to 16 S homomultimers was necessary for efficient deoxycytidine deamination on ssDNA. In addition, we also showed that homodimers of A3G bound to ssDNA were unable to support efficient deaminase activity. We found no evidence that native A3G bound to and deaminated ssDNA as a monomer, nor were free monomers recovered after cross-linking higher-order complexes. Our data showed that the assembly of catalytically active complexes depends on the formation of tetramers and higher-order homo-oligomers on ssDNA substrates.
The role of A3G oligomerization in deaminase activity has been controversial. Prior studies using N-terminal point mutations (C100S, F126A, W127A) have shown that these A3G molecules purify as monomeric species yet were capable of supporting deaminase activity (11, 26). NMR chemical shift analysis of an extensively mutated A3G C-terminal half-molecule bound to ssDNA (15, 16) and a crystal structure of the same A3G half-molecule (27) suggested that the C-terminal ZDD bound ssDNA as a monomer. Subsequent crystallographic analysis of a slightly longer N-terminal extension of the C-terminal half of A3G predicted oligomerization of A3G (13).
Atomic force microscopy of native and mutant A3G also suggested that the oligomeric state of A3G was critical to deaminase activity (7, 11). In these studies, deaminase activity was ascribed to the predominant oligomeric state of A3G, although there was significant size heterogeneity in A3G complexes. A recent high resolution atomic force microscopy re-evaluation of A3G oligomerization concluded that A3G dimers bound to ssDNA; however, the authors were unable to rule out that monomers bound ssDNA first and then became dimers (28). A potential limitation in these studies may have been that the number of A3G subunits in an A3G-ssDNA complex was assumed to remain uniform and invariant in molecular dimensions (solid spheres) following multimerization (i.e. the dimer will be twice the size of a monomer). However, small angle x-ray scattering suggested that this may not be a valid assumption, and in particular, the A3G tetramer is predicted to have less than twice the molecular dimensions of an A3G dimer (e.g. 210 Å versus 140 Å in the long axis, respectively) (18).
In the current study, we would have expected to recover monomeric A3G following cross-linking if monomers alone were in equilibrium with dimers bound to ssDNA. EMSA suggested that complexes migrating faster than the C1 complexes (i.e. an A3G monomer bound to ssDNA) were not detectable. Analytical ultracentrifugation of our A3G preparation also suggested that monomers made up a very low percentage of the total A3G. Taken together with small angle x-ray scattering and analytical ultracentrifugation findings (18–20), we proposed that it is unlikely that A3G monomers persist long enough in cells to be biologically relevant in binding to ssDNA. Our data do not refute the possibility that A3G monomers can bind ssDNA, particularly if, as described above, A3G monomers were produced following extensive mutation with the goal of impairing oligomerization. We also cannot rule out the possibility that rapid assembly of monomers on ssDNA followed by virtually instantaneous dimer formation may have precluded our ability to identify monomeric A3G bound to ssDNA in our cross-linking studies.
There have been numerous studies of higher-order A3G oligomeric species formed through binding to nucleic acids (ssDNA and RNA) (7, 10, 11, 22, 29, 30). The oligomeric state has been proposed to be necessary for the 3′ to 5′ processivity of deaminase activity seen on synthetic ssDNA substrates in vitro (23) and HIV reverse transcripts (31) as well as interstrand transfer of A3G (21). Moreover the ability of A3G deaminase activity to “jump” to single-stranded regions interspersed by double-stranded regions on test substrates has been attributed to A3G oligomers (7).
Although we cannot rule out that at higher A3G concentrations, tetramers formed in solution and then bound to ssDNA, the A3G concentration-dependent assembly of C1, C2, and C3 complexes suggested that A3G tetramers (and higher-order oligomers) were assembled from A3G homodimers bound to ssDNA. In this regard, unlike native A3G homodimers whose assembly is not dependent on nucleic acid binding (18–20), the formation of tetramers and higher-order complexes of A3G may be enhanced (bridged) by nucleic acids binding. This possibility is supported by analytical ultracentrifugation data showing that nucleic acid-depleted A3G protein concentration had to be high in order for homotetramers to form (20).
It is intriguing to consider that initially only A3G homodimers bind to ssDNA, and once they have established a locally high concentration, A3G homodimers interact to form homotetramers (with other dimers bound to different sequences along the ssDNA strand) by bending or looping DNA. The ZDD within each A3G monomer is predicted to be a catalytic domain; however, our data showed that a monomeric domain in native A3G was not enzymatically competent until homomultimers assembled on ssDNA substrates. In this regard, we cannot rule out that an A3G dimer bound to ssDNA becomes active once it interacts with another dimer or tetramer. The interaction of A3G dimers and their orientation relative to one another in the tetramer may also have a role in determining the 3′ to 5′ processivity of A3G deaminase activity or the ability of A3G to jump over dsDNA to adjacent ssDNA regions or carry out interstrand transfer.
Although our studies suggested that A3G tetramers were components of 16 S complexes, the predicted molecular mass of such complexes could accommodate A3G hexamers and octamers bound to multiple ssDNA molecules. We also do not know the A3G composition in the C2 complexes. Given that we did not detect A3G monomers in our study, C2 complexes are unlikely to consist of A3G homotrimers but rather may have been unstable A3G tetramers in which conformational changes may have affected their electrophoretic mobility. The goal for future experiments is to obtain better resolution of the A3G composition of the C2 complexes and their relationship to C1 and C3 complexes.
We have not directly addressed an important question of whether A3G tetramers and higher-order complexes are required in vivo for host defense deaminase activity during viral replication. Current hypotheses consider A3G entering cells with viral particles as the main source of deaminase activity on nascent proviral ssDNA. It has been estimated that there are 7 (±4) A3G subunits in each HIV viral particle (32). A3G within the viral core is bound to viral RNA, and as such, forms high molecular mass ribonucleoprotein complexes (23, 33–36). It is possible that the concentration of A3G in the vicinity of reverse transcription complexes could be sufficiently high to promote A3G homotetramer formation on nascent single-stranded proviral DNA as it is revealed transiently by RNase H (23, 36). Consequently, although our data show that monomers and dimers of native and full-length A3G alone are not sufficient for deaminase activity, the question of whether A3G tetramers and higher-order complexes are required for in vivo deaminase activity remains to be determined.
The methods developed in this study are likely to have broad application considering that all cytidine deaminases have been shown to function as homo- or heteromeric complexes (4). In particular, these methods may apply to the study of the oligomeric state of the APOBEC homolog activation-induced deaminase whose ssDNA deaminase activity is required for immunoglobulin gene class switch recombination and somatic hypermutation (37). The oligomeric state of activation-induced deaminase has been controversial, and its ability to form hetero-oligomers with replication protein A (38) and HSP90 (39) has been implicated in activation-induced deaminase gene targeting, shuttling, and stability. The oligomeric state of APOBEC3 homologs with single and dual deaminase domains (3–5) that function to restrict viral replication and entry of foreign DNA into cells (4, 6, 40) is also unknown. The methods described here may be generally applicable to other enzymes involved in the modification and editing of nucleotides in RNA and DNA.
Supplementary Material
Acknowledgments
We are grateful to Drs. Robert Bambara, Ryan Bennett, Steve Dewhurst, Kim Prohaska, and Jason Salter and Jenny Smith for critical reading of the manuscript and to Michael Terns (University of Georgia, Athens, GA) for the native gel EMSA protocol. The following polyclonal antibody (9906) was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: anti-APOBEC3G antiserum from Drs. Klaus Strebel and Sandra Kao.
This work was supported by a Bill and Melinda Gates foundation Grand Challenges in Exploration grant awarded (to H. C. S.) and by a National Institutes of Health Developmental Center for AIDS Research grant (NIAID Grant P30 078498) awarded to Steve Dewhurst.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. 1–5.
- APOBEC
- apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like
- A3G
- APOBEC3G
- ZDD
- zinc-dependent deaminase domain
- DTBP
- dimethyl 3,3′-dithiobispropionimidate (DTBP)
- nt
- nucleotide.
REFERENCES
- 1. Smith H. C. (2011) Trends Biochem. Sci. 36, 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002) Genomics 79, 285–296 [DOI] [PubMed] [Google Scholar]
- 3. Wedekind J. E., Dance G. S., Sowden M. P., Smith H. C. (2003) Trends Genet. 19, 207–216 [DOI] [PubMed] [Google Scholar]
- 4. Smith H. (2010) in DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (Grosjean H. ed) pp. 181–202, Landes Bioscience, Austin, TX [Google Scholar]
- 5. MacElrevey C., Wedekind J. E. (2008) in RNA and DNA Editing: Molecular Mechanisms and Their Integration into Biological Systems (Smith C. H. ed) pp. 369–419, John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 6. Albin J. S., Harris R. S. (2010) Expert. Rev. Mol. Med. 12, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng Y., Chelico L. (2011) J. Biol. Chem. 286, 11415–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huthoff H., Malim M. H. (2007) J. Virol. 81, 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. (2009) PLoS Pathog. 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Navarro F., Bollman B., Chen H., König R., Yu Q., Chiles K., Landau N. R. (2005) Virology 333, 374–386 [DOI] [PubMed] [Google Scholar]
- 11. Chelico L., Prochnow C., Erie D. A., Chen X. S., Goodman M. F. (2010) J. Biol. Chem. 285, 16195–16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bennett R. P., Presnyak V., Wedekind J. E., Smith H. C. (2008) J. Biol. Chem. 283, 7320–7327 [DOI] [PubMed] [Google Scholar]
- 13. Shandilya S. M., Nalam M. N., Nalivaika E. A., Gross P. J., Valesano J. C., Shindo K., Li M., Munson M., Royer W. E., Harjes E., Kono T., Matsuo H., Harris R. S., Somasundaran M., Schiffer C. A. (2010) Structure 18, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith H. C. (2007) Methods Enzymol. 424, 389–416 [DOI] [PubMed] [Google Scholar]
- 15. Chen K. M., Martemyanova N., Lu Y., Shindo K., Matsuo H., Harris R. S. (2007) FEBS Lett. 581, 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 17. Prochnow C., Bransteitter R., Klein M. G., Goodman M. F., Chen X. S. (2007) Nature 445, 447–451 [DOI] [PubMed] [Google Scholar]
- 18. Wedekind J. E., Gillilan R., Janda A., Krucinska J., Salter J. D., Bennett R. P., Raina J., Smith H. C. (2006) J. Biol. Chem. 281, 38122–38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bennett R. P., Salter J. D., Liu X., Wedekind J. E., Smith H. C. (2008) J. Biol. Chem. 283, 33329–33336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salter J. D., Krucinska J., Raina J., Smith H. C., Wedekind J. E. (2009) Biochemistry 48, 10685–10687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. (2008) Nat. Struct. Mol. Biol. 15, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 22. Iwatani Y., Takeuchi H., Strebel K., Levin J. G. (2006) J. Virol. 80, 5992–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 24. Sowden M. P., Ballatori N., Jensen K. L., Reed L. H., Smith H. C. (2002) J. Cell Sci. 115, 1027–1039 [DOI] [PubMed] [Google Scholar]
- 25. Harris S. G., Hoch S. O., Smith H. C. (1988) Biochemistry 27, 4595–4600 [DOI] [PubMed] [Google Scholar]
- 26. Opi S., Takeuchi H., Kao S., Khan M. A., Miyagi E., Goila-Gaur R., Iwatani Y., Levin J. G., Strebel K. (2006) J. Virol. 80, 4673–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shlyakhtenko L. S., Lushnikov A. Y., Li M., Lackey L., Harris R. S., Lyubchenko Y. L. (2011) J. Biol. Chem. 286, 3387–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chelico L., Sacho E. J., Erie D. A., Goodman M. F. (2008) J. Biol. Chem. 283, 13780–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiu Y. L., Witkowska H. E., Hall S. C., Santiago M., Soros V. B., Esnault C., Heidmann T., Greene W. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15588–15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442 [DOI] [PubMed] [Google Scholar]
- 32. Xu H., Chertova E., Chen J., Ott D. E., Roser J. D., Hu W. S., Pathak V. K. (2007) Virology 360, 247–256 [DOI] [PubMed] [Google Scholar]
- 33. Burnett A., Spearman P. (2007) J. Virol. 81, 5000–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Friew Y. N., Boyko V., Hu W. S., Pathak V. K. (2009) Retrovirology 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khan M. A., Goila-Gaur R., Opi S., Miyagi E., Takeuchi H., Kao S., Strebel K. (2007) Retrovirology 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soros V. B., Yonemoto W., Greene W. C. (2007) PLoS Pathog. 3, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honjo T., Kinoshita K., Muramatsu M. (2002) Annu. Rev. Immunol. 20, 165–196 [DOI] [PubMed] [Google Scholar]
- 38. Chaudhuri J., Khuong C., Alt F. W. (2004) Nature 430, 992–998 [DOI] [PubMed] [Google Scholar]
- 39. Orthwein A., Patenaude A. M., Affar el B., Lamarre A., Young J. C., Di Noia J. M. (2010) J. Exp. Med. 207, 2751–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.