Abstract
Deinococcus radiodurans is among the very few bacterial species extremely resistant to ionizing radiation, UV light, oxidizing agents, and cycles of prolonged desiccation. The proteome of D. radiodurans reflects the evolutionary pressure exerted by chronic exposure to (nonradioactive) forms of DNA and protein damage. A clear example of this adaptation is the overrepresentation of protein families involved in the removal of non-canonical nucleoside triphosphates (NTPs) whose incorporation into nascent DNA would promote mutagenesis and DNA damage. The three-dimensional structure of the DR2231 protein has been solved at 1.80 Å resolution. This protein had been classified as an all-α-helical MazG-like protein. The present study confirms that it holds the basic structural module characteristic of the MazG superfamily; two helices form a rigid domain, and two helices form a mobile domain and connecting loops. Contrary to what is known of MazG proteins, DR2231 protein shows a functional affinity with dUTPases. Enzymatic and isothermal calorimetry assays have demonstrated high specificity toward dUTP but an inability to hydrolyze dTTP, a typical feature of dUTPases. Co-crystallization with the product of hydrolysis, dUMP, in the presence of magnesium or manganese cations, suggests similarities with the dUTP/dUDP hydrolysis mechanism reported for dimeric dUTPases. The genome of D. radiodurans encodes for all enzymes required for dTTP synthesis from dCMP, thus bypassing the need of a dUTPase. We postulate that DR2231 protein is not essential to D. radiodurans and rather performs “house-cleaning” functions within the framework of oxidative stress response. We further propose DR2231 protein as an evolutionary precursor of dimeric dUTPases.
Keywords: Enzyme Structure, Nucleoside Nucleotide Biosynthesis, Nucleoside Nucleotide Metabolism, Oxidative Stress, X-ray Crystallography
Introduction
Bacteria possess a wide variety of enzymes with the common physiological function of sanitizing the cell of wasteful or even toxic endogenous metabolites. Such enzymes also modulate the accumulation of certain intermediates in biochemical pathways. These enzymes have been coined “house-cleaning” enzymes, because they protect the cell from the harmful effects resulting from the unbalanced presence of potentially toxic compounds.
Pyrophosophohydrolysis, the enzymatic cleavage of the α-β phosphodiester bond of nucleoside triphosphates (NTPs) generates the corresponding nucleoside monophosphate and inorganic phosphate (PPi), subsequently hydrolyzed into free phosphate by other enzymes present in the cytosol. These exergonic reactions allow the recycling of nucleoside monophosphates (NMPs) back into metabolic pathways as well as intracellular scavenging for inorganic phosphate. Pyrophosphohydrolysis, however, has yet another role in cellular metabolism and is used in the removal of non-canonical nucleotide triphosphates arising from oxidation, deamination, or other modifications of canonical nucleotides (1), thereby preventing their incorporation into DNA or RNA. Proteins hydrolyzing NTPs with such cleansing functions can be found in structurally different superfamilies, such as the MutT-related hydrolases (Nudix) (α + β), dUTPase (all-β), dITPase (Maf/HAM1) (α/β), all-α-NTP pyrophosphatases (MazG), and phospho-ribosyl-ATP pyrophosphatase (HisE).
Analysis of the complete genome of Deinococcus radiodurans revealed the specific expansion of certain protein families believed to be connected to the organism's response to stress and damage resistance, repair mechanisms, and signal transduction (2). Orthologs of almost all known genes involved in stress response in other bacteria are present in Deinococcus (3). In the case of some missing gene families, function is maintained by nonorthologous proteins with similar functions (4). Among the highly represented protein families are hydrolases, such as the house-cleaning Nudix pyrophosphatases and other pyrophosphohydrolases, the calcineurin-like phosphoesterases, phosphatases, lipase/epoxidase-like (α/β) hydrolases, subtilisin-like proteases, and sugar deacetylases (5). A considerable number of these stress response-related genes are clustered in unusual gene arrays. Some appear to have evolved by tandem duplication (see DR2254/DR2255 or DR0783/DR0784, for example) or gene translocation (DR0675 to DR0677), within the Deinococcus lineage. However, the majority of unusual gene clusters suggest that acquisition took place through horizontal gene transfer from various archael, bacterial, and even eukaryotic sources (4).
Like other bacteria, D. radiodurans seems to lack entirely monomeric, trimeric, and archeal dUTPases, as has been pointed out by Moroz et al. (6). Recently, the crystal structures of dUTPases from Campylobacter jejuni and Trypanosoma cruzi revealed a new all-α-helix fold with a homodimeric arrangement in contrast to the classical trimeric dUTPase reported until then. No homologues of this dimeric dUTPase could be identified in D. radiodurans.
Through sequence analysis restricted to the active site motif, Moroz et al. (6, 7) identified a “basic module” of the dUTPase/dCTPase family in the genomes of several Gram-positive bacteria and respective phages. This basic module, consisting of only five active site-forming helices, is conserved in two other families: the nonspecific NTP-PPase MazG and phosphoribosyl-ATP pyrophosphatase HisE. These three enzyme families, which share similar function, were unified into a single superfamily, the all-α-NTP-PPase superfamily. Following the same criteria, DR2231 was identified by sequence analysis as a putative member of this superfamily, as were two other genes encoding MazG family proteins, DR1022 and DR1183 (7). We performed a broader sequence search using as a query the sequence of Escherichia coli MazG protein, (8) against the D. radiodurans genome and identified only DR1022 and DR1183 as MazG-like proteins. Furthermore, there are no annotated phosphoribosyl-ATP pyrophosphatase HisE entries in the D. radiodurans genome. Determining the three-dimensional structure of DR2231 protein and clarifying its function acquire a particular significance regarding the genetic versatility of D. radiodurans in stress response and DNA damage.
Here, we report the crystal structure of DR2231 from D. radiodurans at 1.8 Å resolution. The enzyme has been identified as the prototype of a subfamily of the NTP pyrophosphohydrolase superfamily (7). It has significant structural resemblance to MazG but is functionally related to the dimeric dUTPases, exhibiting activity exclusively on deoxy-NTPs with a very high specificity toward dUTP and none toward dTTP. The crystal structures of the native protein in the apo form and with Mn2+ coordinated to the active site and DR2231 in complex with its product from dUTP hydrolysis with either Mn2+ or Mg2+ are presented. We propose that the DR2231 protein is a dUTPase with marked specificity in hydrolyzing dUTP into dUMP and thus reduces the dUTP/dTTP ratio, which, otherwise, would compromise DNA integrity.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of DR2231
The open reading frame annotated as DR2231 was amplified by PCR from the genomic DNA of D. radiodurans. The PCR product comprising the 5′-CACC overhang was inserted into the expression pET151/D-TOPO directional vector (Invitrogen) with His6 at the N terminus and a recombinant tobacco etch virus protease cleavage site. The resulting expression vector was used to transform E. coli BL21 (DE3) competent cells (Invitrogen). Transformed cells carrying pET151-DR2231 were grown in Luria-Bertani enriched growth medium containing 100 μg/ml ampicillin; the medium was inoculated with a sample of an overnight culture and grown at 310 K until an optical density of ∼0.65 at 600 nm was reached, at which point isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.3 mm, thus inducing overexpression. Cultures were allowed to grow for a further 4 h, after which the cells were harvested at 7000 × g for 20 min at 277 K and resuspended in lysis buffer (50 mm Tris-HCl, pH 7.0, 300 mm NaCl, 2 mm β-mercaptoethanol, 5% (v/v) glycerol). The cells were frozen at 193 K, thawed, and lysed by two passes through a French press. DNase I was added to a final concentration of 20 μg/ml together with an EDTA-free protease inhibitor tablet (Roche Applied Science). The lysed culture was centrifuged at 40,000 × g for 30 min at 277 K, and the soluble fraction was loaded onto a 5-ml His-Trap column (GE Healthcare) equilibrated with buffer A (50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 5% (v/v) glycerol). The first wash consisted of a step elution to 12% buffer B (buffer A supplemented with 500 mm imidazole) followed by a gradient elution from 12–100% buffer B. DR2231 was eluted and pooled near 250 mm imidazole, as confirmed by SDS-PAGE. The pooled fractions were exchanged into a suitable buffer for His tag cleavage by enzymatic digestion with tobacco etch virus protease. The tag-free protein was again loaded onto a 5-ml His-Trap column, and the flow-through was collected. Further purification was performed, and oligomerization states were confirmed using a Superdex S200 HRHiPrep 16/60 column (GE Healthcare) equilibrated with 25 mm Tris-HCl, pH 8.0, 150 mm NaCl. The oligomerization state and monodispersity of DR2231 in solution were estimated by dynamic light scattering (Malvern Instruments, Zetasizer, Nano series). The purified protein was concentrated to 7.5 mg/ml and cryo-cooled to −70 °C when necessary.
Crystallization, Data Collection, and Structure Determination
DR2231 was successfully crystallized in three different crystallization conditions, each producing a different crystal form. All conditions were obtained through an initial screening (with or without preincubation of substrates or analogues) using the high throughput crystallization robot, Cartesian PixSys, and automated imaging system, both available at the High Throughput Crystallization Laboratory at EMBL (Grenoble, France) (9). The first crystals (crystal type A) appeared in 0.01 m magnesium chloride, 0.05 m sodium cacodylate, pH 6.0, and 1 m lithium sulfate (Natrix screen formulation, Hampton Research). Crystals were reproduced without further optimization and achieved dimensions of ∼0.4 × 0.3 × 0.2 mm within 4–5 days. For data collection, 16% (v/v) glycerol was included in the mother liquor as cryoprotectant, and crystals were immediately flash-cooled in liquid nitrogen (100 K) after soaking for a few seconds. The second crystal form (crystal type B) refers to crystals obtained from 0.18 m lithium acetate and 20% (w/v) PEG 3350 (optimized from the PEG/Ion Formulation Screen, Hampton Research). Prior to setting up of the crystallization drops, protein was incubated with 10 mm dUTP and 10 mm manganese chloride (DR2231_dUMP_Mn) or 10 mm magnesium chloride (DR2231_dUMP_Mg). Single crystals grew in 7–8 days as thick plates. These were cryoprotected by soaking in crystallization solutions progressively increased in glycerol content up to 16% (w/v). The third successful crystallization condition (crystal type C) refers to crystals with a diamond-like shape that grew in 1 m LiCl, 0.1 m citric acid, pH 5, 10% (w/v) PEG 6000 (obtained from Grid Screen PEG6000 KIT, Hampton Research). DR2231 protein was co-crystallized in the presence of 10 mm dUpCpp or dUpNHpp and 10 mm MgCl2 (or MnCl2). These crystals were collected directly from the robot screen and subjected to cryogenic cooling after immersion in paraffin oil.
Type A crystals belong to the space group P212121 with six molecules in the asymmetric unit and were used in three different data collection experiments: DR2231_Apo_1, DR2231_Mn, and DR2231_Gd. For de novo phasing (DR2231_Gd), a heavy atom derivative crystal was prepared by soaking crystal of type A in 20 mm GdCl3 added to the mother liquor for ∼18 h. DR2231_Mn corresponds to a data set collected on a type A crystal that was soaked overnight in a modified precipitant: 250 mm lithium sulfate, 750 mm manganese sulfate.
For the crystal type B form, the space group was P21212 with four molecules in the asymmetric unit, arranged in two dimers. This crystal type has been pursued in order to obtain a protein complexed with the substrate or the product in the active site and with the metal atoms.
Crystal type C was produced in order to obtain DR2231 with a bound non-hydrolyzable analog and followed the unsuccessful trials with type B. Unfortunately, neither crystal form when examined revealed the substrate analog in the postulated active site. Crystals obtained correspond to another apo form (similar to crystal type A) and belong to space group P6122 with one molecule in the asymmetric unit, the dimer being generated by a crystallographic 2-fold axis.
All data collection statistics are summarized in Table 1. Diffraction data from crystal type A were processed with MOSFLM (10) and scaled with SCALA (11). Data from crystal forms B and C were integrated and scaled with XDS (12). In both cases, further data analysis was carried out using the CCP4 suite 6.0.2 (13).
TABLE 1.
Data collection statistics
Numbers in parentheses refer to the highest resolution shell.
| Data collection | Crystal type A |
Crystal type B |
Crystal type C |
|||
|---|---|---|---|---|---|---|
| DR2231_Gd | DR2231_Apo1 | DR2231_Mn | DR2231_dUMP_Mg | DR2231_dUMP_Mn | DR2231_Apo2 | |
| Beamline | ID14-4, ESRF | ID14-4, ESRF | ID23-2, ESRF | ID29, ESRF | ID29, ESRF | ID29, ESRF |
| Space group | P2121 21 | P2121 21 | P2121 21 | P2121 2 | P2121 2 | P6122 |
| Unit cell (Å) | ||||||
| a | 67.8 | 67.8 | 68.1 | 78.1 | 78.7 | 62.9 |
| b | 110.8 | 111.4 | 111.4 | 149.5 | 150.2 | 62.9 |
| c | 166.0 | 166.1 | 166.5 | 52.4 | 52.4 | 167.6 |
| Resolution (Å) | 83-2.00 (2.11-2.00) | 20-1.70 (1.79-1.70) | 50-2.00 (2.11-2.00) | 50-1.77 (1.86-1.77) | 40-1.90 (2.00-1.90) | 45-1.90 (2.00-1.90) |
| Wavelength (Å) | 1.24 | 1.24 | 0.873 | 0.972 | 0.976 | 0.976 |
| No. of unique reflections | 85,194 (12,299) | 137,940 (19,910) | 86,122 (12,456) | 57,356 (6495) | 48,355 (6658) | 16,343 (2266) |
| Multiplicity | 8.1 (8.2) | 5.7 (4.3) | 4.0 (4.0) | 5.1 (4.1) | 3.4 (2.9) | 6.7 (6.8) |
| Completeness (%) | 100 (100) | 99.8 (99.7) | 99.8 (100) | 94.3 (75.0) | 97.8 (93.4) | 99.9 (99.7) |
| Mean (I/σI) | 13.4 (4.1) | 16.0 (7.1) | 15.0 (6.8) | 17.2 (3.1) | 11.2 (2.7) | 12.2 (3.7) |
| Rmergea (%) | 10.7 (45.4) | 7.9 (13.2) | 7.2 (36.9) | 7.5 (46.2) | 7.0 (35.8) | 9.7 (40.2) |
| Rpimb (%) | 5.9 (25.2) | 3.2 (6.8) | 4.0 (21.5) | 3.3 (24.4) | 4.4 (23.1) | 3.6 (15.3) |
| Ranomc (%) | 6.6 (19.3) | |||||
| FOMd (before/after DM) | 0.359/0.706 | |||||
| SHELXD CC/CC (weak) | 48.90/28.90 | |||||
a Rmerge = Σ|Iobs − 〈I〉|/Σ〈I〉.
b Rpim = Σ(1/(N − 1))½Σ|Iobs − 〈I〉|/Σ〈I〉.
c Ranom = Σ|〈I+〉 − 〈I−〉|/Σ(〈I+〉 + 〈I−〉).
d Figure of merit.
The structure of DR2231 was solved by single wavelength anomalous dispersion from a highly redundant data set from the gadolinium derivative collected to 2.0 Å resolution and at a wavelength of 1.245 Å using an x-ray at the European Synchrotron Radiation Facility (ESRF) beam line ID14eh4 (14). After analyzing the structure factors using SHELXC and SHELXD with HKL2MAP version 0.2 (15, 16), the positions of one gadolinium site per monomer could be determined. Initial phases were calculated with SHELXE (17), which also applied density modification. Structure factors and density modified phases were input in ARP/wARP (18), which produced, with the automatic model building feature, a partial starting model. The initial model was then completed by rounds of manual building with Coot (19) and refinement with REFMAC (20). The structure of the native protein was obtained by rigid body refinement using the gadolinium derivative structure as an initial model; for space groups other than P212121, molecular replacement with the program Phaser (21) using the gadolinium derivative structure as a starting model was used for structure determination. Protein/substrate structures were determined again with Phaser using the higher resolution native structure as a search model.
All structures were refined using bulk solvent correction and a maximum likelihood target function. Resulting models were all subject to translation/libration/screw (TLS)4 refinement within REFMAC (20), using the rigid and mobile domains as separate TLS groups except for DR2231_dUMP_Mn and DR2231_dUMP_Mg, which were refined with PHENIX, in which TLS groups were defined differently for the open and closed conformations. TLS groups were chosen after analysis of the structure with TLSMD (22). All of the data measured, apart from a 5% test set used for Rfree calculation, were included in the refinement. The quality of the structures was assessed using PROCHECK (23) and MolProbity (24). Refinement statistics and geometry analysis of the structures are summarized in Table 2. Figures representing protein structures were prepared with PyMOL (25). All six structures were deposited in the Protein Data Bank (PDB) under accession codes 2YEU (DR2231_Gd), 2YF4 (DR2231_Apo1), 2YF9 (DR2231_Apo2), 2YF3 (DR2231_Mn), 2YFD (DR2231_Mg_dUMP), and 2YFC (DR2231_Mn_dUMP).
TABLE 2.
Crystallographic refinement statistics
The overall measure of structure quality was from PROCHECK (21).
| Crystal type A |
Crystal type B |
Crystal type C |
||||
|---|---|---|---|---|---|---|
| DR2231_Gd | DR2231_Apo1 | DR2231_Mn | DR2231_dUMP_Mg | DR2231_dUMP_Mn | DR2231_Apo2 | |
| Rcryst (%)a | 18.8 | 20.9 | 19.1 | 18.4 | 17.8 | 18.8 |
| Rfree (%)b | 23.2 | 23.2 | 23.8 | 21.1 | 21.9 | 22.5 |
| Root mean square deviations | ||||||
| Bond lengths (Å) | 0.017 | 0.013 | 0.020 | 0.010 | 0.007 | 0.02 |
| Angles (degrees) | 1.526 | 1.389 | 1.525 | 1.058 | 1.082 | 1.09 |
| Planar groups (Å) | 0.008 | 0.008 | 0.009 | 0.005 | 0.005 | 0.01 |
| Chiral centers (Å3) | 0.140 | 0.088 | 0.102 | 0.073 | 0.059 | 0.09 |
| Average B-factor (Å2) | ||||||
| Overall (Wilson B) | 25.0 (21.2) | 25.7 (20.8) | 25.6 (19.2) | 21.5 (18.6) | 24.5 (20.3) | 25.4 (23.2) |
| Main chain | 23.2 | 23.4 | 23.8 | 19.1 | 21.5 | 20.6 |
| Side chain | 26.5 | 27.6 | 27.2 | 24.4 | 27.4 | 27.8 |
| Solvent | 28.4 | 30.8 | 28.9 | 26.3 | 26.2 | 35.3 |
| Metal | 23.4 | 19.9 | 28.6 | 21.9 | ||
| Ligand | 13.2 | 13.0 | ||||
| Ramachandran plot (21) (%) | ||||||
| Most favored | 97.0 | 96.9 | 98.1 | 98.9 | 98.1 | 98.5 |
| Allowed | 3.0 | 3.00 | 1.9 | 1.1 | 1.9 | 1.5 |
| Outliers | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 |
a Rcryst = Σhkl‖Fo| − |Fc‖/Σhkl|Fo|.
b Rfree was calculated with a small fraction (5%) of randomly selected reflections.
NTP Pyrophosphatase Assay
NTP pyrophosphatase assays for DR2231 were performed using the malachite green phosphate assay (26, 27). For ELISA plates (200-μl total volume/well) DR2231 (2 μg or 2 ng, as indicated) in reaction buffer (100 mm Tris/HCl, pH 7.2, 20 mm MgCl2) was incubated with varying concentrations of NTPs or dNTPs, ranging from 1.0 to 20 μm at room temperature (delivered from 100-fold stocks) for 10 min. Each reaction contained inorganic pyrophosphatase enzyme (0.05 unit) (Sigma) in order to convert released PPi into Pi and allow full binding to malachite green dye. Reactions were terminated upon the addition of 40 μl of dye reagent, whereupon the dye color changes from orange to green after 10 min. Control reactions lacking DR2231 were performed for each substrate (10 μm) in the presence of inorganic pyrophosphatase, in order to guarantee the absence of β-γ phosphate cleavage. Another set of control reactions lacking inorganic pyrophosphatase produced no color development, further proving that DR2231 releases PPi and not phosphate in free form.
Initial velocities were followed by incubating a 1.8 ml of reaction buffer at 37 °C, with agitation, in the presence of inorganic phosphatase (0.05 unit) and substrate. DR2231 at 0.634 nm (2 ng/μl) was added, and 160-μl aliquots were withdrawn at 0, 1, 2, 4, 6, 8, and 10 min and added to 40 μl of malachite green dye each. After a 10-min incubation, absorbance was measured at 630 nm; phosphate quantification was calculated against a previously determined calibration curve for inorganic phosphate in the buffer conditions used. For metal preference tests, magnesium was replaced by 20 mm MnCl2, CaCl2, ZnCl2, NiCl2, or EDTA.
Isothermal Calorimetry (ITC) Assay
Isothermal calorimetry was carried out using a VP-ITC titration microcalorimeter (Microcal Inc., Northampton, MA). Reaction cells were filled with solutions and equilibrated at the experimental temperature. After equilibration, an additional delay period was allowed to generate the base line used in subsequent data acquisition. All solutions were degassed prior to an experiment. The stirring speed was set at 326 rpm, thermal power was recorded every 2 s, and heat flow (reference power of 15 μcal/s) was recorded as a function of time.
Ligand solutions were prepared in the buffer from the last dialysis change. ITC measurements were routinely performed in 20 mm Tris/HCl, pH 7.0, 5 mm NaCl, 25 mm MgCl2 (or MnCl2). Controls were performed by injecting ligand into buffer under identical experimental conditions. Raw data were collected and corrected for the ligand heat of dilution. Kinetic data analysis was determined by the single injection method, where the change in thermal power as substrate is depleted can be continuously monitored (28, 29).
RESULTS
Overall Fold of DR2231 and Structure of the Dimer
DR2231 is a single chain protein composed of 148 amino acids with an estimated molecular mass of 16,056 Da. The recombinant protein reported here corresponds to the full-length sequence and includes six extra N-terminal residues from the cloning vector (Gly, Ile, Asp, Pro, Phe, Thr). Three different crystal forms belonging to three different space groups, P212121, P21212, and P6122, were obtained, depending on whether a substrate (or substrate analog) was used for co-crystallization or not. All crystal forms show the same basic fold.
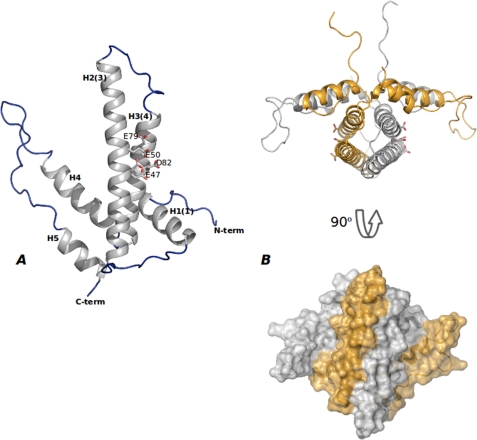
The three-dimensional crystal structure of DR2231 shows that it is mostly composed of α-helices (Fig. 1A) forming a hairpin-like fold. The monomer comprises five helices (H1, residues Thr9–Gly22; H2, residues Pro34–Ala64; H3, residues Ser69–Asp92; H4, residues Asp97–Ser114; H5, residues Asp133–His143) and four connecting loops. H2 packs against H3 in an antiparallel sense, whereas H5 is stacked against H4. Both pairs of stacked helices compose the hairpin topology with the vertices formed by loops 1 and 3 and distant from each other by ∼40°.
FIGURE 1.
Overall shape of DR2231 protein. A, monomeric DR2231 protein structure. DR2231 is an all-α-helix structure with an overall hairpin-like fold. Secondary structure elements are labeled both according to sequence and to the convention of Moroz et al. (7) (in parenthesis) for facility of comparison with other dUTPase/MazG proteins. The EEXX(E/D) motif is presented (Glu47, Glu50, Glu79, and Asp82); Glu47 appears in a double conformation when in the absence of the divalent metal cation. B, DR2231 dimer in open conformation, presented also in orthogonal view. A surface representation illustrates extension of the interface in the dimer.
The dimer (Fig. 1B) is composed of two monomers with essentially the same structure, interacting in a head to tail orientation with overall approximate dimensions of 45 × 49 × 97 Å. Upon dimer formation, helices 2 and 3 from one monomer stack antiparallel to helices 2′ and 3′ of the other monomer, respectively. A stable four-helix bundle is formed in the center. On either side, flanking the central bundle, are the “winged” regions composed of H1 from one monomer and H4′ and H5′ from the other. Helix 1 is anchoring H4′ into its position relative to H3 and gives further stability to the “fork.” The intertwining of the two hairpin structures produces an extensive subunit-subunit interface, burying a total surface of 2832 Å2, where, among the 73 residues participating in the interface, 58% are non-polar and 42% are polar (analyzed using the protein-protein interaction server (30)). This value increases to ∼2922 Å2 for the dimer in complex with dUMP. Interestingly, the long loop (residues 23–33) connecting H1 and H2 that seems to stabilize the residues in the kink (residues 93–96) between H3 and H4, confers further sturdiness to the rigid helix bundle. A considerable array of interactions secures this stabilization through hydrogen bonds: Ala23 (N) with His18 (C=O); Ala24 (C=O) with Gln93 (Nϵ2); Thr25 (C=O) with His15 (Nδ) and Asp92 (Oδ2); Arg28 (C=O) with Thr30 (Oγ1) and Gly95 (C=O); Pro29 (N) with Gly95 (C=O); Thr30 (N) with Leu94 (C=O); Pro33 (C=O) with Leu36 (N) and Glu35 (N). There is yet another hydrogen bond between Pro32 (C=O) and Gln62 (Nϵ2) in helix 2 of the partner monomer.
Structures belonging to the P212121 space group (6 molecules/asymmetric unit) (DR2231_Gd, DR2231_Apo_1, and DR2231_Mn) show very good electron density for the N-terminal residues. These residues (Gly−5 to Pro8), including those provided by the cloning vector, are anchored in the substrate binding pocket of a symmetry-related molecule. The disulfide bond formed between Cys6 residues of each dimer is an artifact induced by crystallographic packing in this crystal form and does not appear in the other two forms reported. The final five residues (Ala144–Asp148) are always unstructured, with the exception of some chains in the DR2231_Mn structure. Structures determined with crystal type B (4 molecules/asymmetric unit; DR2231_dUMP_Mg and DR2231_dUMP_Mn) showed electron density attributable to residue Pro7 and onward. One monomer in each dimer lacks density for residues 116–123 as well as for residues 145–148. Finally, the structure corresponding to space group P6122 (DR2231_Apo_2) holds one protein chain per asymmetric unit, and, similar to the P212121 structures, the first four N-terminal residues from a symmetry-related molecule are imbedded in the active site.
In accordance with the literature (6–8), compared with dUTPases and MazG proteins, DR2231 also holds the Mg2+ (or Mn2+) coordination motif, EXXE12–28EXXD, crucial for biocatalysis. For DR2231, however, the first metal binding conserved motif is EEXXE, similar to all MazG proteins, whereas the second conserved motif, EXXD, does not have the extra glutamate, similar to most dimeric dUTPase conserved motifs (Fig. 2). The Mg2+-coordinating side chains in DR2231 are Glu47, Glu50, Glu79, and Asp82, the first two located on H2 and the latter two on H3, and are directed toward the pocket formed by the central helix bundle and the “winged” helical elements. This crevice composes the substrate binding pocket (described below). The loop between H4 and H5 (Ser114–Glu127) is very mobile and may possibly have a role as a lid for substrate accessibility and recognition. Different structures of DR2231 reported here have shown this loop in an “open” conformation in the P212121 and P6122 structures (DR2231_Apo1, DR2231_Mn, and DR2231_Apo2) and in a “closed” conformation for those complexed with the product, dUMP.
FIGURE 2.
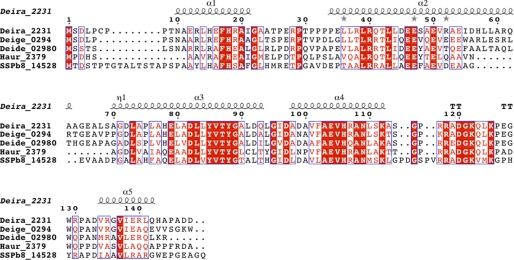
Sequence alignment of DR2231 with members of the dUTPase and MazG subfamilies. The alignment was constructed on the basis of a ClustalW 2.0.12 multiple sequence alignment (58) with minimal manual editing. Experimentally determined crystal structures are available for all listed entries: Bsub_YpjD (PDB code 2GTA), Vibrio_iMazG (PDB codes 2Q5Z and 2Q73), Mmus_RS21-C6 (PDB code 2OIE), Ecol_MazG (PDB code 3CRC), Ssol_MazG (PDB code 1VMG), and Cjej_1451 (PDB code 1W2Y). The proteins are listed by name of the source organism, followed by the protein name; the third column corresponds to UniProt identifiers, and the fourth column shows the percentage of identity with DR2231 sequence. Sequence numbers preceding and following matched segments are indicated. The structural elements shown above the alignment correspond to DR2231 determined in this work. Cylinders indicate α-helices, and the numbering of helices in parenthesis corresponds to the convention established by Moroz et al. (7). The Mg2+-binding residues are shown in red boldface type, and conserved residues are colored as follows: hydrophobic (yellow background), aromatic (blue boldface type), Asp/Asn/Glu/Gln (green boldface type). The E. coli MazG alignment is performed only on the enzymatically active C-terminal domain (residues 136–263); the C. jejuni dUTPase sequence was truncated at residue 143.
Comparison with dUTPase and MazG Structures
As already mentioned, the divalent ion binding motif is the identity signature for dUTPase/dCTPase, MazG, and HisE proteins. Outside this stretch of residues, sequence homology is usually poor even among subfamily members (Fig. 2). However, despite low sequence homology with other family members, the structure similarity is strikingly high when compared with known structures of both dimeric dUTPases and MazG proteins.
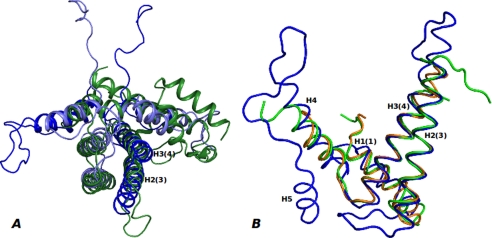
By superimposing the structures of a dimeric dUTPase (C. jejuni dUTPase, PDB code 1W2Y) and two MazG structures (S. solfataricus MazG, PDB code 1VMG; Bacillus subtilis YpjD, PDB code 2GTA) with DR2231_Mn, the structural elements in common become evident, highlighting the basic module as the unifying structural element throughout the all-α-NTP pyrophosphohydrolase superfamily. For C. jejuni dUTPase, the structural overlay is performed with the monomer on the DR2231 protein dimer; the superimposition clearly supports the notion of a gene duplication of the basic module for the dUTPase (Fig. 3A). Within this four-helix bundle, two helices (DR2231 H2(3) and H3(4),5 from monomer A, aligning with C. jejuni dUTPase H3 and H4, respectively) are particularly rigid. The perfect conservation of the Mg2+-binding motif is noteworthy. In contrast to the DR2231 protein dimer, which holds two putative active sites, the C. jejuni dUTPase monomer, despite structural resemblance, has evolved into an enzyme with a single active site. Besides these features, no other similarity between the proteins is found from the superposition.
FIGURE 3.
Overlay of DR2231 structure with known dimeric dUTPase and MazG structures. A, DR2231 dimer (monomer A (dark blue) and monomer B (light blue)) superimposed with C. jejuni dUTPase monomer (green). B, DR2231 structure (blue) overlay with S. solfataricus (PDB code 1VMG; orange) and B. subtilis (PDB code 2GTA; green) MazG structures.
Superposition of the structure of DR2231 with “shorter” versions of MazG structures produced very good overlays (Fig. 3B). For B. subtilis YpjD protein (PDB code 2GTA), the calculated root mean square deviation over all Cα atoms is <2.01 Å, and for S. solfataricus MazG (PDB code 1VMG), the root mean square deviation is <1.85 Å. The overlay includes elements beyond the rigid helices H2(3) and H3(4) and is extended to the entire basic module, including H1, H4, and the loop connecting H3 and H4. In overlaid structures, the magnesium-binding residues are wedged between H2(3) and H3(4) and are structurally coincidental with those of DR2231. No similarity is found in the loops between the helices H1, H2, and H3. There is no structural equivalent for the loop between H4 and H5 (which is involved in substrate recognition) and the H5 helix itself in either MazG structure.
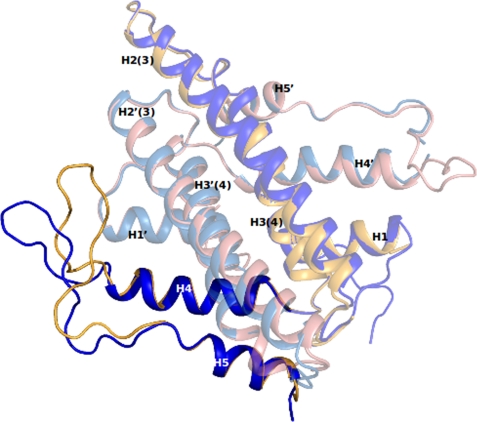
Open and Closed States of DR2231
The superposition of a native structure of DR2231 protein (DR2231_Apo1, DR2231_Apo2, or DR2231_Mn) with a structure in complex with product (DR2231_dUMP_Mg or DR2231_dUMP_Mn) shows major conformational changes concerning H5 and the lidlike loop between H4 and H5 (Fig. 4). The loop region (residues Ser114–Glu127) contains the only β-turns present in DR2231; the first is a type I β-turn and comprises residues Arg118–Gly121, and the second is a type II β-turn encompassing residues Pro126–Trp129.
FIGURE 4.
Open (blue) and closed (yellow) conformations of DR2231. Changes are confined to the “loop” region encompassing residues Lys112 to Pro131.
DR2231_dUMP_Mg and DR2231_dUMP_Mn structures (P21212; with two dimers in the asymmetric unit) configure two situations; in monomers A and C, the active sites are occupied with a dUMP molecule each and the lidlike loops in this conformation that correspond to the closed state. Interestingly, the second situation shows the respective monomers, B and D, with an unoccupied active site and a flexible latch such that no electron density is detected for residue range 116–123. Nevertheless, this conformation suggests the true marked mobility of the lidlike loop when in the open state.
Structural overlay of DR2231_dUMP_Mn and DR2231_Apo1 shows that the latch is displaced ∼10 Å when in the presence of the product, with a rigid rotation of the residue 109–131 fragment of about 30°.
Divergence between open and closed states appears at the end of H4 with residues Asn109–Lys112 (Fig. 4). The Nδ2 from the side chain of Asn109 is bound to a water molecule, whereas the Oδ1 is bound to two waters in the open state; in the closed state, Nδ2 hydrogen binds O3′ of the deoxyribose, and Oδ1 keeps one of the bound water molecules and simultaneously makes a hydrogen bond to the O4′ of the deoxyribose of dUMP and the Nζ of the Lys112 side chain.
The torsion angle (Ψ) of Ser111 changes considerably between open and closed structures, and this implies major modifications on Lys112, changing in particular the direction of its side chain and the number of hydrogen bonds in which this residue is involved. The nitrogen of the peptide bond is hydrogen-bound to the carbonyl oxygen of Ala108 in the open state, but in the closed state, this bond is broken, and the nitrogen becomes involved in two different hydrogen bonds: with the carbonyl oxygen of Asn109 and Leu110, simultaneously. This results in a reorientation of the side chain, and the Nδ2 is directed toward the cavity in order to bind O4′ (of the deoxyribose) and O5′ in dUMP, two water molecules, and Oδ1 of Asn109. The torsion angles of Ala113 and Ser114 (the side chain Oγ, bound to a water molecule in the open state, has a hydrogen bond with the carbonyl oxygen of Ser111 in the closed state) change a great deal as a result of these rearrangements. Curiously, both Gly115 and Pro116 keep their hydrogen bonding intact in both conformations, and only minor rearrangements occur with the residues that follow up to residue 122. Here, both Lys122 and Gln123 play an important role in substrate recognition and participate in the active site arrangement. Lys122 Nζ is pointing outside the substrate cleft in the absence of dUMP, but once in the presence of the product, the peptide nitrogen binds to Oδ1 of Asp120, and Nζ binds in a tetrahedral fashion to a water molecule, Oϵ1 and Oϵ2 of Glu46 of the second subunit, and OP1 (of the α-phosphate coordinated by Mn2) of dUMP. The Oϵ of Gln123 loses a solvent water molecule and becomes bound to Nϵ and Nη1 of Arg117, whereas the Nϵ2 of Gln123 binds the carbonyl oxygen of Gly115 in both states, but in the closed state, it also binds the carbonyl oxygen of Lys112. This observation is particularly relevant because the angle produced by the loop movement upon substrate/product binding seems to be stabilized by this hydrogen bond. Another interesting feature of this residue is that the carbonyl oxygen that binds a bridging water is bound in turn to OP3 of dUMP, whereas the nitrogen of the peptide bond is binding O4 of the uracil in dUMP. Finally, another important residue involved in product binding is Lys125, which normally has the side chain Nζ hydrated by two water molecules but in the closed state binds OP2 (of the α-phosphate) of dUMP.
Comparing DR2231 with the dUTPases and MazG structures available, a lack of structural equivalents to this loop is evident. The closest possible resemblance would come from C. jejuni dUTPase (PDB code 1W2Y) rather than MazG structures. C. jejuni dUTPase reveals a short latch between helices 3 and 4 that is swapped between subunits and reaches into the active site region. This latch contains residues responsible for substrate binding and specificity. iMazG from Vibrio sp. DAT722 (PDB codes 2Q5Z and 2Q73) exhibited an open and closed conformation contributed by a moveable helix, H1, postulated to be involved in substrate capture. Nevertheless, in all structures of DR2231 protein, H1 is rigid and shows no movement or reorientation relative to the central helix bundle.
The structural elements from the lidlike latch in DR2231 protein responsible for substrate recognition and catalysis only partly overlap with structural elements with the same role in C-terminal E. coli MazG (PDB code 3CRC) (namely H10 and the short loop between H6 and H7). This difference strongly suggests that the high structural variability in the vicinity of the binding pocket, other than the rigid helix bundle, conveys to the enzyme its specificity toward substrates and establishes the subfamily identity.
Structure of the Active Site; Mg2+ Versus Mn2+
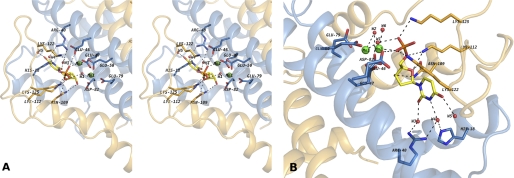
The wild-type DR2231 protein was co-crystallized with the reaction substrate, dUTP, and in the presence of Mg2+ or Mn2+. In both crystal structures obtained (DR2231_dUMP_Mg and DR2231_dUMP_Mn), well defined electron density was located in the ligand binding pocket and subsequently modeled as the hydrolyzed product, dUMP, in one active site per dimer (Fig. 5, A and B).
FIGURE 5.
A, stereo view of the putative binding site of DR2231 in the closed conformation bound to dUMP. The active site shows the contribution of residues from both monomers to substrate binding and coordination. When crystallized in presence of Mg2+, DR2231 presents only one divalent metal-coordinated equivalent to Mn1; a water molecule was modeled in the position of Mn2 (not shown). B, close-up of the binding pocket highlighting the role of structured waters in catalysis (W1 and W2), based on homology with C. jejuni dUTPase and substrate binding (W3, W4, and W5). W6 participates in the hexacoordination shell of Mn2.
In DR2231_dUMP_Mg structure, one magnesium ion is present per subunit, each coordinated in the complexed subunit. The Mg2+ is coordinated by three conserved glutamate residues (Glu47, Glu50, and Glu79) and one conserved aspartate (Asp82) (Fig. 5, A and B). Bond distances correspond well to those expected for the Mg2+ ion (31, 32). A further coordination is established with water W1, which is bridging between the metal and the α-phosphate of the product.
Regarding DR2231_dUMP_Mn, the unoccupied active sites hold only one Mn2+ ion (Mn1) with an identical coordination to DR2231_Mn of crystal type A (and identical to gadolinium coordination in DR2231_Gd). In the dUMP-occupied site, two manganese divalent cations (Mn1 and Mn2) can be modeled. Mn1 is in the same position observed for Mg1, whereas Mn2 is found in the vicinity of the first and is always hexacoordinated to Glu46 (Oϵ2), Glu47 (Oϵ2), and Glu50 (Oϵ1) as well as two water molecules and OP1 of the α-phosphate of dUMP (Fig. 5A). W1 water is located in the same position as in DR2231_dUMP_Mg and is coordinated between the two Mn2+ ions. The two Mn2+ atoms have been confirmed by anomalous difference Fourier map. In the DR2231_dUMP_Mg structure, a water molecule was modeled in the position of Mn2 because the coordination distance was not typical for an Mg2+ ion. An overlay of both structures, DR2231_dUMP_Mg and DR2231_dUMP_Mn, gives an excellent superposition with a minor exception for the residues surrounding Mn2. In the presence of Mg1, Glu50 is bound to this metal in a monodentate manner, through Oϵ1, binding also the “bridging” water and a second water molecule. Glutamate 46 side chain is pointing away from this region. However, for DR2231_dUMP_Mn, when Mn2 is in the position occupied by the second water in DR2231_dUMP_Mg, the side chain of Glu50 rotates in order to to bind Mn1 and Mn2 in a bidentate fashion. The presence of Mn2 reorients Glu46, which in turn is also binding an oxygen atom from the α-phosphate of dUMP.
It is not known whether DR2231 binds manganese in vivo. Enzymatic studies reported here have shown a similar preference for both divalent cations. The first crystallization condition obtained (P212121) produced an apo form of the protein with no Mg2+ in the metal-binding site (DR2231_Apo1), although attempts to produce crystal type A with Mg2+ in the metal-binding site by soaking, co-crystallization, or replacing Li2SO4 with varying concentrations of MgSO4 from 250 to 750 mm, failed. In contrast, the same procedure was reproduced now with varying concentrations of manganese sulfate; all soaks produced structures holding high occupancy of the Mn2+ ion coordinated by the EXXE28EXXD motif. This may be explained by the more labile nature of magnesium coordination when compared with manganese (33).
Specific Substrate-interacting Residues of the Active Site
The environment composing the substrate binding pocket is composed by both hydrophobic and polar residues. The uracyl ring is settled between hydrophobic residues Phe17 and Leu43, each one making van der Waals interactions on both sides of the moiety. Furthermore, O2, N3, and O4 of the uracyl moiety interact with three water molecules, with W3, W4, and W5, respectively (Fig. 5B). W3 bonds to Arg40 (Nη1) and Arg40 (Nη2), and W4 bonds to Arg40 (Nη1) and His18 (Nϵ2), whereas W5 is involved in a hydrogen bond to W4. These two uracil-anchoring residues, His18 and Arg40, are highly conserved within the DR2231 subfamily (Fig. 9).
FIGURE 9.
Sequence alignment of DR2231 with other putative subfamily members: Deira_2231 (D. radiodurans, Q9RS96), Deige_0294 (Deinococcus geothermalis, Q1J1N7), Deide_02980 (Deinococcus deserti, C1CZ79), Haur_2379 (Herpetosiphon aurantiacus, A9AYP1), and SSPb78_14528 (Streptomyces sp. SPB78, UPI0001B553EB). Identical residues are shown in white on a red background, and similar residues are shown in red on a white background. For the secondary structure assignment, α-helices are represented as helices, β-strands are represented as arrows, and β-turns are marked TT. This figure was prepared with ESPript (57). The multiple-sequence alignment was performed with ClustalW (version 2.0.12) (58).
Atom O4 of the uracyl is also bound to the nitrogen of the peptide bond of Gln123. This suggests that there may still be space for dATP or dGTP, depending on the extent of accommodation given by the lidlike latch, although a similar network of interactions as seen with dUMP is not foreseeable with these two nucleotides. On the other hand, the proximity of the backbone of the lidlike loop of the second subunit appears to make steric hindrance toward the methyl group from thymine.
One of the divalent binding residues, Asp82 (through Oδ1), coordinates the O3′ on the deoxyribose moiety, along with Asn109 (from the second subunit). In the vicinity of the C2′ of the deoxyribose are Tyr85 and Val86, whose hydrophobic character would not leave room for the 2′-hydroxyl group of a ribose moiety. Conservation of hydrophobic residues in this region is a feature among dimeric dUTPases, dCTPases, and HisE families, but not the MazG family. In the DR2231 subfamily, Tyr85 and Val86 are included in a highly conserved stretch of residues (positions 85–89). There is another feature quite exclusive to DR2231 concerning the deoxyribose; Lys112 binds both O4′ and O5′, securing a proper orientation of the triphosphate moiety for nucleophilic attack of the catalytic water. This residue is strictly conserved within the subfamily.
Finally, the α-phosphate is bound by residues: Glu46 (when the second divalent cation is present), Lys122, and Lys125. The latter two belong to the swapped lidlike loop, a hallmark motif of this subfamily.
(Deoxy)NTP Pyrophosphatase Activity
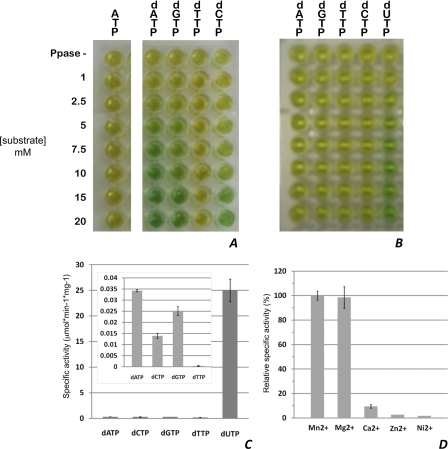
Recent literature relative to enzymatic activity of MazG (nucleoside triphosphate pyrophosphohydrolase)-related proteins has suggested that these enzymes are active on canonical (d)NTPs (8, 34, 35). However, there has also been debate as to whether MazG-like proteins may not be equally involved in the removal of noncanonical nucleotides (36). Purified DR2231 was tested for (d)NTP-PPase activity against ATP, GTP, UTP, CTP, dATP, dGTP, dTTP, dCTP, and dUTP, using an indirect colorimetric malachite green assay. Cleavage of the NTP is expected to occur at the α-β phosphate bond, with the release of PPi as one of the products. Proper binding with the dye and colorimetric measurement requires pyrophosphate to be further cleaved into free Pi. To this end, inorganic phosphate is used, and magnesium must be present in the assay. A simple setup allows quick evaluation of specificity (Fig. 6, A and B).
FIGURE 6.
Enzymatic activity of DR2231. A and B, screening of purified DR2231 for NTP-PPase activity against (d)NTPs in the presence of 0.623 μm (A) and 0.623 nm enzyme (B). The top row (labeled PPase -) in both panels corresponds to screening of each substrate at 10 μm in the absence of inorganic phosphatase. Hydrolysis of ATP is representative of all NTPs. C, specific activities of DR2231 (0.623 nm) at 5 μm substrate; the inset corresponds to determination of specific activities of substrates at 0.623 μm. D, divalent metal preference of DR2231 for 5 μm dUTP hydrolysis; depicted is the percentage of specific activity of the enzyme in the presence of a divalent metal relative to the specific activity in the presence of Mn2+. Enzymatic activity in the absence of divalent metals or in the presence of 20 mm EDTA was not detectable. For details, see “Experimental Procedures,” NTP Pyrophosphatase Assays, Error bars, S.D.
No hydrolysis was observed for canonical NTPs (ATP, GTP, CTP, and UTP). The same amount of enzyme had no specificity toward dTTP. The general order of reactivity for dNTPs is dUTP ≥ dATP > dGTP > dCTP ≥ dTTP. Note that the hydrolysis of dUTP was by far strongest. This screen was repeated in the presence of a 103-fold dilution of enzyme, showing a strong specificity for dUTP and an almost negligible specificity for the others (Fig. 6B). Linear velocities at a 5 μm concentration of each of these dNTPs were performed, confirming the results obtained from the screen (Fig. 6C). As expected, DR2231 cleaves between the α-β phosphate bond, releasing (deoxy)NMP and PPi. Quite unexpected was its utter preference for dUTP, although all reports available on MazG proteins have never included tests for dUTPase activity (8, 34, 35).
Metal preference was also tested by determining initial velocities for dUTP hydrolysis in the presence of Mn2+, Ca2+, Ni2+, and Zn2+ (Fig. 6D). Consistent with the crystallographic data, both manganese and magnesium coordination favor enzymatic activity, whereas calcium and all other metals tested show residual activities.
Isothermal Calorimetric Assays
The requirement of magnesium for inorganic phosphatase activity and the fact that the colorimetric assay is in itself an indirect method led to the need of applying a more direct technique, such as ITC. This technique allows assessment of possible differences in catalysis between manganese and magnesium.
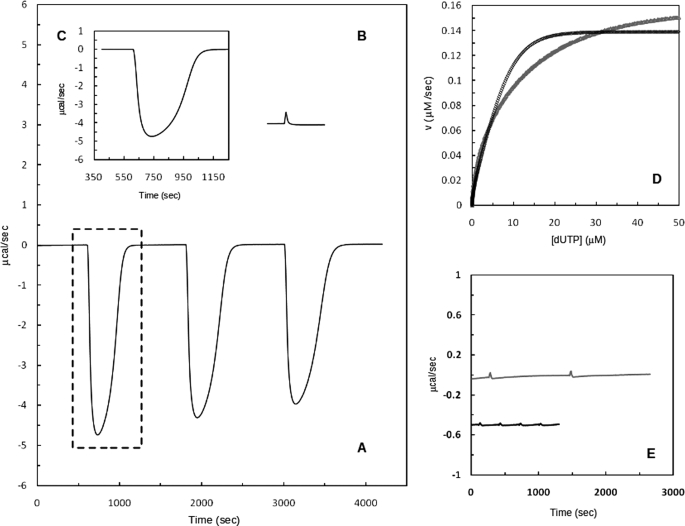
Fig. 7A shows a typical experimental thermogram for the titration of DR2231 protein with dUTP in the presence of 25 mm MgCl2 (at pH 7.0 and 25 °C). Three (single) injections were performed, and the profile of the trace exhibits an enzyme-catalyzed reaction with product inhibition. The raw data from Fig. 7A (indicated by a box) is treated and transformed into a plot, Fig. 7D, representing reaction rates versus substrate concentration. Kinetic parameters were calculated by fitting the curve to the Michaelis-Menten equation using non-linear regression and following the method described by Todd and Gomez (28). The experiment was performed under conditions where product inhibition may be considered negligible for the first injection. The subsequent injections show a shallower peak, due to inhibition by dUMP. Such behavior has been reported for Plasmodium falciparum dUTPase (29) and T. cruzi dUTPase (37). Apparent kinetic parameters (Km(app), kcat, and catalytic turnover) are listed in Table 3. These values fall within a similar range to those reported for P. falciparum (29), a trimeric dUTPase. They show, nonetheless, a weaker specificity and catalytic turnover, when compared with other dimeric dUTPases. Resorting to a continuous spectrophotometric method, studies with C. jejuni dUTPase exhibited a Km of 0.66 μm and a kcat of 12.3 s−1 (38), whereas for T. cruzi dUTPase, Km and kcat are 0.5 μm and 2.8 s−1, respectively (39).
FIGURE 7.
DR2231-catalyzed hydrolysis of dUTP in 20 mm Tris-HCl, 5 mm NaCl, 25 mm MgCl2 at pH 7.0 and 25 °C. A, representation of typical calorimetric trace (μcal versus time) obtained after the addition of three injections of 3 mm dUTP (20-μl injections) to the calorimetric cell containing DR2231 (12.5 nm). Change in instrumental thermal power was monitored until substrate hydrolysis was complete, returning to the original base line. B, thermogram of the dUTP dilution experiment. C, calorimetric trace (first injection) resulting after subtraction of the first peak of the dilution experiment. D, net thermal power (from B) was converted to rate and substrate concentration and fit to the Michaelis-Menten equation (black curve); the gray curve corresponds to a similar assay but in the presence of 25 mm MnCl2 (see “Experimental Procedures”). E, raw data representing the calorimetric trace (gray line) of 20-μl injections of 3 mm dTTP into the calorimetric cell containing enzyme (12.5 nm) and the equivalent dilution experiment in the absence of enzyme (black line). The trace of the dilution experiment is shifted for clarity.
TABLE 3.
Specificity and kinetic parameters of DR2231 protein determined by ITC
| Substrate | kcat | Km | kcat/Km × 10−4 |
|---|---|---|---|
| s−1 | μm | s−1m−1 | |
| dUTP-Mg2+ | 12.1 | 4.4 | 276 |
| dUTP-Mn2+ | 13.5 | 6.9 | 197 |
An identical experiment was conducted with 25 mm MnCl2 in place of MgCl2, and parameters were calculated in the same fashion (Fig. 7D, gray curve). Km(app) and kcat values are similar, although manganese suggests being less efficient catalytically (Table 3). Catalytic efficiency of dUTPase from P. falciparum showed a marked loss of catalytic turnover, ∼17-fold when Mn2+ was exchanged for Mg2+, against a 1.4-fold difference for DR2231 protein.
Activity assays testing for dTTP hydrolysis by DR2231 protein were also implemented (Fig. 7E). Dilution peaks of the substrate in the calorimetric vessel in absence of enzyme were of the same amplitude as those measured in presence of the enzyme. This confirms for DR2231 protein the complete lack of specificity for dTTP, in accordance to what had been observed with the colorimetric assays and as has been reported for true dUTPases.
DISCUSSION
DR2231 protein was characterized structurally and shown to be a variation of the basic module for dimeric d(C/U)TPases, comprising only one copy of the core helical domain. Moroz et al. (7) identified DR2231 as a MazG-like protein and a member of the all-α-NTP-PPase superfamily, and based on sequence analysis, it was assigned to a distinct family. Unpredictable, however, was its striking affinity for dUTP, a feature untested in all reported MazG proteins to date, and it was shown to have a rather broad selectivity for (deoxy)nucleotides. Unique features of operon organization regarding this superfamily open debate as to whether DR2231 is the “missing” dUTPase in the D. radiodurans genome (4) or another MazG-like NTP-PPase in the notably overrepresented hydrolase families of this bacterium.
To date, three entries encoding all-α-NTP-PPase have been identified in the D. radiodurans genome. DR1183 is annotated to encode the MazG protein, and both DR1022 and DR2231 are annotated as genes encoding conserved hypothetical proteins. There is no biochemical or structural information available on these proteins in the literature. A close focus on the genomic context of these genes may provide clues toward their function (40).
In E. coli the genetic module mazEF consists of two adjacent genes, mazE and mazF, located downstream from the relA gene. The mazG gene is the fourth on the operon, located immediately downstream of mazF (41). In D. radiodurans, however, the mazEF operon is organized differently; the addiction module mazE/mazF (corresponding to locus DR0416/DR0417, respectively) is not preceded by the relA gene or rather the spoT gene, as named in Gram-positive bacteria (putatively DR1838), whereas the mazG gene (assigned to DR1183) does not follow the addiction module. This gene dispersion is not unique among bacteria regarding this operon, and for D. radiodurans, in particular, it further highlights horizontal gene transfer as a rich source of genetic diversity (42).
In contrast with other bacteria, D. radiodurans appears to lack MazG(-like) proteins with two tandem domain arrangements; both DR1183 and DR1022 proteins hold, according to sequence analysis, only one copy of the core helical domain, with one active site per monomer, as DR2231 also does. Both DR1183 and DR1022 neighbor a Nudix hydrolase gene each (43, 44). Deducing possible functions based on genomic context has led to the proposal that MazG(-like) domains may be involved in non-canonical NTP processing. Curiously, however, DR2231 does not have any gene neighbors with pyrophosphohydrolase or phosphatase activity that may provide clues toward its cellular function.
The structural data obtained from the crystal structures of DR2231 complexed with dUMP reported here show that the ligand-binding pocket cannot accommodate canonical nucleotides. This not only explains the lack of hydrolytic activity on all nucleotide triphosphates tested but also rules out, in particular, the stringent response molecule, ppGpp, as a substrate for DR2231 protein. In E. coli MazG, nonpolar aromatic residues constitute a hydrophobic pocket for bases without any interaction with the 2′-OH of the ribose ring, explaining how both oxy- and deoxyribonucleotides could be substrates to this enzyme (8). Although DR2231 has a remarkable structural and topological resemblance to MazG proteins, structural details within the active site relative to the binding of the (deoxy)ribose ring reveal its strong affinity to dUTPases.
Through their catalytic activity, dUTPases provide the precursor for the formation of dTMP by thymidylate synthase but simultaneously have a crucial role in maintaining a low dUTP/dTTP ratio in the cell in order to limit the incorporation of deoxyuridylate into DNA by DNA polymerases. Misincorporation of deoxyuridine can impair DNA integrity by producing a base pair change of C:G into U(T):A and, thus, a stable point mutation (for reviews, see Refs. 45 and 46).
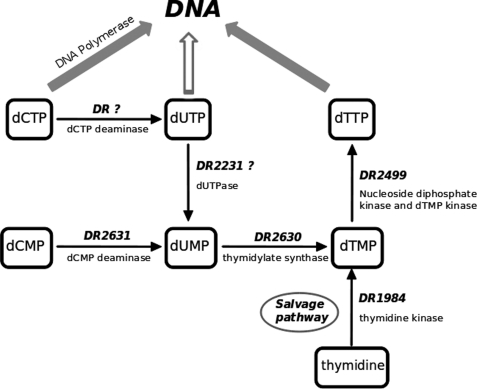
Regarding dTTP synthesis, the precursor for de novo pyrimidine biosynthesis is the cytosine ring that must undergo deamination to result in uracil, upon which the methyl group can be added to produce thymine. Cytosine deamination may occur at different levels; one is through the direct input provided by dCMP deaminases, directly producing dUMP, widespread in eukaryotes and most Gram-positive bacteria. The other pathway, common to Gram-negative bacteria, is at the dCTP level, catalyzed by dCTP deaminases, producing dUTP that must be converted subsequently into dUMP by a dUTPase. For some organisms, such as Mycobacteria and Plasmodia, this is the only course in obtaining dUMP (47, 48). In the first path, however, dUMP supply from the dUTPase-catalyzed reaction has a minor role.
Fig. 8 illustrates the de novo and salvage pathways for dTTP biosynthesis. D. radiodurans genes that could be identified with the various enzymes participating in these pathways are represented. Although D. radiodurans has many features reminiscent of Gram-negative bacteria and many others that are entirely unique, it is classified as Gram-positive (49, 50). Accordingly, we could not identify a gene that encodes a dCTP deaminase with significant homology with those known to exist in Gram-negative bacteria. This suggests that in D. radiodurans, the de novo production of dTMP relies mostly on the dCMP pathway or on the salvage alternative directly from thymidine. We propose that the dUTPase activity detected in DR2231 protein may correspond to the house-cleaning task of removing dUTP arising from deamination of dCTP but also unidentified non-canonical NTPs occasionally. It would be interesting to assert the latter experimentally with naturally occurring oxidative products of cytosine, such as 5-hydroxycytosine and 5-hydroxyuracil, for example.
FIGURE 8.
De novo and salvage pathways for dTTP biosynthesis. Identifiable homologous loci from D. radiodurans are represented beside their respective arrows.
There are reasons to believe that the catalytic mechanism of DR2231 protein is similar, if not identical, to that of C. jejuni dUTPase (6). The orientation of the product relative to the metal-binding site is very similar to that observed in the structure of C. jejuni dUTPase complexed with dUpNHpp. By comparing DR2231_dUMP_Mn structure with C. jejuni structure, more information may be inferred; W1 possibly occupies the position of the γ-phosphate of dUTP after catalysis, and another water molecule, W2 (water molecule 107), bound to Oδ2 of Asp82 may correspond to the catalytic water identified in C. jejuni dUTPase, thus regenerating the active site. A third manganese ion was never observed in our co-crystallizations with dUMP. Still, the residues that bind Mg3 in C. jejuni dUTPase (Glu49 and Glu79) are aligned with those of DR2231 protein, Glu79 and Glu50, leaving open the possibility of a third, albeit labile, coordination site, probably occupied when substrate is bound.6 There are other elements in the pocket that provide clues as to being involved in β- and γ-phosphate binding and stabilization. These residues are possibly His78 and Lys125, the latter from the second subunit. We further postulate that one of the most crucial functional residues is Lys112, located at the end of H4. Along with Asn109, these two residues have a dual role; they provide both orientation and stabilization of the deoxynucleotide. As the substrate is bound, it induces conformational alterations upon the hinge, forcing the entire latch to move and close upon the binding pocket.
From sequence alignment analysis of a non-redundant database, it is clear that only putative DR2231 family members show high homology for the “lid” region (Fig. 9). This structural feature is unique to the DR2231 subfamily and can be considered as its identity signature.
The affinity of DR2231 for Mn2+ is not entirely understood. It has been reported that D. radiodurans accumulates very high intracellular manganese levels and that such accumulation may be radiation- and dessication resistance-related (51). This accumulation suggests that Mn(II) facilitates recovery from radiation injury. In particular, it is essential for the detoxification of reactive oxygen species in most bacteria and also by preventing the production of iron-dependent reactive oxygen species (52). More recently, other studies showed that the extraordinary robustness to proteome oxidation (i.e. protein carbonylation) of D. radiodurans depends on efficient proteome protection (not DNA protection) conferred by low molecular weight cytosolic compounds (molecular species of less than 3 kDa) (53). Daly et al. (54) showed that ultrafiltered, protein-free preparations of D. radiodurans cell extract prevent protein oxidation at massive doses of ionizing radiation compared with those of radiation-sensitive bacteria, with no protective effect at all. The D. radiodurans ultrafiltrate is enriched in manganese, phosphate, nucleosides, bases (of which the most highly represented are uridine, uracil, adenosine, and inosine), and peptides.
As a putative house-cleaning enzyme, we may speculate whether DR2231 protein expression may not be connected to situations of oxidative stress when Mn2+ intracellular levels are high. It should be noted, however, that according to reported transcriptome dynamics and proteomic analysis studies, DR2231 protein is not among those gene products identified to overexpress when exposed to prolonged irradiation (55, 56).
The gene product of DR2231 may not be an essential protein for D. radiodurans concerning the pyrimidine metabolic pathway, but it ensures cell survival by balancing the dUTP availability in the cell pool and delivering dUMP, processed also into uridine and uracil elsewhere, as well as inorganic phosphate, important in proteome radioprotection (53). It stands out in the genome of D. radiodurans as one among many in the set of NTP pyrophophatases that have been possibly acquired through horizontal gene transfer. The findings reported here on the structure and function of DR2231 protein support the view that the MazG-like subunit may be considered as the common ancestor for other family members, in particular, dimeric dUTPases.
Acknowledgments
We acknowledge the High Throughput Crystallization (HTX) Laboratory (EMBL Grenoble, France) for access to the Cartesian PixSys and the support provided by the ESRF Structural Biology group at the ESRF beamlines and in the laboratory housed in the Carl-Ivar Branden building.
The atomic coordinates and structure factors (codes 2YEU, 2YF4, 2YF9, 2YF3, 2YFD, and 2YFC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Henceforth, structural elements of DR2231 protein crystal structure are annotated both according to sequence and according to the Moroz et al. convention (7) in parentheses.
Note that Mn1 will correspond to Mg2 from C. jejuni dUTPase and Mn2 will correspond to Mg1; in the DR2231 protein structures presented here, there is no correspondence to Mg3.
- TLS
- translation/libration/screw
- ITC
- isothermal calorimetry
- PDB
- Protein Data Bank.
REFERENCES
- 1. Kamiya H. (2003) Nucleic Acids Res. 31, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bessman M. J., Frick D. N., O'Handley S. F. (1996) J. Biol. Chem. 271, 25059–25062 [DOI] [PubMed] [Google Scholar]
- 3. Koonin E. V., Aravind L., Galperin M. Y. (2000) in Bacterial Stress Responses (Storz G., Hengge-Aronis R. eds) pp. 417–444, American Society for Microbiology Press, Washington, D. C [Google Scholar]
- 4. Makarova K. S., Aravind L., Wolf Y. I., Tatusov R. L., Minton K. W., Koonin E. V., Daly M. J. (2001) Microbiol. Mol. Biol. Rev. 65, 44–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarova K. S., Aravind L., Daly M. J., Koonin E. V. (2000) Genetica 108, 25–34 [DOI] [PubMed] [Google Scholar]
- 6. Moroz O. V., Harkiolaki M., Galperin M. Y., Vagin A. A., González-Pacanowska D., Wilson K. S. (2004) J. Mol. Biol. 342, 1583–1597 [DOI] [PubMed] [Google Scholar]
- 7. Moroz O. V., Murzin A. G., Makarova K. S., Koonin E. V., Wilson K. S., Galperin M. Y. (2005) J. Mol. Biol. 347, 243–255 [DOI] [PubMed] [Google Scholar]
- 8. Lee S., Kim M. H., Kang B. S., Kim J. S., Kim G. H., Kim Y. G., Kim K. J. (2008) J. Biol. Chem. 283, 15232–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dimasi N., Flot D., Dupeux F., Márquez J. A. (2007) Acta Crystallogr. F 63, 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Powell H. R. (1999) Acta Crystallogr. D 55, 1690–1695 [DOI] [PubMed] [Google Scholar]
- 11. Evans P. (2006) Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 12. Kabsch W. (2010) Acta Crystallogr. D 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 14. McCarthy A. A., Brockhauser S., Nurizzo D., Theveneau P., Mairs T., Spruce D., Guijarro M., Lesourd M., Ravelli R. B., McSweeney S. (2009) J. Synchrotron Radiat. 16, 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 16. Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 17. Sheldrick G. M. (2010) Acta Crystallogr. D 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Painter J., Merritt E. A. (2006) Acta Crystallogr. D 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 23. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 24. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeLano W. L. (2003) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 26. Baykov A. A., Evtushenko O. A., Avaeva S. M. (1988) Anal. Biochem. 171, 266–270 [DOI] [PubMed] [Google Scholar]
- 27. Geladopoulos T. P., Sotiroudis T. G., Evangelopoulos A. E. (1991) Anal. Biochem. 192, 112–116 [DOI] [PubMed] [Google Scholar]
- 28. Todd M. J., Gomez J. (2001) Anal. Biochem. 296, 179–187 [DOI] [PubMed] [Google Scholar]
- 29. Quesado-Soriano I., Leal I., Casas-Solvas J. M., Varagas-Berenguel A., Barón C., Ruiz-Pérez L. M., González-Pacanowska D., García-Fuentes L. (2008) Biochim. Biophys. Acta 1784, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 30. Jones S., Thornton J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13–208552589 [Google Scholar]
- 31. Harding M. M. (2001) Acta Crystallogr. D 57, 401–411 [DOI] [PubMed] [Google Scholar]
- 32. Harding M. M. (2006) Acta Crystallogr. D 62, 678–682 [DOI] [PubMed] [Google Scholar]
- 33. Bock C. W., Kaufman-Katz A., Markham G. D., Glusker J. P. (1999) J. Am. Chem. Soc. 121, 7360–7372 [Google Scholar]
- 34. Zhang J., Zhang Y., Inouye M. (2003) J. Biol. Chem. 278, 21408–21414 [DOI] [PubMed] [Google Scholar]
- 35. Robinson A., Guilfoyle A. P., Harrop S. J., Boucher Y., Stokes H. W., Curmi P. M., Mabbutt B. C. (2007) Mol. Microbiol. 66, 610–621 [DOI] [PubMed] [Google Scholar]
- 36. Galperin M. Y., Moroz O. V., Wilson K. S., Murzin A. G. (2006) Mol. Microbiol. 59, 5–19 [DOI] [PubMed] [Google Scholar]
- 37. Téllez-Sanz R., Yassin Z., Bernier-Villamor V., Ortiz-Salmerón E., Musso-Buendia J. A., Barón C., Ruíz-Pérez L. M., González-Pacanowska D., García-Fuentes L. (2007) Biochimie 89, 972–980 [DOI] [PubMed] [Google Scholar]
- 38. Musso-Buendía J. A., Vidal A. E., Kasinthan G., Nguyen C., Carrero-Lérida J., Ruiz-Pérez L. M., Wilson K., Johansson N. G., Gilbert I. H., González-Pacanowska D. (2009) J. Enzyme Inhib. Med. Chem. 24, 111–116 [DOI] [PubMed] [Google Scholar]
- 39. Bernier-Villamor V., Camacho A., Hidalgo-Zarco F., Pérez J., Ruiz-Pérez L. M., González-Pacanowska D. (2002) FEBS Lett. 526, 147–150 [DOI] [PubMed] [Google Scholar]
- 40. Galperin M. Y., Koonin E. V. (2000) Nat. Biotechnol. 18, 609–613 [DOI] [PubMed] [Google Scholar]
- 41. Engelberg-Kulka H., Glaser G. (1999) Annu. Rev. Microbiol. 53, 43–70 [DOI] [PubMed] [Google Scholar]
- 42. Boucher Y., Labbate M., Koenig J. E., Stokes H. W. (2007) Trends Microbiol. 15, 301–309 [DOI] [PubMed] [Google Scholar]
- 43. Kang L. W., Gabelli S. B., Bianchet M. A., Xu W. L., Bessman M. J., Amzel L. M. (2003) J. Bacteriol. 185, 4110–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ranatunga W., Hill E. E., Mooster J. L., Holbrook E. L., Schulze-Gahmen U., Xu W., Bessman M. J., Brenner S. E., Holbrook S. R. (2004) J. Mol. Biol. 339, 103–116 [DOI] [PubMed] [Google Scholar]
- 45. Nyman P. O. (2001) Curr. Protein Pept. Sci. 2, 27–285 [DOI] [PubMed] [Google Scholar]
- 46. Vértessy B. G., Tóth J. (2009) Acc. Chem. Res. 42, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chan S., Segelke B., Lekin T., Krupka H., Cho U. S., Kim M. Y., So M., Kim C. Y., Naranjo C. M., Rogers Y. C., Park M. S., Waldo G. S., Pashkov I., Cascio D., Perry J. L., Sawaya M. R. (2004) J. Mol. Biol. 341, 503–517 [DOI] [PubMed] [Google Scholar]
- 48. Whittingham J. L., Leal I., Nguyen C., Kasinathan G., Bell E., Jones A. F., Berry C., Benito A., Turkenburg J. P., Dodson E. J., Ruiz Perez L. M., Wilkinson A. J., Johansson N. G., Brun R., Gilbert I. H., Gonzalez Pacanowska D., Wilson K. S. (2005) Structure 13, 329–338 [DOI] [PubMed] [Google Scholar]
- 49. Battista J. R. (1997) Annu. Rev. Microbiol. 51, 203–224 [DOI] [PubMed] [Google Scholar]
- 50. Al-Bakri G. H., Mackay M. W., Whittaker P. A., Moseley B. E. (1985) Gene 33, 305–311 [DOI] [PubMed] [Google Scholar]
- 51. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Venkateswaran A., Hess M., Omelchenko M. V., Kostandarithes H. M., Makarova K. S., Wackett L. P., Fredrickson J. K., Ghosal D. (2004) Science 306, 1025–1028 [DOI] [PubMed] [Google Scholar]
- 52. Daly M. J. (2009) Nat. Rev. Microbiol. 7, 237–245 [DOI] [PubMed] [Google Scholar]
- 53. Krisko A., Radman M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14373–14377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Kiang J. G., Fukumoto R., Lee D. Y., Wehr N. B., Viteri G. A., Berlett B. S., Levine R. L. (2010) PLoS ONE 5, e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu Y., Zhou J., Omelchenko M. V., Beliaev A. S., Venkateswaran A., Stair J., Wu L., Thompson D. K., Xu D., Rogozin I. B., Gaidamakova E. K., Zhai M., Makarova K. S., Koonin E. V., Daly M. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4191–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang C., Wei J., Zheng Z., Ying N., Sheng D., Hua Y. (2005) Proteomics 5, 138–143 [DOI] [PubMed] [Google Scholar]
- 57. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 58. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]