Abstract
The fibrillar collagen types I, II, and V/XI have recently been shown to have partially 3-hydroxylated proline (3Hyp) residues at sites other than the established primary Pro-986 site in the collagen triple helical domain. These sites showed tissue specificity in degree of hydroxylation and a pattern of D-periodic spacing. This suggested a contributory role in fibril supramolecular assembly. The sites in clade A fibrillar α1(II), α2(V), and α1(I) collagen chains share common features with known prolyl 3-hydroxylase 2 (P3H2) substrate sites in α1(IV) chains implying a role for this enzyme. We pursued this possibility using the Swarm rat chondrosarcoma cell line (RCS-LTC) found to express high levels of P3H2 mRNA. Mass spectrometry determined that all the additional candidate 3Hyp substrate sites in the pN type II collagen made by these cells were highly hydroxylated. In RNA interference experiments, P3H2 protein synthesis was suppressed coordinately with prolyl 3-hydroxylation at Pro-944, Pro-707, and the C-terminal GPP repeat of the pNα1(II) chain, but Pro-986 remained fully hydroxylated. Furthermore, when P3H2 expression was turned off, as seen naturally in cultured SAOS-2 osteosarcoma cells, full 3Hyp occupancy at Pro-986 in α1(I) chains was unaffected, whereas 3-hydroxylation of residue Pro-944 in the α2(V) chain was largely lost, and 3-hydroxylation of Pro-707 in α2(V) and α2(I) chains were sharply reduced. The results imply that P3H2 has preferred substrate sequences among the classes of 3Hyp sites in clade A collagen chains.
Keywords: Bone, Cartilage, Chondrocytes, Collagen, Enzymes, Hydroxylase, Mass Spectrometry (MS), Osteoblasts, Osteosarcoma, Protein Self-assembly
Introduction
Collagen, the major structural protein of vertebrates has evolved a range of post-translational modifications that are essential for triple helix assembly and stability, intermolecular cross-linking, and strength of fibrils and tissue function. Enzymes from the family of 2-oxo-glutarate-dependent dioxygenases are responsible for most of these modifications. For example, prolyl 4-hydroxylase catalyzes proline 4-hydroxylation, a modification necessary for the secondary and tertiary structure of collagen (1, 2), and the lysyl hydroxylase isoenzymes catalyze lysine hydroxylation, a modification essential for the formation of intermolecular cross-links in collagen (2–4).
Prolyl 3-hydroxlase 1 (P3H1),2 another member of the 2-oxo-glutarate-dependent dioxygenase family, catalyzes the post-translational 3-hydroxylation of certain proline residues in fibril-forming collagens. This enzyme was only recently cloned from chicken tissues (5) after being partially purified >25 years ago (6). P3H1 was identified as the chick homologue of rat leprecan, a glycoprotein, which was first isolated from a rat parietal yolk sac tumor cell line (7). Within the past few years, novel mutations in the human P3H1 gene and genes encoding P3H1-associated proteins were shown to cause recessive forms of osteogenesis imperfecta (8–11). By mass spectrometry, we and other investigators have shown that Pro-986 in the α1(I) collagen chain of bone and skin from such patients can be severely under-3-hydroxyated. The P3H1 enzyme therefore is required for the 3-hydroxylation of Pro-986 in the α1(I) chain of type I collagen (8, 10). Although the presence of 3-hydroxyproline (3Hyp) in collagen was discovered >50 years ago (12), the function of this relatively rare but important modification in fibril-forming collagens is unknown. Jenkins et al. (13) observed a small destabilizing effect of 3Hyp on the collagen triple helix formed by synthetic peptides, but more recently, further studies revised this to a slight increase in stability (14).
Genomic database analyses indicate the presence of three isoenzymes in the vertebrate prolyl 3-hydroxylase family, P3H1, P3H2, and P3H3, encoded by the genes LEPRE1, LEPREL1, and LEPREL2, respectively, in humans (15). The presence of conserved catalytic domains in the 2-oxo-glutarate-dependent dioxygenase superfamily (cl01206, NCBI Conserved Domain Database) suggest that P3H2 and P3H3 may well be involved in the 3-hydroxylation of proline residues in additional chains of the collagen family, for example, type IV collagen chains (16). Expression profiles of human and mouse P3H2 by Northern blots and quantitative PCR showed expression in placenta, lung, heart, and kidney (15, 17). Immunoelectron microscopy verified that P3H2 was expressed in mouse kidney tubular cells, the acinar cells of the pancreas, and in the Schawnn cells of nerve (16). Because these tissues were rich in basement membranes containing type IV collagen, further in vitro studies using recombinant P3H2 and synthetic peptides corresponding to sequences in α1(IV) collagen chains showed that these peptides can be 3-hydroxylated more efficiently than synthetic peptides corresponding to the 3Hyp site (Pro-986) in the α1(I) chain of type I collagen, implying that P3H2 could 3-hydroxylate specific proline residues in the α1(IV) collagen chain (16).
P3H2 is co-expressed with P3H1 and P3H3 in various tissues of the developing mouse embryo coincident with regions of fibrillar collagen expression. This is especially noticeable in areas of mesenchymal cartilage condensation, cartilages of the limbs, mandible, developing and adult eye, bone, and skin (17, 18). The widespread expression pattern of P3H isoenzymes in collagen fibril-containing tissues of mouse embryos rather than just basement membranes suggested a more general function in processing fibrillar collagens (18). This premise is supported by our recent findings that the fibrillar collagens I, II, and V/XI have partially modified 3-hydroxyproline residues at sites other than at the primary A1 site (Pro-986) in the collagen triple helical domain (19, 20). Furthermore, these sites are within three residues of the collagen D-period molecular stagger (234 amino acid residues) suggesting a role in collagen fibril assembly. For example, in the evolutionary group of clade A collagen chains (Fig. 1A), which includes α1(I), α2(I), α1(II), and α2(V), we have shown that the A2 site at Pro-944, the A3 site at Pro-707, and the A4 site at Pro-470 are all partially 3-hydroxylated (19). These sites are candidate substrates for P3H2 and P3H3. We pursued this experimentally using a Swarm rat chondrosarcoma cell line (RCS-LTC) known to synthesize and post-translationally cross-link collagen types II, IX, and XI heteropolymeric fibrils in long term monolayer culture (21). Mass spectrometry determined that in contrast to control cartilage type II collagen, all of the candidate 3Hyp sites in the RCS-LTC type II collagen chains were highly hydroxylated, providing a system for studying the effects of manipulating P3H isoenzyme activity on the prolyl 3-hydroxylation pattern. Using RNA interference, we demonstrate that a knockdown of P3H2 protein coordinately reduces 3-hydroxylation at Pro-944, Pro-707, and the GPP repeat at the C terminus of the triple helix but not at Pro-986 of the clade A pNα1(II) collagen chain. We further show that when P3H2 expression is turned off, 3-hydroxylation at residue Pro-944 in the α2(V) chain is nearly completely lost, and 3Hyp occupancy at Pro-707 in α2(V) and α2(I) chains is significantly reduced. The results identify a role for P3H2 in 3-hydroxylation of non-A1 proline residues in clade A collagen chains.
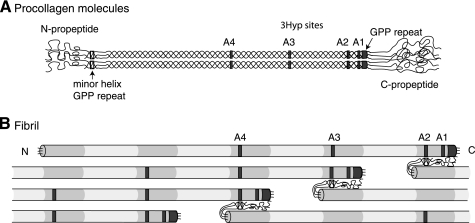
FIGURE 1.
Relative molecular positions of prolyl 3-hydroxylation sites in aligned procollagen molecules and a microfibril. A, occupied 3Hyp sites are indicated by gray boxes within the triple helical regions of the molecule. From right to left, these are the C-terminal (GPP)4 repeat, A1 at Pro-986, A2 at Pro-944, A3 at Pro-707, and A4 at Pro-470. The GPP repeat in the minor triple helix of the N-propeptide is indicated by an open box. B, axial relationships required for trifunctional intermolecular cross-link formation in a pN type II collagen microfibril. Collagen molecules are aligned in the typical collagen D-periodic stagger of 234 amino acid residues. Note the position of the N-propeptide GPP repeat (open box) relative to an A2 site in an adjacent pN type II collagen molecule and alignment of D-staggered A2, A3, and A4 3Hyp sites.
EXPERIMENTAL PROCEDURES
Cell Culture
The RCS-LTC cell line was maintained in monolayer culture in high glucose DMEM (Hyclone), 10% iron-supplemented bovine calf serum (Hyclone), 10 μg/ml l-ascorbate at 37 °C, and 5% CO2 for 4 weeks (22). Some cultures were additionally supplemented with β-aminoproprionitrile (Sigma) to inhibit lysyl oxidase. The SAOS-2 cell line (ATCC catalog no. HTB-85) was maintained as monolayer cultures for a month in McCoy's medium containing 10% FBS (Invitrogen) and 50 μg/ml ascorbate (23, 24). The human chondrosarcoma cell line, CH1.2 (24), human breast cancer cell lines MDA 231 (ATCC catalog no. HTB-26), and MDA 361 (ATCC catalog no. HTB-27) were cultured in high glucose DMEM containing 10% FBS and 50 μg/ml ascorbate.
Collagen Extraction and Purification
The RCS-LTC cell layer was extracted with 1 m NaCl, 50 mm Tris, pH 7.5, containing, 1 mm PMSF, 1 mm benzamidine, and 5 mm EDTA for 20 h at 4 °C, to solubilize newly synthesized, non-cross-linked collagen. After centrifugation for 30 min at 4 °C and 25,000 × g, the supernatant was adjusted to 4.5 m NaCl and stirred at 4 °C for 20 h. Following centrifugation, the pellet was denatured in Laemmli sample buffer, and the collagen chains were run on SDS-PAGE (25). After a month in culture, the SAOS-2 cell layers were extracted with 1 m NaCl, 50 mm Tris, pH 7.5, containing 1 mm PMSF, 1 mm benzamidine, and 5 mm EDTA for 2 days at 4 °C. The NaCl extract and the insoluble residue were separated by centrifugation. The residue was further digested with 0.1 mg/ml pepsin in 0.5 m acetic acid for 24 h at 4 °C to solubilize the cross-linked collagen. Type I and V collagens were precipitated from the pepsin digest at 0.8 m and 2.2 m NaCl, respectively, and harvested by centrifugation.
Gel Electrophoresis and Mass Spectrometry
Collagen chains were resolved by SDS-PAGE gel electrophoresis under reducing conditions and identified by staining with Coomassie Blue. Individual collagen α-chains were cut out and subjected to in-gel trypsin digestion (26). Electrospray MS was performed on the tryptic peptides using an LCQ Deca XP ion trap mass spectrometer equipped with in-line LC (ThermoFinnigan) using a C8 capillary column (300 × 150 mm; Grace Vydac 208MS5.315) eluted at 4 μl/min. The LC mobile phase consisted of Buffer A (0.1% formic acid in MilliQ water) and Buffer B (0.1% formic acid in 3:1 acetonitrile:n-propanol, v/v). Sequest search software (ThermoFinnigan) was used for peptide identification using the NCBI protein database. Many large collagenous peptides not found by Sequest had to be identified manually by calculating the possible MS/MS ions and matching these to the actual MS/MS. The percentage 3-hydroxylation at a particular site was determined from the abundance of the 3Hyp-containing ion as a fraction of the sum of both 3Hyp and Pro versions of the same tryptic peptide.
RT-PCR Analysis of PPIB, CRTAP, P3H1, P3H2, and P3H3 Gene Expression
Normal adult rat cartilage RNA was purchased from Zyagen (San Diego, CA). SAOS-2 cell total RNA was obtained from the cells using the RNeasy kit (Qiagen) as we have described (27). For RCS-LTC cells, total cell RNA was isolated using the PureLink RNA Mini Kit (Invitrogen) according to the manufacturer's specifications. cDNA was generated from 2 μg of RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's specifications in a thermocycler (Bio-Rad). cDNA, as generated above from various rat and human cells or tissue RNA samples, was used to generate gene products for prolyl cis-trans isomerase, PPIB (rat, 100 bp; human, 171 bp); cartilage-associated protein, CRTAP (rat, 189 bp; human, 213 bp); P3H1/LEPRE1 (rat, 178 bp; human, 226 bp); P3H2/LEPREL1 (rat, 233 bp; human, 164 bp); and P3H3/LEPREL2 (rat, 130 bp; human, 270 bp). The primers used had sequences as follows (from 5′ to 3′): rat GAPDH (forward, ATGACTCTACCCACGGCAAG; reverse, TACTCAGCACCAGCATCACC) and human GAPDH (forward, GGCCTCCAAGGAGTAAGACC; reverse, AGGGGTCTACATGGCAACTG), rat PPIB (forward, GGTGGATGCTGCGCCTCTCG; reverse, ACGGAGGGTCCAGGCAGCAA) and human PPIB (forward, GCAAGATCGAGGTGGAGAAG; reverse, CTGTGGAATGTGAGGGGAGT), rat CRTAP (forward, GCGCGCAGTATGAGCGCTAC; reverse, AGTTGCGGTGGCAGAAGGCC), human CRTAP (forward, GGTTTGAAGGGCAGTCTTCTCTGGC; reverse, GTGAAGACCATTGTGAGGCTGGAG), rat P3H1 (forward, GTGAAGAGCTGGACCTGGAG; reverse, ACCCCAGACATGGTTTGGTA), human P3H1 (forward, GACTTCCTCCCATCGCATTA; reverse, TTTCCAGTAGGCTTCGCTGT), rat P3H2 (forward, AAGCCACACCTGGAAAGCTA; reverse, TGCTGACAGACCAGAACCTG), human P3H2 (forward, GTGCAACTGTCCTGAAAGCA; reverse, TCGGCAGACCATGTGTGTAT), rat P3H3 (forward, CCCCTCATAGTCCTCACGAA; reverse, AAGGTGCGTACTCGCTCACT), and human P3H3 (forward, CGGACTCCTCTACCTCAACG; reverse, TCTTCCTCCTCCTCCTGTGA).
Each PCR amplification cycle consisted of 20 s of denaturation at 94 °C, 20 s of annealing at 55 °C, and 1 min of extension at 72 °C for a total of 30 cycles. Samples were run on 2% agarose gels (Research Products International Corp.) stained in Tris acetate-EDTA buffer containing ethidium bromide (Invitrogen).
Real-time PCR (Quantitative PCR) Analysis of P3H Gene Expression
The identification of mRNA for rat P3H1, P3H2, and P3H3 was performed in triplicate for each cDNA sample using the TaqMan gene expression assay probe and primer set (Applied Biosystems) for each respective gene, P3H1 (Rn01642789_m1), P3H2 (Rn01404939_m1), and P3H3 (Rn01459473_m1). Samples were analyzed using an Applied Biosystems 7900HT real-time PCR system. The ΔΔ threshold cycle (ΔΔCT) method was used to analyze gene amplification, using eukaryotic 18 S rRNA (Hs03928990_g1, Applied Biosystems) as the reference gene, with pooled cDNA from RCS-LTC cells and normal adult rat cartilage serving as the calibrator.
Gene Silencing
Predesigned rat Stealth RNAiTM siRNA Select RNAi constructs (Invitrogen) were used to specifically knock down P3H1 (leprecan 1, catalogue no. RSS300139), P3H2 (leprecan-like 1,catalogue no. RSS305239), P3H3 (leprecan-like 2, catalogue no. RSS355963) in RCS-LTC cells. The Stealth RNAiTM siRNA Negative Control Med GC (catalogue no. 12935-300, Invitrogen) was included as a control. RCS-LTC cells in serum-free OptiMEM containing 25 μg/ml ascorbate and 100 μg/ml βAPN were transfected with 200 nm siRNA/5 × 105 cells, using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol for transfecting the Rat1 cell line. Transfection was carried on for 72 h, with fresh medium and siRNA added every 24 h. Following this period pN type II collagen was extracted from the cell layers and analyzed for proline 3-hydroxylation by mass spectrometry. P3H2 protein was detected by Western blotting as described below.
Immunoblot Analysis
Total lysates from plates of cultured cells were prepared by rinsing with D-PBS (Hyclone), adding radioimmune precipitation assay buffer (Sigma) containing protease inhibitors (Roche Applied Science) and freeze-thawing for three cycles with occasional vortexing. Lysates were cleared of insoluble material by centrifugation at 10,000 × g. Equal aliquots of protein in samples were separated by SDS-PAGE on 6% polyacrylamide gels and transferred to PVDF membranes. Blots were blocked and probed according to standard procedures using P3H2 antibody (Sigma HPA007890, 1:1000). A proliferating cell nuclear antigen antibody (Stressgen Biotechnologies, 1:1000) was used to control for protein loading (28). Goat anti-rabbit or goat anti-mouse IgG-horseradish peroxidase (Bio-Rad, 1:5000) were used as secondary antibody. Detection was by chemiluminescence using SuperSignal West Pico substrate (Thermo Scientific) (21).
RESULTS
Gene Expression of P3H Isoenzymes and Prevalence of 3Hyp in Clade A Collagen from RCS-LTC Cells
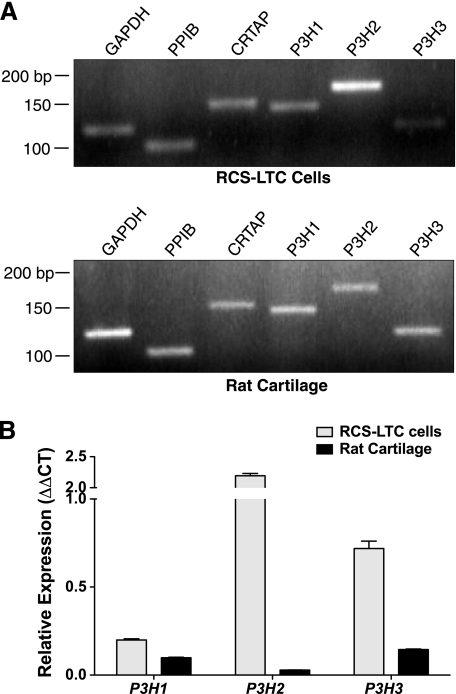
RT-PCR analyses showed that all the components of the proline 3-hydroxlation complex (P3H1, CRTAP, and PPIB) as well as the isoenzymes P3H2 and P3H3 were expressed by both normal adult rat cartilage and the RCS-LTC cells (Fig. 2A). Quantitative PCR of normal rat cartilage for the three P3H isoenzymes showed lowest expression for P3H2, but the RCS-LTC cells expressed 8-fold higher P3H2 mRNA than normal rat cartilage. Moreover, the RCS-LTC cells express all P3H isoenzymes at relatively higher levels compared with rat cartilage (Fig. 2B).
FIGURE 2.
Expression of PPIB, CRTAP, P3H1, P3H2, and P3H3 in the RCS-LTC cell line and rat cartilage. A, RCS-LTC cells transcribe a full complement of the genes assayed. When compared with adult normal rat cartilage, the RCS-LTC cells express an abundance of P3H2. B, quantitative comparison by quantitative PCR of P3H1, P3H2, and P3H3 mRNA expression between RCS-LTC cells and normal adult rat cartilage yielded modestly increased expression of P3H1, an ∼2-fold increase in P3H3 expression and an excess of 8-fold increase in P3H2 in the RCS-LTC cells. Data represents n = 3 for each isoenzyme.
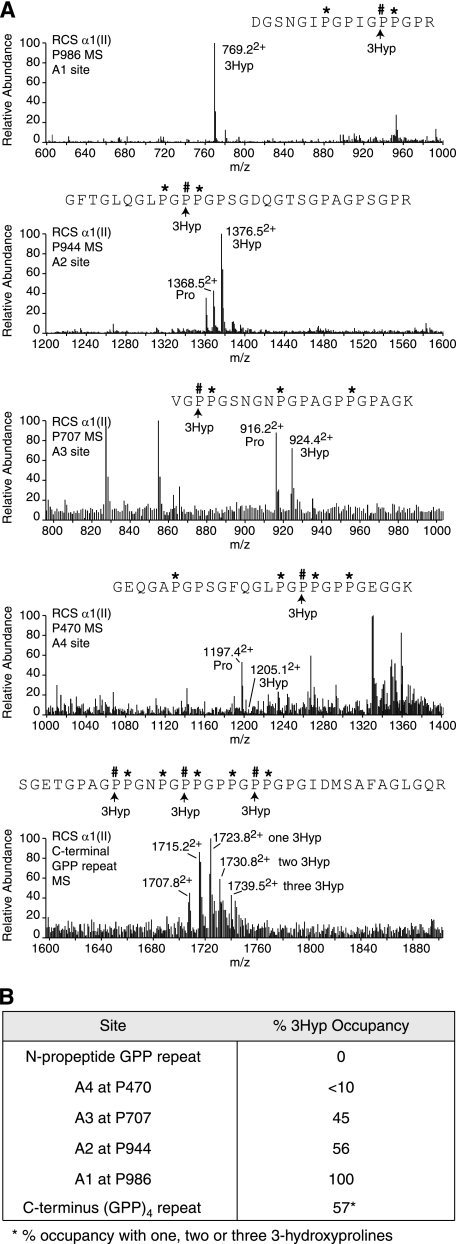
To determine whether high P3H isoenzyme expression had an effect on the 3-hydroxylation of proline residues in clade A collagen synthesized by the RCS-LTC cells, the matrix pNα1(II) collagen chains were analyzed by ion-trap tandem mass spectrometry after in-gel trypsin digestion. The primary (A1) site at Pro-986 showed 100% 3Hyp occupancy (Fig. 3A) consistent with the RT-PCR data showing that all three components of the P3H1 protein complex were expressed. In addition, 3Hyp occupancy was high at the A2 site, Pro-944 (56%), moderate at the A3 site, Pro-707 (45%), and low at the A4 site Pro-470 (<10%). The candidate proline residues in the (GPP)4-repeat containing peptide from the C terminus of the collagen triple helix showed masses indicating up to three 3Hyp residues per individual peptide as seen in the bottom panel of Fig. 3A. Occupancy ranged from zero to three residues of 3Hyp per individual peptide based on the parent ion ladder. The estimated mean occupancy for this multiple site and the other individual sites is summarized in Fig. 3B.
FIGURE 3.
Tandem mass-spectrometric analysis of the RCS-LTC pNα1(II) collagen chain. A, full scan mass spectra from the tryptic peptides LC-MS profiles of pNα1(II) across the elution window of the post-translational variants containing Pro-986, Pro-944 and Pro-707, Pro-470, and the C-terminal (GPP)4 repeat. The relative abundance of the ions shown provides an index of the degree of 3-hydroxylation at the A1, A2, A3, A4, and C-terminal GPP repeat sites. As shown, Pro-944 is 56% 3Hyp occupied and Pro-986 is 100% 3Hyp occupied. B, summary of the relative abundance (%) of 3-hydroxylation of proline residues at sites in pNα1(II) collagen chains from RCS-LTC extracellular matrix.
Because RCS-LTC cells fail to process pN type II collagen molecules and deposit a pN type II microfibrillar polymer in the matrix (22), this enabled a search for the presence of 3Hyp in the N-propeptide minor helix (see Fig. 1A). The results showed no detectable 3Hyp in any of the four candidate prolines within the (GPP)4 repeat that occurs at the C terminus of the minor helix.
Gene Silencing Provides Clues on Preferred Substrate for P3H2
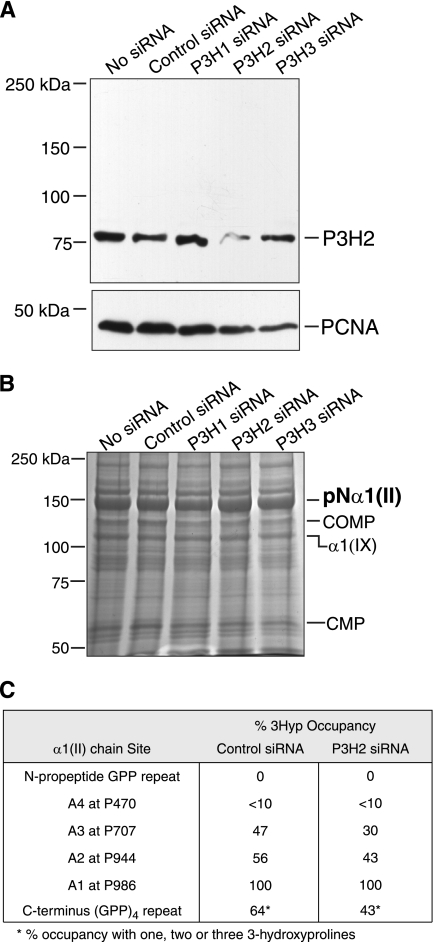
The high relative abundance of P3H2 in the RCS-LTC cells and the abnormally high content of 3-hydroxylated proline residues in the pN-α1(II) collagen chains suggested this enzyme may be responsible for prolyl 3-hydroxylation at the non-A1 sites. To investigate this, RCS-LTC cells were cultured for 72 h in the presence of siRNA oligonucleotides designed specifically to interfere and silence the P3H2 isoenzyme. As seen in Fig. 4A, a marked knockdown in P3H2 protein was observed when cell lysates were probed with an antibody specific to P3H2. This decrease is clear on comparing cultures treated with control siRNA or P3H1 and P3H3 siRNA, which show no major change in P3H2 levels.
FIGURE 4.
RNA interference of P3H2 in cultures of the RCS-LTC cell line. A, Western blot of equal aliquots of cell lysates from siRNA-treated cultures probed for P3H2 protein. The P3H2 antibody robustly reacted with an 80-kDa band in siRNA-untreated and control siRNA-treated cultures. A clear reduction of band intensity was specifically observed only in P3H2 siRNA-treated cultures. Proliferating cell nuclear antigen protein was detected as a loading control. Globular protein molecular weight standards were used. B, Coomassie Blue stained gel showing similar amounts of pNα1(II) collagen chains extracted from P3H2 siRNA-treated and untreated cultures. This band from control siRNA and P3H2 siRNA-treated lanes was analyzed by mass spectrometry. Cartilage oligomeric matrix protein (COMP), α1(IX) collagen chain and cartilage matrix protein 1 (CMP) synthesized by the cell line were also identified by mass spectrometry. Globular protein molecular weight standards were run. C, degree (%) of 3-hydroxylation of proline residues in pNα1(II) collagen chains determined by tandem mass spectrometry. The pNα1(II) chains from P3H2 siRNA-treated and control siRNA-treated lanes were subjected to in-gel trypsin digestion, and peptides containing 3Hyp sites were analyzed. 3Hyp occupancy fell at all non-A1 sites on suppression of P3H2 protein.
SDS-PAGE of total collagen in extracts of P3H2 siRNA- treated, control siRNA, P3H1, and P3H3 siRNA-treated cultures showed no obvious differences in the amount of type II collagen extracted from the cell layers (Fig. 4B). Mass spectrometry of tryptic digests of the pN-α1(II) collagen chains showed a reduced degree of 3Hyp occupancy at all the substrate proline other than Pro-986 residues from the P3H2-targeted siRNA cultures compared with control cultures (Fig. 4C). Pro-944, Pro-707, and the C-terminal GPP repeat were reduced 56–43%, 47–30%, and 64–43% respectively. All three peptide ions at 1723.82+, 1730.82+, 1739.52+, for one, two, and three 3Hyp in the GPP repeat peptide were suppressed from the P3H2-targeted siRNA cultures compared with control (Fig. 4C). P3H3-targeted siRNA cultures showed no change in 3Hyp occupancy at Pro-944, Pro-470, and Pro-986 compared with control cultures. Pro-707 and the C-terminal GPP repeat were reduced 47–38% and 64–52%, respectively. Because Pro-986 (substrate of the P3H1 isoenzyme) remained unchanged from the control, the effect of reduced P3H2 enzyme was exclusively at non-A1 sites, implicating P3H2 in the 3-hydroxlation of Pro-944, Pro-707, and the C-terminal GPP repeat.
P3H2 Is Responsible for 3-Hydroxylation of Specific Proline Residues in α2(V) and α2(I) Collagen Chains
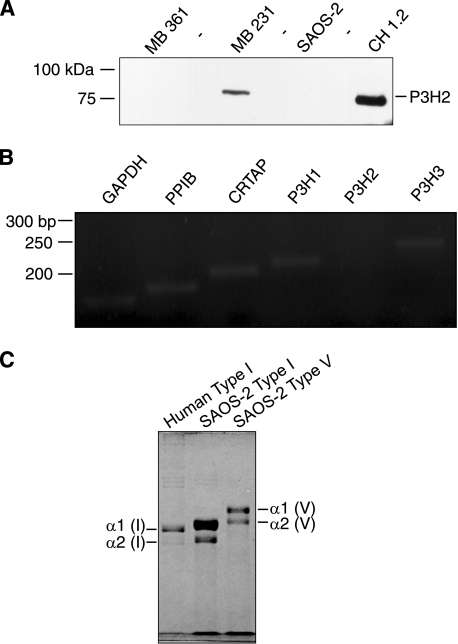
To confirm the finding that P3H2 3-hydroxylated non-A1 sites, the other clade A collagen chains were interrogated. Because RNAi gene silencing was unable to completely knock-out P3H2 activity in RCS-LTC cells, a different approach was pursued. Based on a recent report that P3H2 is epigenetically silenced in some cancer cells (29), cell lysates of cultured human breast cancer, chondrosarcoma, and osteosarcoma cells were screened for P3H2 protein on Western blots. Fig. 5A shows P3H2 protein detected in a lysate of the MB 231 breast cancer cells as expected as these cells are reported to express this gene (29). P3H2 was also detected in CH1.2 cells, a human chondrosarcoma cell line. Fig. 5A also shows that P3H2 protein was not detected in a lysate of SAOS-2 osteosarcoma cells. The specificity of the antibody to P3H2 was confirmed when P3H2 was not detected in a lysate of MB 361 breast cancer cells (Fig. 5A). The P3H2 gene is reported to be epigenetically silenced in these cells (29). RT-PCR confirmed the absence of P3H2 message in SAOS-2 cells, whereas P3H1, PPIB, CRTAP and P3H3 were robustly expressed (Fig. 5B).
FIGURE 5.
Expression of PPIB, CRTAP, P3H1, P3H2, and P3H3 in the SAOS-2 cell line. A, cell lysates from MBA 361 and MBA 231 human breast cancer cells, CH1.2 human chondrosarcoma cells, and SAOS-2 human osteosarcoma cells were probed with P3H2 antibody on Western blots. The antibody failed to detect P3H2 protein in the MBA 361 and SAOS-2 cells. B, RT-PCR confirmed the lack of expression of P3H2 by SAOS-2 cells. P3H1, CRTAP, PPIB, and P3H3 were strongly expressed. C, Coomassie Blue-stained gel showing type I collagen (middle lane) and type V collagen (right lane) extracted from the extracellular matrix of cultured SAOS-2 cells. Human bone type I collagen was run as a control (left lane). The α1(I), α2(I), and the α2(V) bands were cut out and analyzed for 3Hyp occupancy by mass spectrometry.
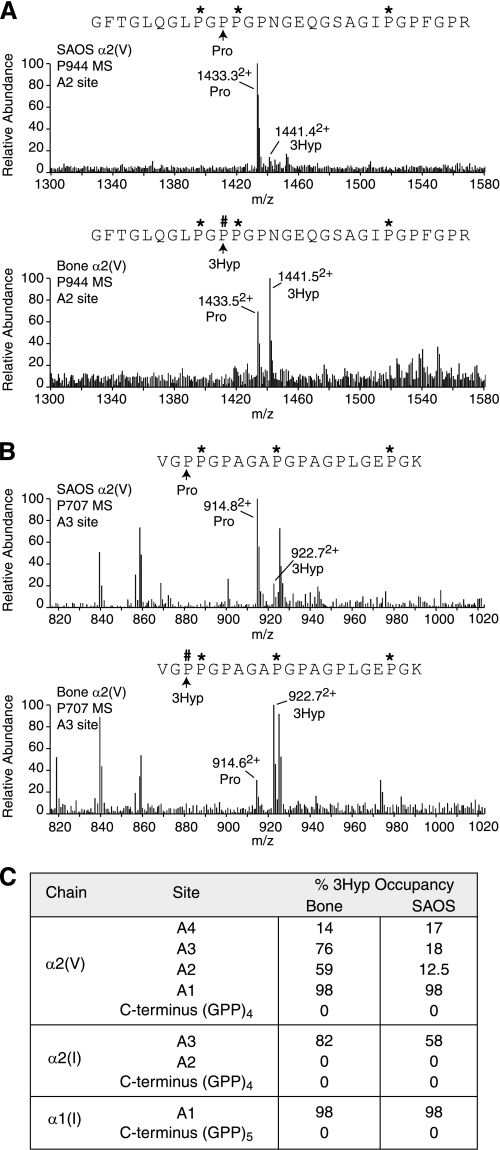
Purified collagen from the matrix laid down by SAOS-2 cells revealed type I and type V collagen chains on electrophoresis as expected for an osteoblast phenotype (Fig. 5C). Because the α2(V) collagen chain (from human bone) had the most complete pattern of occupation of 3Hyp residues at sites A1, A2, A3, and A4 of all clade A collagen chains (19), the pattern of occupancy of α2(V) from SAOS-2 cultures was assessed. Fig. 6A shows 12.5% 3-hydroxylation of Pro-944 and 18% of Pro-707 hydroxylated compared with α2(V) from normal human bone (59 and 76%, respectively). Fig. 6B summarizes the results and shows that Pro-986 and Pro-470 in α2(V) was unaffected compared with normal bone. As in normal bone α1(I) from SAOS-2 cells was fully hydroxylated at Pro-986, and no 3Hyp was detected in the GPP repeat at its C terminus or that of α2(V) chains. Only tendon α1(I) chains are heavily 3-hydroxylated at this site (20).
FIGURE 6.
Tandem mass spectrometric analysis of 3Hyp in matrix collagen deposited by SAOS-2 cells. A, B, α2(V) chains from the SAOS-2 cell line were compared with human bone. Full scan mass spectra from the tryptic peptide LC-MS profiles of α2(V) across the elution window of the post-translational variants containing Pro-986 (A1), Pro-944 (A2), and Pro-707 (A3) are shown. C, summary of % prolyl 3-hydroxylation levels at sites in α2(V), α2(I), and α1(I) collagen chains from SAOS-2 cells and normal human bone.
DISCUSSION
The findings indicate that the P3H2 isoenzyme can 3-hydroxylate prolines at certain Gly-Pro-4Hyp sites in fibrillar collagen chains. Previous studies using recombinant enzyme in vitro had shown that P3H2 can 3-hydroxylate synthetic peptides matching the sequences of known 3-hydroxylation sites in α1(IV) collagen chains (16). Additional substrates for P3H2 were presumed to exist for two reasons. First, mRNA for this isoenzyme is expressed in embryonic and adult tissues that express fibrillar collagens as well as in basement membrane-rich tissues (17, 18). Second, the recently discovered non-A1 sites in clade A fibrillar collagen chains share sequence features with known prolyl 3-hydroxylation sites in type IV collagen notably a phenylalanine N-terminal to the proline substrate residue at sites A2 and A3. Similar 3Hyp-containing sequence occurs in both α1(IV) and α2(IV) chains (30).
The results are consistent with P3H2 being responsible at least in part for 3-hydroxylation of Pro-944, Pro-707, and the C-terminal (GPP)4 repeat sites in the α1(II) chain. The high level of expression of P3H2 by RCS-LTC cells, (eight times that of normal rat chondrocytes; Fig. 2), supports that. Because the level is so high, it may explain why only a modest suppression of 3Hyp occupancy at the A2 site in pNα1(II) collagen was achieved in knockdown experiments (Fig. 4C). The lack of P3H2 protein in SAOS-2 cells, associated with low 3Hyp at the A2 site in α2(V) (Fig. 6) is also consistent with P3H2 being primarily responsible for α1(II) Pro-944 hydroxylation. The residual 3Hyp presumably was produced by P3H1 or P3H3. It is well documented that α1(I) collagen chains made by skin fibroblasts from patients with P3H1 null mutations have low but significant 3Hyp occupancy at Pro-986 (8–10). The lack of P3H2 enzyme expressed in SAOS-2 cells and low occupancy of 3Hyp at Pro-707 in α2(V) and α2(I) collagen chains compared with normal bone (Fig. 6) supports P3H2 being the primary enzyme responsible for this hydroxylation. Similar 3Hyp levels at α2(V) Pro-470 compared with bone suggests that P3H2 may not modify this site. Unchanged 3Hyp levels at pNα1(II) Pro-470 in the siRNA-treated RCS-LTC cultures (Fig. 4C) is consistent with this. Residual 3Hyp at Pro-707 in SAOS-2 is probably due to P3H3. Marginally reduced 3Hyp levels at pNα1(II) Pro-707 in P3H3 siRNA-treated RCS-LTC cultures is consistent with this. The P3H1 enzyme in complex with CRTAP and cyclophillin B appears to be solely responsible for hydroxylation of Pro-986 in α1(I), α1(II), and α2(V) chains (8, 11), whereas P3H2 and P3H3 may only have activity at the other sites. Also, the amino acid sequence motif of the A1 site is different from that of the non-A1 sites (19). The differential substrate specificities of P3H2 and P3H3 and the nature of any protein complexes that regulate their activities are largely unexplored.
Because neither SAOS-2 nor RCS-LTC cells synthesize type IV collagen, they provide no insight on the role of P3H2 in 3Hyp formation in type IV collagen. The relative roles of P3H1, P3H2, and P3H3 in collagen type IV prolyl 3-hydroxylation are unknown, though all three are expressed in type IV collagen-rich basement membranes (7, 16, 17). In fact, P3H1 was first isolated as leprecan, a basement membrane-associated proteoglycan from a rat parietal yolk sac tumor cell line (7).
From normal mammalian cartilages, no 3Hyp is present at Pro-707 or Pro-470 of α1(II) (19). Chicken cartilage α1(II) Pro-707 was 18% 3-hydroxylated (31). The progressively higher 3Hyp levels going from Pro-470 to Pro-707 to Pro-944 in RCS α1(II) chains, we conclude, is a consequence of overexpression of P3H2 as is the 3Hyp of the (GPP)4 repeat at the C terminus. The latter site has comparable levels with those in rat tail tendon α1(I) and α2(I) collagen chains (20).
The matrix deposition of entirely unprocessed pN type II collagen by the RCS-LTC cells (22) allowed us to examine whether candidate proline residues in the minor triple-helix of the N-propeptide were 3-hydroxylated. In cartilage, this domain is normally removed from the procollagen molecule on fibrillogenesis. Purifying the type II N-propepetide from normal cartilage is a challenge so we have not been able to examine the (GPP)4 repeat at the C terminus of this domain for 3Hyp by mass spectrometry until now. The results clearly showed no prolyl 3-hydroxylation in this GPP-repeat despite high levels of all three P3H isoenzymes in RCS-LTC cells as compared with normal rat cartilage (Fig. 2B). The progressively lower 3Hyp content observed from the C terminus to the N terminus at sites in the main helix is consistent with 3-hydroxylation beginning at the C terminus and shutting down as the triple helix folds (Fig. 3).
We have reported that the variation in the degree of 3-hydroxylation at the A2 site (Pro-944) in α1(II) collagen is tissue-specific, ranging from <10% in bovine articular cartilage, 40% in nucleus pulposus of the intervertebral disc to 90% in vitreous of the eye (19). In this study, we have observed that the level of 3Hyp at the A2 site in pNα1(II) from the RCS-LTC cells (57%) is also high. Expression of P3H2 is also high in adult mouse eye tissue (17). Because type II collagen processing by the RCS-LTC cell line is arrested at the stage of pN type II collagen (22), and the functional form of type II collagen in vitreous is mostly pN type II collagen (32), it is tempting to speculate that retention of the N-propeptide and the distinctive prolyl 3-hydroxylation pattern may be related. It may be relevant that the N-propeptide is folded back on the main triple helix of the collagen molecule as a structural prerequisite for cleavage by ADAMTS-3, the procollagen N-propeptidase for type II collagen in cartilage (33–37). In such a conformation within a fibril, the N-propeptide cleavage site is likely to be closely aligned with Pro-944 in an adjacent molecule staggered by 4D-periods, as illustrated in Fig. 1B. Could prolyl 3-hydroxylation at Pro-944, Pro-707, and Pro-470 modify susceptibility to N-propeptide cleavage or, conversely, could failed removal of N-propeptides result in continuing prolyl 3-hydroxylation of the triple-helical domains in a nacent fibril? This is highly speculative but within the range of possibilities given the scope of the current understanding of regulatory mechanisms of collagen assembly and prolyl 3-hydroxylation.
In conclusion, the tumor-derived cell lines RCS-LTC and SAOS-2 have proven to be useful systems in which to study control mechanisms of collagen post-translational modifications. In addition to the prolyl 3-hydroxylations reported on here, previous work has shown overexpression of PLOD1 (lysyl hydroxylase 1) by SAOS-2 cells relative to normal osteoblasts (23) and high levels of hydroxylysyl pyridinoline cross-links in the pN type II polymer deposited by RCS-LTC cells (21).
Acknowledgments
We thank Dr. David M. Hudson for critical review of the manuscript, Dr. James Wu for helpful discussions, and Lammy Kim for technical expertise.
This work was supported, in whole or in part, by National Institutes of Health, NIAMS Grants AR057025 (to R. J. F.) and AR036794 and AR037318 (to D. R. E.). This work was also supported by National Center for Research Resources Grant TL1 RR025016 (to A. W. F.) and the Ernest M. Burgess Endowed Chair research program at the University of Washington.
- P3H1
- prolyl 3-hydroxlase 1
- 3Hyp
- 3-hydroxyproline.
REFERENCES
- 1. Olsen B. R., Berg R. A., Kivirikko K. T., Prockop D. J. (1973) Eur. J. Biochem. 35, 135–147 [DOI] [PubMed] [Google Scholar]
- 2. Risteli J., Kivirikko K. I. (1974) Biochem. J. 144, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kivirikko K. I., Ryhänen L., Anttinen H., Bornstein P., Prockop D. J. (1973) Biochemistry 12, 4966–4971 [DOI] [PubMed] [Google Scholar]
- 4. Myllylä R., Alitalo K., Vaheri A., Kivirikko K. I. (1981) Biochem. J. 196, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vranka J. A., Sakai L. Y., Bächinger H. P. (2004) J. Biol. Chem. 279, 23615–23621 [DOI] [PubMed] [Google Scholar]
- 6. Tryggvason K., Majamaa K., Risteli J., Kivirikko K. I. (1979) Biochem. J. 183, 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wassenhove-McCarthy D. J., McCarthy K. J. (1999) J. Biol. Chem. 274, 25004–25017 [DOI] [PubMed] [Google Scholar]
- 8. Cabral W. A., Chang W., Barnes A. M., Weis M., Scott M. A., Leikin S., Makareeva E., Kuznetsova N. V., Rosenbaum K. N., Tifft C. J., Bulas D. I., Kozma C., Smith P. A., Eyre D. R., Marini J. C. (2007) Nat. Genet. 39, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baldridge D., Schwarze U., Morello R., Lennington J., Bertin T. K., Pace J. M., Pepin M. G., Weis M., Eyre D. R., Walsh J., Lambert D., Green A., Robinson H., Michelson M., Houge G., Lindman C., Martin J., Ward J., Lemyre E., Mitchell J. J., Krakow D., Rimoin D. L., Cohn D. H., Byers P. H., Lee B. (2008) Hum. Mutat. 29, 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willaert A., Malfait F., Symoens S., Gevaert K., Kayserili H., Megarbane A., Mortier G., Leroy J. G., Coucke P. J., De Paepe A. (2009) J. Med. Genet. 46, 233–241 [DOI] [PubMed] [Google Scholar]
- 11. Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., Fernandes R. J., Eyre D. R., Yao Z., Boyce B. F., Lee B. (2006) Cell 127, 291–304 [DOI] [PubMed] [Google Scholar]
- 12. Ogle J. D., Arlinghaus R. B., Lgan M. A. (1962) J. Biol. Chem. 237, 3667–3673 [PubMed] [Google Scholar]
- 13. Jenkins C. L., Bretscher L. E., Guzei I. A., Raines R. T. (2003) J. Am. Chem. Soc. 125, 6422–6427 [DOI] [PubMed] [Google Scholar]
- 14. Mizuno K., Peyton D. H., Hayashi T., Engel J., Bächinger H. P. (2008) FEBS J. 275, 5830–5840 [DOI] [PubMed] [Google Scholar]
- 15. Järnum S., Kjellman C., Darabi A., Nilsson I., Edvardsen K., Aman P. (2004) Biochem. Biophys. Res. Commun. 317, 342–351 [DOI] [PubMed] [Google Scholar]
- 16. Tiainen P., Pasanen A., Sormunen R., Myllyharju J. (2008) J. Biol. Chem. 283, 19432–19439 [DOI] [PubMed] [Google Scholar]
- 17. Vranka J., Stadler H. S., Bächinger H. P. (2009) Cell Struct. Funct. 34, 97–104 [DOI] [PubMed] [Google Scholar]
- 18. Capellini T. D., Dunn M. P., Passamaneck Y. J., Selleri L., Di Gregorio A. (2008) Genesis 46, 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weis M. A., Hudson D. M., Kim L., Scott M., Wu J. J., Eyre D. R. (2010) J. Biol. Chem. 285, 2580–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eyre D. R., Weis M., Hudson D. M., Wu J. J., Kim L. (2011) J. Biol. Chem. 286, 7732–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandes R. J., Schmid T. M., Eyre D. R. (2003) Eur. J. Biochem. 270, 3243–3250 [DOI] [PubMed] [Google Scholar]
- 22. Fernandes R. J., Schmid T. M., Harkey M. A., Eyre D. R. (1997) Eur. J. Biochem. 247, 620–624 [DOI] [PubMed] [Google Scholar]
- 23. Fernandes R. J., Harkey M. A., Weis M., Askew J. W., Eyre D. R. (2007) Bone 40, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller A. D., Vigdorovich V., Strong R. K., Fernandes R. J., Lerman M. I. (2006) Osteoarthritis Cartilage 14, 1315–1317 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Fernandes R. J., Weis M., Scott M. A., Seegmiller R. E., Eyre D. R. (2007) Matrix Biol. 26, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goto T., Matsui Y., Fernandes R. J., Hanson D. A., Kubo T., Yukata K., Michigami T., Komori T., Fujita T., Yang L., Eyre D. R., Yasui N. (2006) J. Bone Miner. Res. 21, 661–673 [DOI] [PubMed] [Google Scholar]
- 28. Wang B., Liu K., Lin F. T., Lin W. C. (2004) J. Biol. Chem. 279, 54140–54152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shah R., Smith P., Purdie C., Quinlan P., Baker L., Aman P., Thompson A. M., Crook T. (2009) Br. J. Cancer 100, 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schuppan D., Glanville R. W., Timpl R. (1982) Eur. J. Biochem. 123, 505–512 [PubMed] [Google Scholar]
- 31. Hudson D. M., Weis M., Eyre D. R. (2011) PLoS One 6, e19336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bishop P. N., Reardon A. J., McLeod D., Ayad S. (1994) Biochem. Biophys. Res. Commun. 203, 289–295 [DOI] [PubMed] [Google Scholar]
- 33. Kadler K. E. (1993) Int. J. Exp. Pathol. 74, 319–323 [PMC free article] [PubMed] [Google Scholar]
- 34. Mould A. P., Hulmes D. J. (1987) J. Mol. Biol. 195, 543–553 [DOI] [PubMed] [Google Scholar]
- 35. Fernandes R. J., Hirohata S., Engle J. M., Colige A., Cohn D. H., Eyre D. R., Apte S. S. (2001) J. Biol. Chem. 276, 31502–31509 [DOI] [PubMed] [Google Scholar]
- 36. Holmes D. F., Watson R. B., Steinmann B., Kadler K. E. (1993) J. Biol. Chem. 268, 15758–15765 [PubMed] [Google Scholar]
- 37. Arnold W. V., Fertala A., Sieron A. L., Hattori H., Mechling D., Bächinger H. P., Prockop D. J. (1998) J. Biol. Chem. 273, 31822–31828 [DOI] [PubMed] [Google Scholar]