Abstract
Hepatocellular carcinoma (HCC) is a heterogeneous and highly aggressive malignancy, for which there are no effective cures. Identification of a malignant stemlike subtype of HCC may offer patients with a dismal prognosis a potential targeted therapy using c-MET and Wnt pathway inhibitors. MicroRNAs (miRNAs) show promise as diagnostic and prognostic tools for cancer detection and stratification. Using a TRE-c-Met-driven transgenic HCC mouse model, we identified a cluster of 23 miRNAs that is encoded within the Dlk1-Gtl2 imprinted region on chromosome 12qF1 overexpressed in all of the isolated liver tumors. Interestingly, this region is conserved among mammalian species and maps to the human DLK1-DIO3 region on chromosome 14q32.2. We thus examined the expression of the DLK1-DIO3 miRNA cluster in a cohort of 97 hepatitis B virus-associated HCC patients and identified a subgroup (n = 18) of patients showing strong coordinate overexpression of miRNAs in this cluster but not in other cancer types (breast, lung, kidney, stomach, and colon) that were tested. Expression levels of imprinted gene transcripts from neighboring loci in this 14q32.2 region and from a subset of other imprinted sites were concomitantly elevated in human HCC. Interestingly, overexpression of the DLK1-DIO3 miRNA cluster was positively correlated with HCC stem cell markers (CD133, CD90, EpCAM, Nestin) and associated with a high level of serum α-fetoprotein, a conventional biomarker for liver cancer, and poor survival rate in HCC patients. In conclusion, our findings suggest that coordinate up-regulation of the DLK1-DIO3 miRNA cluster at 14q32.2 may define a novel molecular (stem cell-like) subtype of HCC associated with poor prognosis.
Keywords: Gene Expression, Liver, MicroRNA, Stem Cells, Tumor Marker, Hepatocellular Carcinoma, Prognosis
Introduction
Hepatocellular carcinoma (HCC)5 is the fifth leading cause of cancer deaths worldwide, and its incidence is increasing steadily in both the United States and China (1). The presence of stemlike cancer cells in liver tumors is considered to cause the aggressive and malignant phenotypes of HCC as well as conferring resistance to various chemotherapeutics (2, 3). Although considerable progress in understanding the molecular pathogenesis of HCC has been made in recent years, definitive molecular markers for identifying liver cancer stem cells remain limited and poorly characterized. Nevertheless, there are a handful of reports showing that CD133, CD90, and EpCAM-positive subpopulations in HCC tumors displayed stemlike phenotypes, as characterized by serial clonal passages, invasive and metastatic properties, as well as chemoresistance capacity (4–6). However, the underlying genetic causes and/or molecular signatures for HCC stemness characteristics remain largely unknown.
MicroRNAs (miRNAs) are ∼22-nucleotide-long non-coding RNAs as promising diagnostic and prognostic tools for cancer detection and stratification (7, 8) and also play essential roles in gene transcript stability and translation efficiency (9) and cancer metabolism (10), having significant functions as both tumor suppressors and oncogenes (11). As a new class of genomic information, miRNA dysregulation can provide insight for identifying new pathways of carcinogenesis and providing the opportunities for biomarker and therapeutic target discovery.
Overexpression of the c-MET oncogene has been shown to drive liver carcinogenesis through activation of the Wnt signaling pathway in transgenic mice (12) and is linked to the progenitor stem-like subtypes of HCC. To address whether microRNA dysregulation was involved in this c-MET-driven HCC model, we analyzed genome-wide miRNA alterations in tumor and adjacent non-tumor liver tissues and compared these alterations with those of normal liver tissues from wild-type FVB mice. The microRNA signatures identified in mice were subsequently compared with the expression profiles in human HCC clinical samples. The present study demonstrates a DLK1-DIO3 genomic imprinted microRNA cluster highly enriched in mouse liver tumors representing a stemlike subtype of human HCC associated with poor prognosis in patients.
EXPERIMENTAL PROCEDURES
Patient Cohort and Samples
Patients with hepatitis B virus-associated HCC underwent curative hepatectomy at Queen Mary Hospital (Pokfulam, Hong Kong) between 1993 and 2007 (13). This study was approved by the Institutional Review Board for Human Ethics and each patient gave his/her written informed consent for the use of the clinical specimens for research. Tumor and adjacent non-tumor samples were collected at the time of the curative surgery, immediately snap-frozen in liquid nitrogen, and stored at −80 °C until use.
Transgenic Mice
The c-Met mouse HCC model has been described previously (14). All mice had an FVB genetic background. Mice overexpressing human c-MET carried one copy of the LAP-tTa transgene (the liver-specific LAP promoter driving the Tet-VP16 transactivator) and one copy of the TRE-met transgene (Tet operator-regulated human c-MET gene). The presence of both transgenes results in expression of the human c-MET gene specifically in and throughout the liver (referred to henceforth as the TRE-met strain). Seven mice of each strain were sacrificed at 6 (TRE-met), 7 (LAP-tTa), or 14 (TRE-met) weeks of age. Normal liver or liver tumor tissue (two per mouse) was collected and processed for RNA analysis.
MicroRNA and mRNA Analyses
Total RNA was purified using an RNeasy kit (Qiagen, Valencia, CA). Expression levels of 220 human miRNAs were measured by custom quantitative PCR assays, as described previously (15, 16). Transcript expression levels were detected by microarray, as described previously (17, 18). Seven mice were analyzed for each treatment or control group. Microarray hybridizations were performed as described previously (19) and following the manufacturer's recommendations. Profiling data for human samples are available in the GEO database (NCBI, National Institutes of Health) under accession number GSE22058. Mouse mRNA samples were profiled on a custom Affymetrix array (RM-MG01Aa520487), whereas mouse miRNA samples were profiled on a standard Agilent miRNA array (G4471A-019119). Mouse microarray data transformation and analysis was performed as described previously (17–19). The log10(ratio) of each gene in each sample was computed by subtracting the mean of log10(intensity) of that gene across all adjacent non-tumor samples to make them comparable with the TRE-met mouse model data. Microarray data on gene expression profiling were available in the GEO database with the following accession numbers: GSE25142 (for TRE-met mice) and GSE25097 (for human HCC). Raw profiling data on miRNAs in human HCC and mice are also available in the GEO database under accession number GSE22058 (released on June 4, 2010).
miRNA Expression Constructs
Expression vectors of miR-127 (pc-miR-127), miR-431 (pc-miR-431), and miR-433 (pc-miR-433) were constructed by PCR amplification of genomic DNA from PLC/PRF/5 hepatoma cell line. The primer pairs are as follows: miR-127 (forward), 5′-GGCCTCGAGAGCACAAAGAACCCTAGCATGTCCT-3′ and 5′-GGCGATATCGCTCTACACGGAGCCCCTGGT-3′ (reverse); miR-431, 5′-GGCCTCGAGGGCTGAGCAGGTGCAGCTGGCCAT-3′ (forward) and 5′-GGCGGATCCCCCAGCTGCTCACCCAGATGCCCG-3′ (reverse); miR-433, 5′-GGCCTCGAGGGAGGCCTCGGAAGAAGTGCA-3′ (forward) and 5′-GCTAAGATCTCTGGTGCGGCAGCTGCTGAG-3′ (reverse). The PCR products were cloned into pcDNA-3.1/myc-His (−) expression vector plasmid (Invitrogen), respectively, and subsequently verified by Sanger DNA sequencing.
Wound Healing Assay
PLC/PRF/5 cells were seeded onto a 6-well plate, and transfection was performed when cells reached ∼90% confluence. 4 μg of either miRNA expression vectors or empty vector (pcDNA3.1) was transfected using Lipofectamine 2000 (Invitrogen) (20). 24 h post-transfection, a wound was made using a pipette tip, and photographs were taken at time 0 and 24 h to measure cell.
In Vitro Knockdown of c-Met in HCC
Hep3B cells grown on 6-well plate were transfected with c-Met siRNA (catalogue no. HSS106477, Invitrogen) or control siRNA (catalogue no. AM4635, Ambion Inc., Austin, TX) at a final concentration of 40 nm using Lipofectamine RNAiMax (Invitrogen). 48 h post-transfection, transcript levels of c-Met and miR-127, miR-431, miR-433, and miR-154* were measured by real-time quantitative PCR or using the corresponding TaqMan miRNA assay (Applied Biosystems, Foster City, CA). Expressions of c-Met and miRNAs were normalized to 18 S RNA and compared among different treatment groups.
Immunohistochemistry
Immunohistochemistry staining was performed in paraffin-embedded HCC tumor and adjacent non-tumor tissues from 14 patients, following a protocol described previously (21). Mouse monoclonal anti-EpCAM antibody (Cell Signaling Technology, Danvers, MA) (1:250), rabbit monoclonal anti-DLK antibody (OriGene Technologies, Rockville, MD) (1:50), rabbit polyclonal anti-MEST antibody (Novus Biologicals, Littleton, CO) (1:200), mouse polyclonal anti-CD133 antibody (Sigma-Aldrich) (1:200), and mouse monoclonal anti-CD90 antibody (BD Biosciences) (1:50) were incubated with the sections at 4 °C overnight. Horseradish peroxidase (HRP)-conjugated goat anti-mouse (1:200; Zymed Laboratories Inc., Invitrogen) or goat anti-rabbit (1:200; Zymed Laboratories Inc.) secondary antibodies were used for detecting the primary antibodies. Signal was visualized by incubating with liquid DAB+ reagent (Dako, Glostrup, Denmark), and images were captured using a digital camera (22).
Data Analysis
Data were analyzed using Rosetta ResolverTM and MATLABTM software. Using the hypergeometric distribution, regulated transcripts were tested for enrichment of transcripts belonging to gene sets in the GO biological process as annotation sources (23).
RESULTS AND DISCUSSION
Landscape of MicroRNA Expression Profiles in c-MET Mouse Liver Tumors and Human HCC Clinical Samples
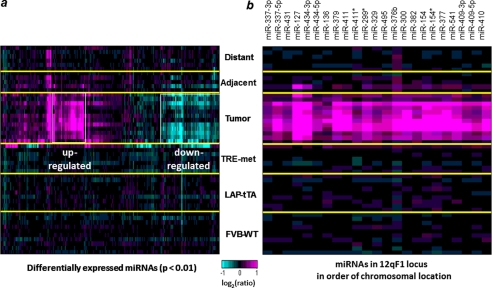
As shown in Fig. 1a, the greatest miRNA expression changes occurred in the tumors, whereas the adjacent non-tumor liver tissues from tumor-bearing mice did not show any consistent miRNA expression changes relative to normal liver tissues from wild-type FVB mice. 39 miRNAs were found to be significantly (p < 0.01) down-regulated, and 42 miRNAs were up-regulated in the tumor tissues (supplemental Table S1). We examined the chromosomal locations of the up-regulated miRNAs and, strikingly, found that over half of them (23 of 42 miRNAs) were encoded within the 12qF1 mouse chromosomal region (Fig. 1b). The mouse 12qF1 region, also known as Dlk1-Gtl2, is an imprinted locus; expression of the miRNAs in this region is restricted to the maternal chromosome, primarily in the developing embryo (24, 25). Most interestingly, this genomic imprinted region is activated in fully pluripotent embryonic stem cells but aberrantly silenced in mouse induced pluripotent stem cells that show low pluripotency and reprogramming efficiency (26, 27). This chromosomal locus is conserved among mammals and is mapped to the human DLK1-DIO3 region on chromosome 14q32.2 (25), raising the possibility that expression of these miRNAs might be conserved in human HCC. The miRNAs that were assayed comprise a subset of a cluster of ∼60 putative mature miRNAs that are spread over less than 200 kilobases in the DLK1-DIO3 region. The miRNAs that cluster in the 14q32.2 region are thought to be encoded by two precursor transcripts and/or long polycistronic transcripts, which are separated by a cluster of putative C/D box small nucleolar RNAs (28).
FIGURE 1.
miRNA expression profile in the c-Met mouse model of HCC. a, tumor samples show the greatest changes in miRNA expression. miRNAs with significant expression changes in at least one sample (p < 0.01) are shown. Yellow lines separate the sample groups, and white boxes highlight the up- and down-regulated miRNAs in tumors. FVB-WT, liver tissue from control animals; LAP-tTA and TRE-met, liver tissue from single transgene parental strains; Tumor, tumor tissue from double transgene, tumor-bearing animals; Adjacent/Distant, non-tumor liver tissue from double transgene, tumor-bearing animals. b, expression pattern of 23 miRNAs from the mouse 12qF1 chromosomal cluster.
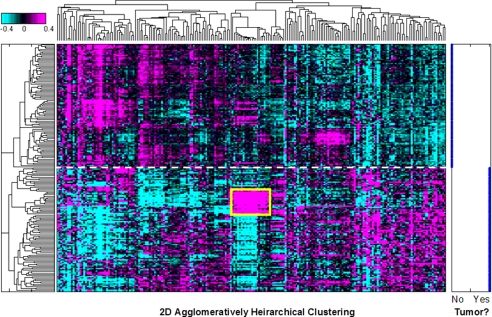
To investigate the role of miRNAs in human HCC, we examined the expression of 220 miRNAs from 97 pairs of tumor and matched adjacent non-tumor tissues from hepatitis B virus-associated HCC patients. The expression profile of miRNAs perfectly distinguished tumor from non-tumor tissues (Fig. 2), suggesting that carcinogenesis in liver involves a large scale disruption of miRNA expression. Accordingly, 21 miRNAs of 22 from chromosome 14q32.2 that we measured were coordinately up-regulated in the tumor tissues from a subset of HCC patients (n = 18; Fig. 2), which was about 6–7-fold higher than the expression level in adjacent non-tumor samples (supplemental Table S2). In short, strong coordinate up-regulation of miRNAs from the DLK1-DIO3 genomic imprinted region (mouse 12qF1; human 14q32.2) was observed in liver tumors from both human and mouse HCC.
FIGURE 2.
Global view of 220 miRNA expression patterns in tumor and non-tumor clinical tissues from human HCC. A heat map shows two-dimensional unsupervised clustering of log10 expression ratios for 220 human miRNAs in 97 pairs of tumor and adjacent non-tumor HCC tissues. miRNA expression levels were normalized to the mean of all samples. A cluster of coordinately up-regulated miRNAs from the DLK1-DIO3 region is boxed in yellow. The degree of up-regulation is shown in magenta, and down-regulation is shown in cyan on a log scale as indicated by the color bar. Separation by clustering of tumor and adjacent non-tumor samples is indicated by the dashed white line. Right, tumor status of each sample. No, adjacent non-tumor; Yes, HCC.
To test whether this 14q32.2 miRNA cluster is regulated by c-MET in human HCC, we knocked down c-MET by siRNA in the Hep3B hepatoma cell line and observed significant down-regulation of the miR-127, miR-431, and miR-154*, but not miR-433, of the microRNA cluster as determined by a real-time quantitative PCR assay (supplemental Fig. 1). The exact mechanism of how c-MET regulates the 14q32.2 miRNA cluster remains to be elucidated.
Concomitant and Coordinate Up-regulation of MicroRNA Cluster and Genomic Imprinted Gene Transcripts in the DLK1-DIO3 Region at 14q32.2
To further determine the cross-species relevance of up-regulation of the DLK1-DIO3 miRNA cluster, we examined the correlation of human and mouse miRNA expression in all pairs of human and mouse tumor miRNA profiles. On average, there was no significant correlation (r ∼ 0), but a subset of pairs showed significant correlation. This subset corresponded to human tumors up-regulating DLK1-DIO3 miRNAs (median correlation, r = 0.36; significance of shift in correlation, p < 1 × 10−114 by one-tailed Student's t test). Strong up-regulations of miRNAs were well correlated between mouse and human tumors, whereas little correspondence was seen between strong down-regulations of miRNAs. It is suggested that the DLK1-DIO3 miRNAs cluster is driving the correlation. Representative pairwise comparisons of human and mouse tumor miRNA profiles are shown in supplemental Fig. 2. miRNAs up-regulated by >2 times in at least 15% of both human and mouse tumors are listed in supplemental Table S3.
Next we explored the transcriptional regulatory role of this 14q32.2 miRNAs cluster by genome wide expression profiling using the same set of 97 pairs of HCC and adjacent non-tumor tissues. There were 15 gene probes available for multiple transcripts encoded within the DLK1-DIO3 region, and strikingly, all of the genes were highly correlated (0.75 < r < 0.96), with the mean regulation of the 14q32.2 miRNA cluster in HCC samples, suggesting coordinate control of a large transcribed region. Our array comparative genomic hybridization data on the same set of samples ruled out the possibility that the overexpression was due to DNA amplification.
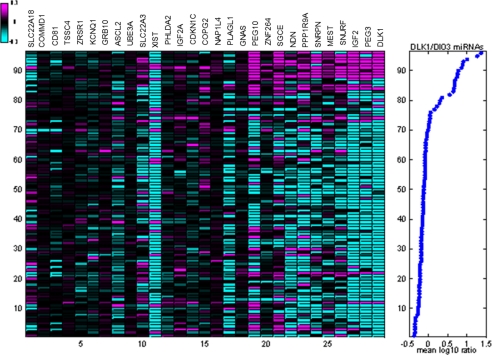
Loss of imprinting is often seen in early oncogenesis (29). We hypothesize that the coordinate overexpression of the 14q32.2 miRNAs in HCC samples is related to a change in the imprinting status of the locus. To test this hypothesis, we compared regulation of the 14q32.2 miRNAs with regulation of known imprinted genes (30). A subset of imprinted genes was clearly coordinately regulated with the 14q32.2 miRNAs (Fig. 3). These imprinted genes included DLK1. We found coordinate regulation of additional imprinted genes from other loci in the human genome, such as paternally expressed genes IGF2, PEG3, PEG10, SGCE, SNURF, and MEST, and maternally expressed genes PPP1R9A and ZNF264. Information on these imprinted genes in HCC is limited. Recent studies showed gain of imprinting of IGF2 found in 47% (8 of 17) of HCC cases and suppressing IGF2 expressions by DNA methylation could enhance survival in HCC (31), whereas PEG10 is a progression-related and diagnostic biomarker for HCC (32, 33). In our study, clinical correlation analysis of transcript expression levels among these genes in the present cohort of HCC samples revealed that PPP1R9A and SGCE were significantly associated with poor prognostic outcomes (i.e. shorter overall survival rate (p = 0.003 for PPP1R9A and p = 0.027 for SGCE) and disease-free survival rate (p = 0.001 for PPP1R9A and p = 0.022 for SGCE)), as shown by the log-rank test (supplemental Fig. 3).
FIGURE 3.
Overexpression of DLK1-DIO3 miRNA cluster at 14q32.2 is associated with up-regulation of subsets of imprinted gene transcripts. Left, log10 expression ratios in 96 HCC and matched adjacent non-tumor samples of all transcripts of genes detectably expressed in these samples and described as imprinted in humans and/or mice (17). The genes were sorted by their correlation with DLK1-DIO3 miRNA expression in matched miRNA profiles. Experiments were sorted by mean expression of DLK1-DIO3 miRNAs in matched miRNA profiles. Ratios shown are relative to mean expression in adjacent non-tumor samples. Right, the average log10 expression ratio of 22 14q32.2 miRNAs is shown for each sample. One sample pair was excluded for low quality of gene expression data.
Transcripts correlated (r > 0.4) with the mean regulation of the 14q32.2 miRNAs were enriched for significantly imprinted genes (p < 1 × 10−9). Similarly, genes up-regulated in the mouse tumor model were also significantly enriched for imprinted genes (p < 2 × 10−3). This finding suggests that overexpression of the 14q32.2 miRNAs is related to a change in imprinting status of the locus and could potentially contribute to early detection of HCC. Other studies have demonstrated that aberrant imprinting is involved in HCC tumor progression (29, 34, 35); thus, up-regulation of 14q32.2 miRNAs could also be associated with changes in expression of tumor-promoting genes. We analyzed the transcripts correlated with expression of 14q32.2 miRNAs. In our HCC samples, correlated transcripts were enriched in GO annotation for embryonic development. Genes up-regulated in the mouse tumor model were also significantly enriched for embryonic development in GO annotation, with the five most significant terms all relating to development. In an atlas of tumor and adjacent non-tumor tissues from kidney, gastric, colon, lung, and breast cancer patients, biological annotations of the transcripts correlated with 14q32.2 miRNA expression were enriched in organogenesis and morphogenesis according to GO and KEGG pathway annotation sources.
Association of DLK1-DIO3 Imprinted MicroRNA Cluster with HCC Stem Cell Markers and Aggressive Phenotypes and Poor Survival of Patients
In this mouse model, overexpression of the c-MET oncogene drives liver carcinogenesis through activation of the Wnt signaling pathway. It is also known that Wnt/β-catenin signaling plays an important role in regulating cancer stem cell activation. Therefore, it is important to know the activity of Wnt/β-catenin signaling in human HCC with 14q32.2 overexpression. Indeed, we find that this microRNAs cluster is significantly correlated with the Wnt/β-catenin signaling downstream targets as represented by LGR5 (r = 0.42, p = 1.7 × 10−5) and DKK1 (r = 0.54, p = 9.5 × 10−9). Further investigation of the molecular mechanisms and biological implications is under way.
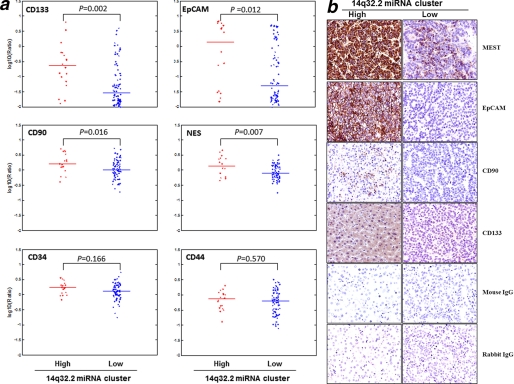
To assess whether overexpression of the DLK1-DIO3 miRNA cluster at 14q32.2 is linked to embryonic or stem cell phenotype in an HCC subpopulation, we tested the 14q32.2 miRNA expression status against expression of a panel of documented stem cell markers. We found that the DLK1-DIO3 miRNA cluster was positively correlated to the expression levels of CD133, CD90, EpCAM, and nectin, previously reported as markers of HCC tumor-initiating or progenitor stem cells (Fig. 4a). Furthermore, an immunohistochemistry study on the corresponding 14q32.2 high expressing samples also confirmed expression of these HCC stem cell markers (CD133, CD90, and EpCAM), but expression of these markers was weak or undetectable in the 14q32.2 miRNA low expressing samples. Imprinted gene MEST up-regulated by the microRNA cluster was also found highly expressed in the HCC tissues. Representative findings are shown in Fig. 4b.
FIGURE 4.
DLK1-DIO3 miRNA cluster positively correlates with HCC-specific stem cell markers. a, gene expression of putative cancer stem cell markers is shown in 14q32.2 overexpression (n = 18) and no overexpression (n = 78) subgroups of HCC tumors. CD133, CD90, EpCAM, and Nestin, which have been reported as stem cell markers in HCC, were significantly correlated to the 13q32.2 miRNA expression. However, other stem cell markers, such as CD34 and CD44 (including CD29 and Nanog), did not show significant correlation. The expression levels of each marker between the 14q32.2 high and low expression groups were compared, and the p value was computed by the Wilcoxon rank sum test/Mann-Whitney U test. b, expression and localization of molecules encoded by DLK1-DIO3 imprinted microRNA cluster and stem cell markers in HCC patients. Immunohistochemistry was performed to investigate the relative expression and localization of molecules regulated by the DLK1-DIO3 imprinted microRNA cluster (MEST) in tumor tissues isolated from patients positive or negative with this microRNA cluster. In addition, the expression of stem cell markers, such as EpCAM, CD90, and CD133, was also investigated in parallel. Original magnification, ×400.
These putative stemlike HCC cells were shown to be associated with high α-fetoprotein (AFP) level (36), tumor recurrence, and poor prognostic outcome of patients (3, 4, 6, 37). Therefore, we examined whether up-regulation of the DLK1-DIO3 miRNAs was associated with any clinicopathologic conditions in HCC patients. Our data showed that there was no association between DLK1-DIO3 microRNA expression in tumors and patient variables by one-way analysis of variance, including tumor stage, tumor grade, smoking, drinking, family history of HCC, and gender of the patients, except for the patient age (correlation coefficient, r = −0.21, p = 4 × 10−2) and serum SGOT level (r = 0.32, p = 1.3 × 10−3).
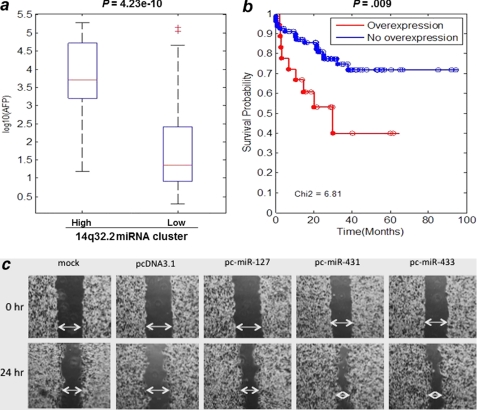
AFP, a fetal growth marker in serum, is a tumor marker (cut-off level >400 ng/ml) for HCC diagnosis but is expressed only in a subgroup of all HCC patients. Strikingly, all 18 patients with 14q32.2 miRNA overexpression also exhibited extraordinarily high levels of serum AFP (mean >7000 ng/ml) (Fig. 5a), differing significantly from those patients with negative DLK1-DIO3 miRNA expression (approximately at 40 ng/ml) (p = 4.23 × 10−10). Albeit a weak association, tumors with high DLK1-DIO3 miRNA expression correlated with the appearance of satellite lesions surrounding the main nodular tumor and with venous infiltration. These clinicopathologic features associated with elevated expression of 14q32.2 miRNAs suggested that these patients may experience higher metastatic tendency than patients not overexpressing these miRNAs. In fact, patients with lower expression levels of 14q32.2 miRNAs had longer survival times, whereas patients with overexpression of this miRNA cluster showed significantly poorer overall survival (p = 0.009) (Fig. 5b). By Cox proportional hazard model analysis, we also found that the hazard ratio was 2.812 with a 95% confidence interval of (1.255, 6.299).
FIGURE 5.
Overexpression of 14q32.2 miRNA cluster in HCC is associated with high serum AFP value and poor overall survival of liver cancer patients. a, analysis of variance box-and-whisker plots comparing serum AFP levels (ng/ml) (y axis) in two groups of patients. 97 HCC patients are divided into two groups by 14q32.2 microRNA expression levels; 79 show no overexpression, and 18 show overexpression. The significance of the difference in AFP level distributions is given above the plot. b, Kaplan-Meier plots comparing survival times of two groups of patients with overexpression and without overexpression of 14q32.2 miRNAs. c, in vitro wound healing assay showing that miRNAs from the DLK1-DIO3 cluster enhanced HCC cell migration potential. The PLC/PRF/5 hepatoma cell line was transfected with the selected miRNAs (miR-127, miR-431, and miR-433) as indicated or control vector. The migration of cells toward the wound was monitored, and images were captured at time 0 and 24 h. Magnification, ×100.
It is known that high expression levels of putative HCC stem cell markers affect tumor aggressiveness and prognosis of patients (2, 38). This prompted us to further investigate the tumorigenic properties of three most abundant microRNAs (miR-127, miR-433, and miR-431) selected from this cluster. As shown by an in vitro wound healing assay (Fig. 5c), PLC/PRF/5 hepatoma cells transfected with the DLK1-DIO3 miRNAs demonstrated higher metastatic potential than the vector and mock controls.
A recent study observed up-regulation of several miRNAs from the 14q32.2 region in a subtype of acute myeloid leukemia that bears a t(15;17) translocation (39). To examine whether 14q32.2 miRNAs are overexpressed in other types of tumors, we analyzed the expression profiles of miRNAs from kidney, gastric, colon, lung, and breast cancer samples and found that the 14q32.2 miRNAs were overexpressed in only a small subset of cases, and the overexpression was not as strong as in HCC.
Taken together, these data demonstrate that a physical cluster of DLK1-DIO3 genomic imprinted miRNAs at 14q32.2 is coordinately up-regulated in a subset of HCC patients with poor clinical outcomes. Our findings also imply that tumors overexpressing this imprinted miRNA cluster appear to be aggressive and very likely give rise to satellite lesions and vascular invasion. Also, additional characteristic genes, including AFP, HCC stem cell markers, and a subset of imprinted genes, were expressed in these tumors. Patients with overexpressed 14q32.2 miRNAs showed short survival time, and they may be candidates for more aggressive therapy or deserve targeted therapies designed to act on unique embryonic pathway-related signaling or molecular mechanisms. Therefore, this novel 14q32.2 miRNA cluster is suggested to define a new molecular subtype of HCC, arising from imprinted genomic loci and associated with poor clinical outcomes. A recent study has showed that a histone deacetylase inhibitor can reactivate the Dlk1-Dio3 locus in mouse induced pluripotent stem cells and subsequently changes stem cell development programming (11). Therefore, reprogramming the expression of the DLK1-DIO3 region in human HCC tumor epigenetically or by RNAi silencing should be explored as a therapeutic approach for this “stemlike” HCC subpopulation, as defined by overexpression of the 14q32.2 miRNA cluster. To this end, detection of the 14q32.2 miRNAs in plasma or serum samples as biomarkers for this HCC subtype warrants further investigation and clinical evaluation.
Supplementary Material
Acknowledgments
We gratefully acknowledge Douglas Bassett, Alan Sachs, and Stephen Friend (Merck & Co., Inc.) for guidance and support of these studies. We also thank Irene Ng (Hong Kong University) and Lee Lim, Ke Hao, Tao Xie, Steven Bartz, Scott Martin, Robert Phillips, and Peter Linsley (Merck & Co., Inc.) for scientific input and critical discussions.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. 1–3.
- HCC
- hepatocellular carcinoma
- AFP
- α-fetoprotein
- miRNA
- microRNA.
REFERENCES
- 1. Parkin D. M., Bray F., Ferlay J., Pisani P. (2005) CA Cancer J. Clin. 55, 74–108 [DOI] [PubMed] [Google Scholar]
- 2. Lee T. K., Castilho A., Ma S., Ng I. O. (2009) Liver Int. 29, 955–965 [DOI] [PubMed] [Google Scholar]
- 3. Sell S., Leffert H. L. (2008) J. Clin. Oncol. 26, 2800–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Z. F., Ho D. W., Ng M. N., Lau C. K., Yu W. C., Ngai P., Chu P. W., Lam C. T., Poon R. T., Fan S. T. (2008) Cancer Cell 13, 153–166 [DOI] [PubMed] [Google Scholar]
- 5. Ma S., Lee T. K., Zheng B. J., Chan K. W., Guan X. Y. (2008) Oncogene 27, 1749–1758 [DOI] [PubMed] [Google Scholar]
- 6. Yamashita T., Ji J., Budhu A., Forgues M., Yang W., Wang H. Y., Jia H., Ye Q., Qin L. X., Wauthier E., Reid L. M., Minato H., Honda M., Kaneko S., Tang Z. Y., Wang X. W. (2009) Gastroenterology 136, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slack F. J., Weidhaas J. B. (2008) N. Engl. J. Med. 359, 2720–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calin G. A., Croce C. M. (2006) Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 9. He L., Hannon G. J. (2004) Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 10. Burchard J., Zhang C., Liu A. M., Poon R. T., Lee N. P., Wong K. F., Sham P. C., Lam B. Y., Ferguson M. D., Tokiwa G., Smith R., Leeson B., Beard R., Lamb J. R., Lim L., Mao M., Dai H., Luk J. M. (2010) Mol. Syst. Biol. 6, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 12. Tward A. D., Jones K. D., Yant S., Cheung S. T., Fan S. T., Chen X., Kay M. A., Wang R., Bishop J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14771–14776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hao K., Luk J. M., Lee N. P., Mao M., Zhang C., Ferguson M. D., Lamb J., Dai H., Ng I. O., Sham P. C., Poon R. T. (2009) BMC Cancer 9, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang R., Ferrell L. D., Faouzi S., Maher J. J., Bishop J. M. (2001) J. Cell Biol. 153, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raymond C. K., Roberts B. S., Garrett-Engele P., Lim L. P., Johnson J. M. (2005) RNA 11, 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu A. M., Zhang C., Burchard J., Fan S. T., Wong K. F., Dai H., Poon R. T., Luk J. M. (2011) Omics 15, 187–191 [DOI] [PubMed] [Google Scholar]
- 17. Eklund A. C., Turner L. R., Chen P., Jensen R. V., deFeo G., Kopf-Sill A. R., Szallasi Z. (2006) Nat. Biotechnol. 24, 1071–1073 [DOI] [PubMed] [Google Scholar]
- 18. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. (2003) Nat. Biotechnol. 21, 635–637 [DOI] [PubMed] [Google Scholar]
- 20. Liu A. M., Poon R. T., Luk J. M. (2010) Biochem. Biophys. Res. Commun. 394, 623–627 [DOI] [PubMed] [Google Scholar]
- 21. Lee N. P., Chen L., Lin M. C., Tsang F. H., Yeung C., Poon R. T., Peng J., Leng X., Beretta L., Sun S., Day P. J., Luk J. M. (2009) J. Proteome Res. 8, 1293–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mok B. W., Yeung W. S., Luk J. M. (1999) FEBS Lett. 453, 243–248 [DOI] [PubMed] [Google Scholar]
- 23. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin S. P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A. C. (2003) Nat. Genet. 35, 97–102 [DOI] [PubMed] [Google Scholar]
- 25. Seitz H., Royo H., Bortolin M. L., Lin S. P., Ferguson-Smith A. C., Cavaillé J. (2004) Genome Res. 14, 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L., Luo G. Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H. J., Wang L., Wang X. J., Zhou Q. (2010) J. Biol. Chem. 285, 19483–19490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. (2010) Nature 465, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tierling S., Dalbert S., Schoppenhorst S., Tsai C. E., Oliger S., Ferguson-Smith A. C., Paulsen M., Walter J. (2006) Genomics 87, 225–235 [DOI] [PubMed] [Google Scholar]
- 29. Jelinic P., Shaw P. (2007) J. Pathol. 211, 261–268 [DOI] [PubMed] [Google Scholar]
- 30. Thorvaldsen J. L., Bartolomei M. S. (2007) Cell 130, 958. [DOI] [PubMed] [Google Scholar]
- 31. Yao X., Hu J. F., Daniels M., Shiran H., Zhou X., Yan H., Lu H., Zeng Z., Wang Q., Li T., Hoffman A. R. (2003) J. Clin. Invest. 111, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ip W. K., Lai P. B., Wong N. L., Sy S. M., Beheshti B., Squire J. A., Wong N. (2007) Cancer Lett. 250, 284–291 [DOI] [PubMed] [Google Scholar]
- 33. Jia H. L., Ye Q. H., Qin L. X., Budhu A., Forgues M., Chen Y., Liu Y. K., Sun H. C., Wang L., Lu H. Z., Shen F., Tang Z. Y., Wang X. W. (2007) Clin. Cancer Res. 13, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 34. Wu J., Qin Y., Li B., He W. Z., Sun Z. L. (2008) Genomics 91, 443–450 [DOI] [PubMed] [Google Scholar]
- 35. Huang J., Zhang X., Zhang M., Zhu J. D., Zhang Y. L., Lin Y., Wang K. S., Qi X. F., Zhang Q., Liu G. Z., Yu J., Cui Y., Yang P. Y., Wang Z. Q., Han Z. G. (2007) Carcinogenesis 28, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 36. Yamashita T., Forgues M., Wang W., Kim J. W., Ye Q., Jia H., Budhu A., Zanetti K. A., Chen Y., Qin L. X., Tang Z. Y., Wang X. W. (2008) Cancer Res. 68, 1451–1461 [DOI] [PubMed] [Google Scholar]
- 37. Ma S., Chan K. W., Hu L., Lee T. K., Wo J. Y., Ng I. O., Zheng B. J., Guan X. Y. (2007) Gastroenterology 132, 2542–2556 [DOI] [PubMed] [Google Scholar]
- 38. Yang X. R., Xu Y., Yu B., Zhou J., Qiu S. J., Shi G. M., Zhang B. H., Wu W. Z., Shi Y. H., Wu B., Yang G. H., Ji Y., Fan J. (2010) Gut 59, 953–962 [DOI] [PubMed] [Google Scholar]
- 39. Dixon-McIver A., East P., Mein C. A., Cazier J. B., Molloy G., Chaplin T., Andrew Lister T., Young B. D., Debernardi S. (2008) PLoS ONE 3, e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.