Abstract
DNA damage response is an important surveillance mechanism used to maintain the integrity of the human genome in response to genotoxic stress. Histone variant H2AX is a critical sensor that undergoes phosphorylation at serine 139 upon genotoxic stress, which provides a docking site to recruit the mediator of DNA damage checkpoint protein 1 (MDC1) and DNA repair protein complex to sites of DNA breaks for DNA repair. Here, we show that monoubiquitination of H2AX is induced upon DNA double strand breaks and plays a critical role in H2AX Ser-139 phosphorylation (γ-H2AX), in turn facilitating the recruitment of MDC1 to DNA damage foci. Mechanistically, we show that monoubiquitination of H2AX induced by RING finger protein 2 (RNF2) is required for the recruitment of active ataxia telangiectasia mutated to DNA damage foci, thus affecting the formation of γ-H2AX. Importantly, a defect in monoubiquitination of H2AX profoundly enhances ionizing radiation sensitivity. Our study therefore suggests that monoubiquitination of H2AX is an important step for DNA damage response and may have important clinical implications for the treatment of cancers.
Keywords: DNA Damage, Phosphorylation Enzymes, Protein Phosphorylation, Signal Transduction, Ubiquitination, ATM, H2AX
Introduction
DNA damage response (DDR)3 involves serial molecular events that are activated during DNA double strand breaks (DSBs) and are important for the repair of damaged DNA (1, 2). Histone variant H2AX is a central player for DDR. Upon DNA damage, H2AX is phosphorylated by ataxia telangiectasia mutated (ATM) and ATM-related kinases at serine 139, known as γ-H2AX, which serves as a docking site to recruit the mediator of DNA damage checkpoint protein 1 (MDC1) to sites of DNA damage, named DNA damage foci (3, 4). MDC1 is then phosphorylated by ATM within the DNA damage foci and facilitates the recruitment of RING finger protein 8 (RNF8) ubiquitin ligase (E3) to the DNA damage foci (1, 2). RNF8 then recruits RING finger protein 168 (RNF168) E3 ligase to promote polyubiquitination of proteins near the DNA damage foci, in turn recruiting breast cancer 1, early onset (BRCA1)-receptor-associated protein 80 (RAP80) complex to DNA damage sites for DNA repair.
H2AX is thought to be a critical sensor that can detect DSBs to initiate the early DDR. H2AX deficiency results in defects in the recruitment of MDC1 and other repair proteins to DNA damage foci for DNA repair and G2/M checkpoint, in turn enhancing the sensitivity to ionizing radiation (IR) (5, 6). In addition to its Ser-139 phosphorylation, H2AX has been recently shown to be phosphorylated by Williams-Beuren syndrome transcription factor at tyrosine 142, and this phosphorylation negatively regulates H2AX Ser-139 phosphorylation (γ-H2AX) (7, 8). Although it is known that H2AX also undergoes polyubiquitination by RNF8 and RNF168 E3 ligase upon DSBs (9–12), the functional significance of this polyubiquitination is currently unclear. Interestingly, H2AX also undergoes monoubiquitination, but the functional significance of this ubiquitination is once again unclear (10, 13).
Ubiquitination is thought to regulate protein degradation; however, it has recently emerged to play a nonproteolytic role involved in various biological processes (1, 14–16). We hypothesize that monoubiquitination of H2AX may play an important role in DDR. In this study, we aimed to identify E3 ligases responsible for H2AX monoubiquitination and its potential role in DDR.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Mouse embryonic fibroblasts (MEFs) and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). The source of ionizing radiation (IR) is from cesium 137 in the Mark 1 irradiator (J. L. Shepherd and Associates). pcDNA3-HA-FLAG-H2AX-K6R, pcDNA3-HA-FLAG-H2AX-K10R, pcDNA3-HA-FLAG-H2AX-K14R, pcDNA3-HA-FLAG-H2AX-K16R, pcDNA3-HA-FLAG-H2AX-K37R, pcDNA3-HA-FLAG-H2AX-K75/76R, pcDNA3-HA-FLAG-H2AX-K96R, pcDNA3-HA-FLAG-H2AX-K119R/K120R, pcDNA3-HA-FLAG-H2AX-K128R, and pcDNA3-HA-FLAG-H2AX-K134/135R constructs were generated by using a site-directed mutagenesis kit (Stratagene) according to the manufacturer's standard procedures using pcDNA3-HA-FLAG-H2AX-wild type (WT) as a template. pWZL-H2AX or pWZL-H2AX-K119R/K120R was generated by subcloning H2AX or H2AX-K119R/K120R into EcoRI and BamHI sites of pWZL vector. pcDNA3-HA-FLAG-H2AX-WT, pcDNA3-HA-FLAG-H2AX-S139A, and pcDNA3-HA-FLAG-RNF8-WT were described previously (10). The RING finger protein 2 (RNF2) plasmid was a gift from Dr. S. Y. Lin (M.D. Anderson Cancer Center, Houston). WT and H2AX−/− MEFs were kindly provided by Dr. B. Wang (M.D. Anderson Cancer Center, Houston).
Viral Infection
For Mock, H2AX, or H2AX-K119R/K120R restoration, H2AX−/− MEFs were infected with Mock, pWZL-H2AX, or pWZL-H2AX-K119R/K120R and selected by 50 μg/ml hygromycin for 4 days.
Small Interfering RNA (siRNA) Transfection
U2OS cells were transfected with siRNA against control (Dharmacon Research) or RNF2 (sequence, GAGAAAUACUGGAAAGUGA, Dharmacon Research) using Oligofectamine (Invitrogen) for 48 h and treated with or without IR (10 Gy). After 20 min of IR treatment, U2OS cells were harvested for immunofluorescence assay (IFA).
Immunoprecipitation (IP), Immunoblotting (IB), and IFA
IP, IB, and IFA were done essentially as described with mild modification (17, 18). For IP, cells were lysed by E1A lysis buffer (250 mm NaCl, 50 mm Hepes, pH 7.5, 0.1% Nonidet P-40 (v/v), 5 mm EDTA, protease inhibitor mixture (Roche Applied Science)). For IFA, cells were grown on chamber slides. After treating with or without IR, cells were extracted with cold cytoskeleton buffer (10 mm Hepes, pH 7.4, 300 mm sucrose, 100 mm NaCl, 3 mm MgCl2, 0.5% Triton X-100 (v/v), and protease inhibitor mixture (Roche Applied Science)). Cells were then fixed with 4% paraformaldehyde and permeabilized in 0.2% Triton X-100 in phosphate-buffered saline (PBS) at room temperature. Cells were immunolabeled using specific antibodies and observed on Zeiss Axioplan 2 imaging and FV300 Olympus confocal microscopy. The following antibodies were used for IP, IB, and IFA: anti-H2AX antibody (IP, 1:100; IB, 1:1000, Cell Signaling, Calbiochem, and Bethyl Laboratories); anti-MDC1 antibody (IB, 1:1000; IFA, 1:50–200, Bethyl Laboratories and Santa Cruz Biotechnology); anti-RNF8 antibody (IB, 1:1000, Abnova and Santa Cruz Biotechnology); anti-RNF2 antibody (IB, 1:1000; IFA, 1:100, Santa Cruz Biotechnology); anti-phospho-(Ser-139)-H2AX antibody (IB, 1:1000; IFA, 1:200, Cell Signaling and Millipore); anti-H2A antibody (IB, 1:1000, Cell Signaling); anti-H3 antibody (IB, 1:5000, Abcam); anti-ATM antibody (IB, 1:1000, Cell Signaling); anti-phosphorylated (Ser-1981)-ATM antibody (IB, 1:1000; IFA, 1:200, Rockland); anti-α-tubulin antibody (IB, 1:10,000, Sigma); anti-β-actin antibody (IB: 1:10,000, Sigma); and anti-FLAG antibody (M2, IP, 1:200; IB, 1:3000, Sigma).
Ubiquitination Assay
In vivo ubiquitination assays were performed as described previously (19). In brief, 293T cells were transfected with the plasmids for 48 h and harvested by denatured buffer (6 m guanidine HCl, 0.1 m Na2HPO4/NaH2PO4, 10 mm imidazole), followed by nickel bead purification and IB analysis. Proteins were eluted in SDS sample buffer, subjected to SDS-PAGE, transferred to nitrocellulose membrane, and immunoblotted with antibody.
Chromatin Fractionation
Chromatin fractionation was performed as described previously (20). In brief, cells with or without IR were washed twice by cold PBS. Cell pellets were resuspended in buffer A (50 mm Hepes, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol (v/v), 1 mm dithiothreitol (DTT), protease inhibitor mixture (Roche Applied Science), 0.1% Triton X-100 (v/v), and phosphatase inhibitor mixture I and II (Sigma)) on ice. After centrifuging, pellets were lysed by buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitor mixtures I and II (Sigma)). After centrifuging, pellets were washed twice by washing buffer I (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 150 mm NaCl, protease inhibitor mixture (Roche Applied Science)) and buffer II (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 250 mm NaCl, protease inhibitor mixture (Roche Applied Science)). After washing, pellets were sonicated and lysed by E1A lysis buffer. The proteins were then analyzed by IB analysis.
Colony Formation Assay
For colony formation assay, H2AX−/− MEFs infected with mock, H2AX, or H2AX-K119R/K120R (3000 cells per well) were split into 12-well plates and then treated with various doses of IR. After a 7-day culture, the viable cells were fixed and stained with crystal violet. The number of colonies (more than 50 cells for each colony) was calculated.
Foci Quantification
Foci quantification was performed as described previously (21, 22). The number of foci per cell was calculated on the microscope using the oil lens (63×). The captured images were analyzed for foci using AlphaEase® FC software. At least 40 cells were randomly collected from each group. The captured images were transformed into the intensity scale (8-bit resolution) by using AlphaEase® FC software. The number of radiation-induced DNA damage protein foci, representing the higher density thresholds of gray level as compared with background with lower intensities, was counted by the automatic counting routine separately using the AlphaEase® FC software.
Statistical Analyses
All values were the means ± S.D. of replicate samples (n = 3–6, depending on the different experiments), and experiments were repeated a minimum of three times. Differences between the two groups were assessed using the unpaired two-tailed Student's t test or by analysis of variance if more than two groups were analyzed. The Tukey test was used as a post hoc test in analysis of variance for testing the significance of pairwise group comparisons. p values <0.05 were considered statistically significant in all comparisons. SPSS version 13.0 for Windows (LEAD Technologies, Inc.) was used for all calculations.
RESULTS
Monoubiquitination of H2AX Is Required for the Recruitment of MDC1 to DNA Damage Foci upon IR
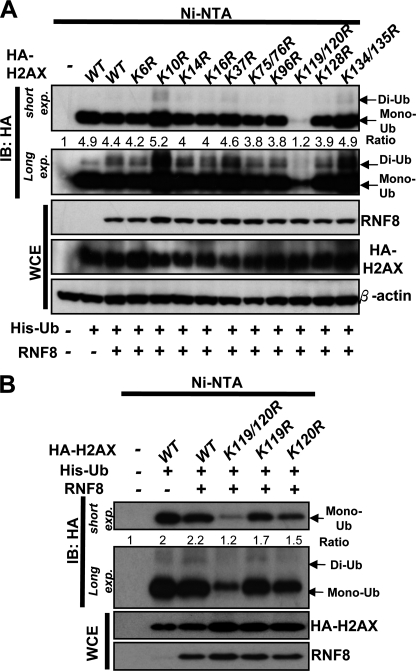
To study the role of monoubiquitination of H2AX in DDR, we first determined which residues of H2AX are the monoubiquitination sites. To this end, we mutated all the lysine residues of H2AX to arginine and tested these mutants in in vivo ubiquitination assay. We found that H2AX displayed monoubiquitination (Fig. 1, A and B). RNF8 E3 ligase did not affect the level of H2AX monoubiquitination but promoted di-ubiquitination of H2AX, consistent with the previous results showing that RNF8 is not required for H2AX monoubiquitination but for di-ubiquitination and H2AX (10). We found that mutations on Lys-119 or Lys-120 partly reduced monoubiquitination of H2A, whereas mutations on other sites have little effect on monoubiquitination of H2AX (Fig. 1B). This result is consistent with the previous report showing that Lys-119 is a monoubiquitination site for H2AX (13). Strikingly, mutations on both Lys-119 and Lys-120 residues of H2AX completely abolished monoubiquitination of H2AX, suggesting that Lys-119 and Lys-120 are two major monoubiquitination sites for H2AX (Fig. 1, A and B), consistent with a recent report (10). Interestingly, RNF8 also failed to induce di-ubiquitination of the H2AX-K119R/K120R mutant (Fig. 1A), suggesting that monoubiquitination of H2AX is a prerequisite for H2AX di-ubiquitination induced by RNF8.
FIGURE 1.
Lys-119/Lys-120 are the major monoubiquitination sites for H2AX. A and B, 293T cells were transfected with histidine-ubiquitin (His-Ub), RNF8 along with HA-H2AX or various H2AX mutants for 48 h, treated with IR (10 gray), and harvested for in vivo ubiquitination assay (see “Experimental Procedures” for details). Ni-NTA, nickel-nitrilotriacetic acid; WCE, whole-cell extracts; IB, immunoblotting; exp, exposure.
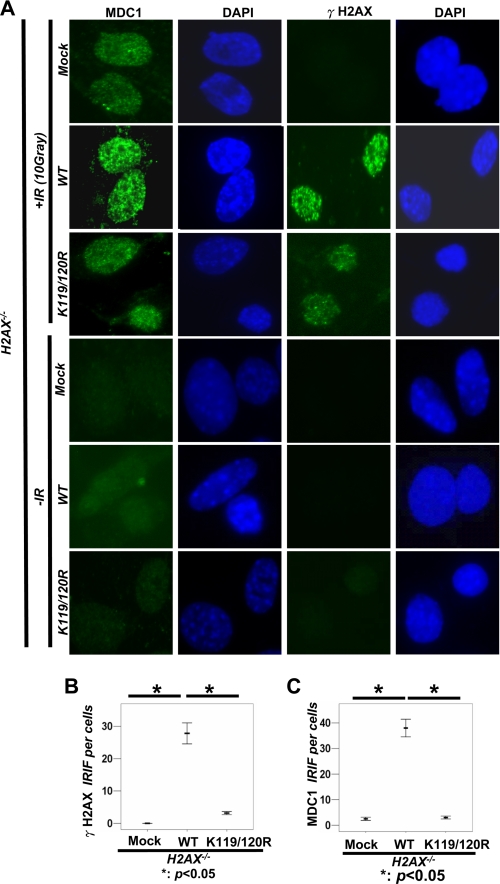
To study the role of monoubiquitination of H2AX in DDR, we examined the effect of WT-H2AX and H2AX-K119R/K120R mutant on the recruitment of MDC1 to DNA foci upon IR. As expected, we found that H2AX−/− MEFs had a defect in the recruitment of MDC1 to DNA damage foci after 20 min of IR treatment using IFA (Fig. 2, A and C, and supplemental Fig. S1A). Although restoration of WT-H2AX into H2AX−/− MEFs by using the retroviral infection system rescued the ability of MDC1 to DNA damage foci, H2AX-K119R/K120R mutant failed to do so (Fig. 2, A and C, and supplemental Fig. S1A).
FIGURE 2.
Monoubiquitination of H2AX regulates γ-H2AX and MDC1 foci formation. A, H2AX−/− MEFs infected with mock, HA-H2AX, or HA-H2AX-K119R/K120R were treated with or without IR (10 gray). After 20 min, cells were fixed and stained with γ-H2AX and MDC1 antibodies for IFA. B and C, number of foci per cell was calculated under the microscope using the oil lens (63×) (see “Experimental Procedures” for details). The mean number of γ-H2AX foci (B) and MDC1 foci (C) per each cell is shown from 40 cells calculated. The horizontal line shows the mean number of foci per cell. These differences are statistically significant (p < 0.005).
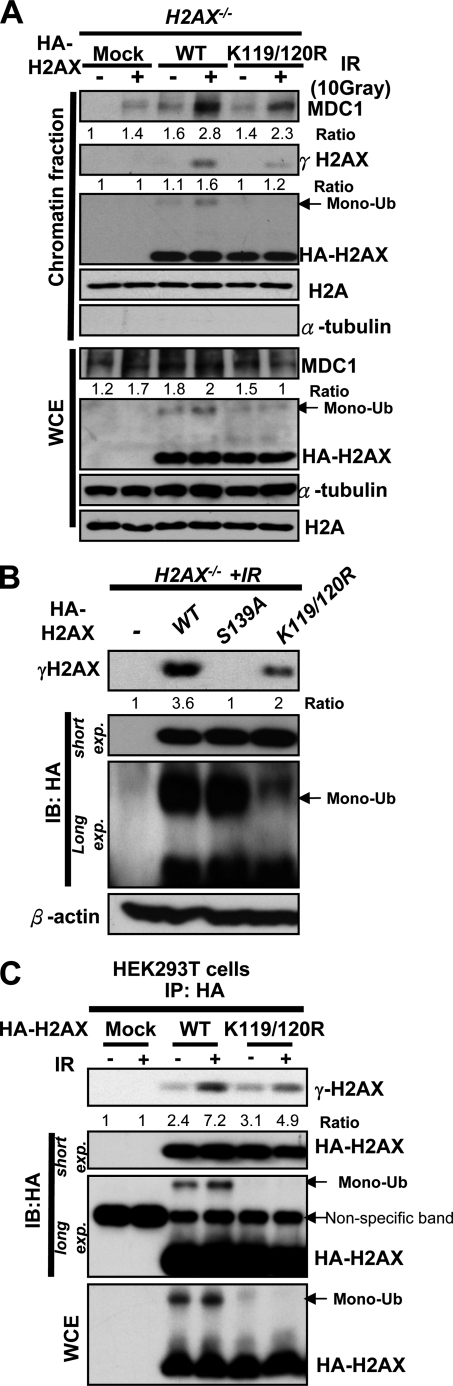
To further confirm this phenomenon, we used a biochemical approach to isolate the chromatin fraction where the DNA damage sites are located, and we examined the recruitment of MDC1 to these regions upon IR. Consistent with the IFA results, Western blot analysis revealed that WT-H2AX restored the recruitment of MDC1 to the chromatin fraction in H2AX−/− MEFs in response to IR, but the H2AX-K119R/K120R mutant compromised this effect (Fig. 3A), which correlates with the fact that monoubiquitination of WT-H2AX in the chromatin fraction was enhanced by IR, but the K119R/mutant lost its ability to become monoubiquitinated (Fig. 3, A–C). These results suggest that monoubiquitination of H2AX is required for the recruitment of MDC1 to DNA damage foci.
FIGURE 3.
Monoubiquitination of H2AX regulates γ-H2AX formation and the recruitment of MDC1 to chromatin fraction. A, H2AX−/− MEFs infected with mock, HA-H2AX, or HA-H2AX-K119R/K120R were treated with or without IR (10 gray). After incubation for 20 min, the chromatin fraction and WCE were collected for IB analysis. B, H2AX−/− MEFs transfected with mock, HA-H2AX, HA-H2AX-S139A, or HA-H2AX-K119R/K120R were treated with or without IR (10 gray). After incubation for 20 min, WCE was collected for IB analysis. C, 293T cells transfected with mock, HA-H2AX-WT, or HA-H2AX-K119R/K120R for 48 h were treated with or without IR (10 gray) and harvested 1 h after IR for IP with HA antibody, followed by IB analysis.
Monoubiquitination of H2AX Is Required for γ-H2AX Formation upon IR
Because the binding of MDC1 to H2AX requires the formation of γ-H2AX upon IR (3, 4), we therefore determined whether H2AX monoubiquitination regulates the γ-H2AX formation upon IR. As expected, restoration of H2AX, but not of H2AX-K119R/K120R mutant, into H2AX−/− MEFs rescued the formation of γ-H2AX foci after 20 min of IR treatment using IFA (Fig. 2, A and B). We also performed the biochemical fractionation experiment to isolate the chromatin fraction and found that H2AX-K119R/K120R mutant drastically reduced its ability to induce the formation of γ-H2AX in both total cell extracts and chromatin fractions after 20 min of IR treatment compared with WT-H2AX and correlated with its defect in monoubiquitination (Fig. 3, A–C). These results suggest that monoubiquitination of H2AX positively regulates the formation of γ-H2AX in response to IR treatment.
Monoubiquitination of H2AX Regulates IR Sensitivity
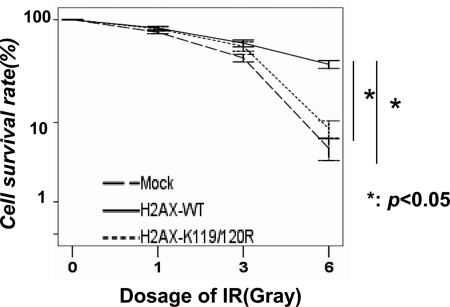
H2AX and MDC1 are known to play a critical role in IR sensitivity (3, 4). As we have revealed the critical role of H2AX monoubiquitination in γ-H2AX formation and the recruitment of MDC1 to sites of DNA breaks, we next determined whether monoubiquitination of H2AX regulates IR sensitivity. Notably, we found that although WT-H2AX restoration in H2AX−/− MEFs using the retroviral system caused the resistance of the cells to IR compared with mock restoration, H2AX-K119R/K120R mutant compromised this effect (Fig. 4). This result strongly suggests that monoubiquitination of H2AX plays a critical role in IR sensitivity.
FIGURE 4.
Monoubiquitination of H2AX regulates IR sensitivity. H2AX−/− MEFs infected with mock, H2AX, or H2AX-K119R/K120R were treated with various doses of IR, and the survival rate of these cells was determined by colony formation assay after 7 days of IR treatment. The results are shown as means ± S.D. of one representative experiment (from three independent experiments) performed in triplicate. *, p < 0.05.
Monoubiquitination of H2AX Regulates the Recruitment of ATM to DNA Foci
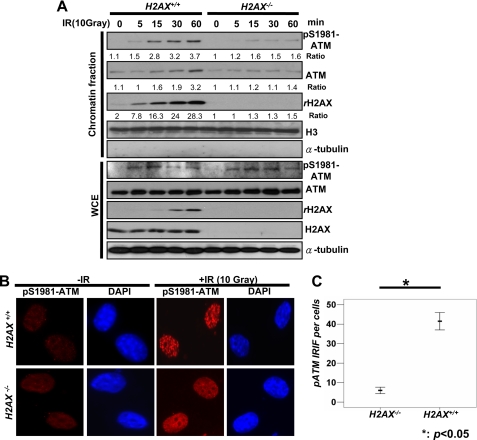
ATM is a critical kinase responsible for γ-H2AX upon IR, and it is highly possible that monoubiquitination of H2AX may either regulate ATM activation or the recruitment of ATM to the sites of DNA damage to induce γ-H2AX. To test this notion, we at first compared the degree of ATM activation and the recruitment of ATM to the sites of DNA damage between WT and H2AX−/− MEFs upon DNA damage. We found that the ATM phosphorylation (p-ATM) level in whole cell extracts upon IR was no different between WT and H2AX−/− MEFs, suggesting that H2AX is not required for ATM activation in response to IR treatment (Fig. 5A).
FIGURE 5.
H2AX is required for the recruitment of ATM to DNA damage foci. A, WT and H2AX−/− MEFs were treated with or without IR (10 Gy), and cells were harvested for the isolation of the chromatin fraction and WCE after 20 min of IR incubation, followed by IB analysis. B and C, WT and H2AX−/− MEFs were treated with or without IR (10 Gy). After incubation for 20 min, cells were fixed and stained with p-ATM antibody for IFA. The number of foci per cell was calculated on the microscope using the oil lens (63×). The mean number of foci per each cell is shown from 40 cells calculated.
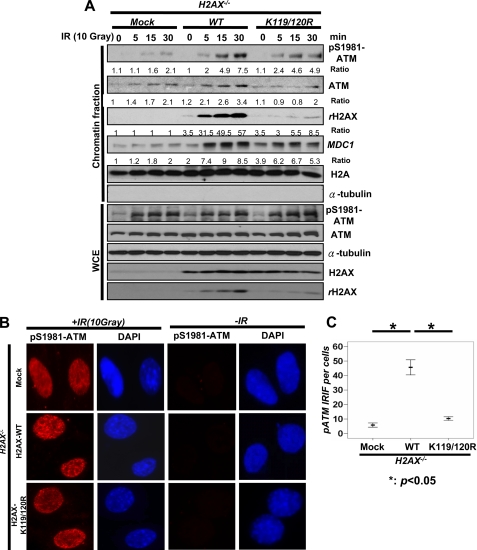
However, the recruitment of ATM to the chromatin fraction upon IR was markedly impaired in H2AX−/− MEFs, which was correlated with a profound reduction in p-ATM levels in chromatin fractions (Fig. 5A). Similarly, we found that p-ATM foci induced by IR in H2AX−/− MEFs, as determined by IFA, was also drastically reduced compared with that in WT MEFs (Fig. 5, B and C, and supplemental Fig. S1B). We next determined whether monoubiquitination of H2AX regulates ATM recruitment to DNA foci in response to DNA damage, thereby facilitating the formation and γ-H2AX and MDC1 foci. Notably, we found that WT-H2AX restoration in H2AX−/− MEFs rescued the recruitment of ATM and p-ATM to the chromatin fraction and p-ATM foci, but restoration of H2AX-K119R/K120R mutant failed to do so (Fig. 6, A–C, and supplemental Fig. S1C). However, WT-H2AX and H2AX-K119R/K120R did not affect p-ATM level in whole cell extracts (Fig. 6B). Thus, monoubiquitination of H2AX orchestrates the recruitment of ATM to sites of DNA damage, in turn regulating γ-H2AX formation and the recruitment of MDC1 to the sites of DNA breaks.
FIGURE 6.
Monoubiquitination of H2AX is required for the recruitment of ATM to DNA damage foci. A, H2AX−/− MEFs infected with mock, H2AX, or H2AX-K119R/K120R were treated with or without IR (10 Gy) and harvested for the isolation of the chromatin fraction and WCE after 20 min of IR treatment, followed by IB analysis. B and C, H2AX−/− MEFs infected with mock, H2AX, or H2AX-K119R/K120R were treated with or without IR (10 Gy). After incubation for 20 min, cells were fixed and stained with p-ATM antibody for immunofluorescence assay. The number of foci per cell was calculated manually on the microscope using the oil lens (63×). The mean number of foci each cell is shown from 40 cells calculated.
RNF2 Induces Monoubiquitination of H2AX and Regulates γ-H2AX Formation, the Recruitment of MDC1 and ATM to DNA Foci
The polycomb repressive complex 1 (PRC1), including BMI1 polycomb RING finger oncogene (BMI1) and RING finger protein 1 (RING1) and RNF2 (also known as RING2), was shown to display an E3 ligase for monoubiquitination of H2A and negatively regulates gene expression (23, 24). Interestingly, BMI1 and RNF2 were recently shown to participate in DNA damage response and IR sensitivity by regulating the recruitment of tumor suppressor p53-binding protein 1 (53BP1), BRCA1, and RAP80 to DNA damage sites (25). Moreover, BMI1 and RNF2 were also shown to contribute to monoubiquitination of H2AX (25), although the role of monoubiquitination of H2AX in DNA damage response was not examined.
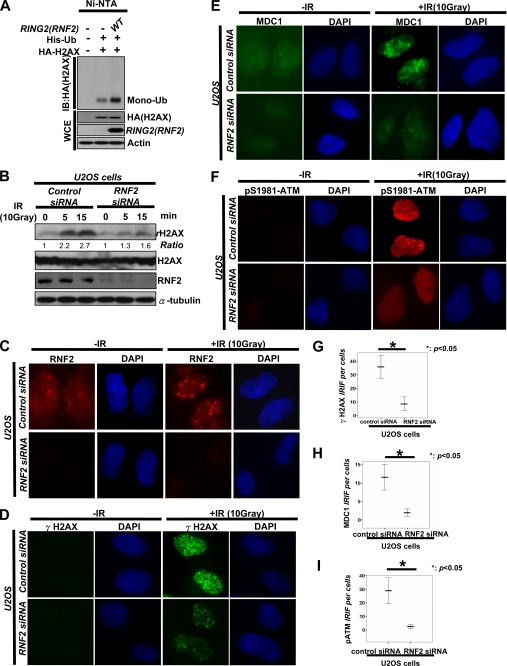
Consistent with this recent report (25), we found that RNF2 was recruited to DNA damage foci upon IR treatment, and its overexpression promoted monoubiquitination of H2AX (Fig. 7, A and C). Given that monoubiquitination of H2AX plays an important role in γ-H2AX formation and that RNF2 triggers monoubiquitination of H2AX, RNF2 should also regulate the formation of γ-H2AX as monoubiquitination of H2AX does. To test this hypothesis, we generated control and RNF2 -knockdown U2OS cells by using siRNA against green fluorescent protein (GFP) and RING2. As expected, we found that the expression level of γ-H2AX and the formation of γ-H2AX foci were profoundly impaired in RNF2-knockdown U2OS cells compared with that in GFP knockdown cells, as determined by Western blot analysis and IFA (Fig. 7, B, D, and G, and supplemental Fig. S1D). Because monoubiquitination of H2AX regulates the recruitment of MDC1 to DNA foci upon IR, we next determine whether RNF2 recapitulates the function of monoubiquitination of H2AX. Although MDC1 was recruited to DNA damage foci in control U2OS cells upon IR, it was impaired in RNF2-knockdown U2OS cells (Fig. 7, E and H, and supplemental Fig. S1D). Likewise, we found that RNF2-knockdown U2OS cells displayed the defects in p-ATM foci compared with control U2OS cells (Fig. 7, F and I, and supplemental Fig. S1D). Thus, RNF2-mediated monoubiquitination of H2AX participates in the recruitment of ATM to DNA damage sites, in turn facilitating the formation of γ-H2AX and MDC1 foci.
FIGURE 7.
RNF2-mediated monoubiquitination of H2AX regulates p-ATM foci, γ-H2AX, and MDC1 foci. A, 293T cells were transfected with ubiquitin (His-Ub), RNF2 along with HA-H2AX, treated with IR (10 Gy) for 48 h, and harvested for in vivo ubiquitination assay. B, U2OS cells were transfected with siRNA against GFP or RNF2 for 48 h, treated with or without IR (10 Gy), and harvested for WCE after 20 min of IR treatment, followed by IB analysis. C–I, control- and RNF2-knockdown U2OS cells were treated with or without IR (10 Gy). After incubation for 20 min, cells were fixed and stained with indicated antibodies for IFA. The number of foci per cell was calculated under the microscope using the oil lens (63×). The mean number of foci per each cell is shown from 40 cells calculated.
DISCUSSION
The formation of γ-H2AX by ATM kinase and other ATM-related kinases upon DSBs is known to provide a docking site for recruiting MDC1 to DNA damage foci, in turn facilitating the recruitment of BRCA1-RAP80 complex to DNA foci important for DNA repair (3, 4). Our study reveals for the first time that monoubiquitination of H2AX at Lys-119/Lys-120, which is induced in the chromatin fraction by IR, plays a critical role in DDR by facilitating γ-H2AX formation and the recruitment of MDC1 to DNA damage foci. Mechanistically, we further show that monoubiquitination of H2AX is required for the recruitment of ATM to DNA damage foci, thus affecting the formation of γ-H2AX upon IR. Importantly, we demonstrate that monoubiquitination of H2AX plays a critical role in IR sensitivity.
The recruitment of ATM to DNA damage foci represents an early event important for γ-H2AX formation and subsequent recruitment of DNA repair proteins to sites of DNA breaks for DNA repair. How exactly ATM is recruited to the DNA damage foci in response to DSBs remains unclear. Our finding that H2AX deficiency and H2AX monoubiquitination-dead mutant (H2AX-K119R/K120R) both display a defect in ATM recruitment to DNA damage foci but not ATM activation suggests that ATM activation may not occur in DNA damage foci during DSBs. Thus, the role of H2AX monoubiquitination induced by DSBs is to recruit active ATM to sites of DNA breaks to trigger γ-H2AX formation and subsequent recruitment of repair proteins to DNA foci.
Recent studies identified RNF8 E3 ligase as a novel regulator important for DDR (10, 11, 26). RNF8 was shown to regulate IR sensitivity by facilitating the recruitment of the RAP80-BRCA1 complex to DNA damage foci without affecting the formation of γ-H2AX and MDC1 foci. Thus, RNF8 functions downstream of MDC1 but upstream of RAP80. Although RNF8 plays an important role in DDR, how it participates in this process remains largely unclear. Interestingly, RNF8 was recently shown to trigger and contribute to di-ubiquitination and tri-ubiquitination of H2AX (10), although the functional significance of H2AX ubiquitination remains to be determined. However, RNF8 is not required for monoubiquitination of H2AX and γ-H2AX, as RNF8 knockdown does not affect monoubiquitination of H2AX and γ-H2AX (10). Consistent with this, we found that RNF8 overexpression did not promote monoubiquitination of H2AX (Fig. 1, A and B), suggesting that RNF8 is not an E3 ligase for monoubiquitination of H2AX.
Importantly, we show that monoubiquitination of H2AX is also required for H2AX di-ubiquitination (Fig. 1A). We rationalize that monoubiquitination of H2AX may affect the recruitment of RNF8 to DNA damage foci upon IR, in turn regulating H2AX di-ubiquitination and polyubiquitination. Although H2AX di-ubiquitination and polyubiquitination are elicited during DDB, their role in DDR is currently unclear. Given that the loss of RNF8 only affects the recruitment of the RAP80-BRCA1 complex to DNA damage sites, but not MDC1 recruitment and γ-H2AX foci (10, 11), it is conceivable that di-ubiquitination and tri-ubiquitination of H2AX does not participate in ATM recruitment, γ-H2AX formation, and the formation of MDC1 foci. We show here, however, that monoubiquitination of H2AX indeed plays an important role in these processes. Thus, it is unlikely that monoubiquitination of H2AX regulates ATM recruitment, γ-H2AX formation, and the formation of MDC1 foci indirectly through affecting di-ubiquitination and tri-ubiquitination of H2AX.
What is the E3 ligase responsible for monoubiquitination of H2AX? One recent study suggests that a polycomb-repressive complex-1, including BMI1, RING1, and RING2 (RNF2), is likely an E3 ligase for monoubiquitination of H2AX, as BMI1 deficiency or knockdown inhibits H2AX monoubiquitination (25). Consistent with this notion, we found that RNF2 overexpression promotes in vivo H2AX monoubiquitination (Fig. 7A). Moreover, we provide the evidence that monoubiquitination of H2AX elicited by RNF2 is critical for the recruitment of ATM to sites of DNA breaks, in turn facilitating γ-H2AX formation and the recruitment of MDC1 to DNA damage foci. Thus, it is likely that RNF2 regulates the recruitment of DNA repair proteins, such as RAP80, BRCA1, and 53BP1, to DNA damage sites and IR sensitivity by regulating monoubiquitination of H2AX, in turn facilitating ATM recruitment, γ-H2AX formation, and MDC1 foci formation. In a previous article, Lin and co-workers (32) demonstrated that H2AX monoubiquitination is also induced by BMI1 and RNF2 and plays a critical role in facilitating γ-H2AX formation and the recruitment of MDC1 to DNA foci, supporting our conclusion that H2AX monoubiquitination is critical for DNA damage response.
However, it should be noted that two earlier reports revealed that H2AX monoubiquitination at Lys-119/Lys-120 does not obviously affect IR-induced γ-H2AX levels (10, 27), which is different from our current observation that monoubiquitination of H2AX at Lys-119/Lys-120 plays a positive role in regulating γ-H2AX formation. Although such discrepancy remains unclear, the possible explanation for it may be due to the distinct time course analyzed in the experiments. In our case, we analyzed γ-H2AX at an early time point (within 30 min), whereas these two studies (10, 27) examined it at late time point (4–6 h). These results are indeed consistent with the current model indicating that ATM positively regulates γ-H2AX only at early time points (within 30 min), but not at late time points (>1 h) (28–31). Thus, monoubiquitination of H2AX may affect γ-H2AX only at early time points, but not at late time points, by regulating the recruitment of ATM to DNA damage foci.
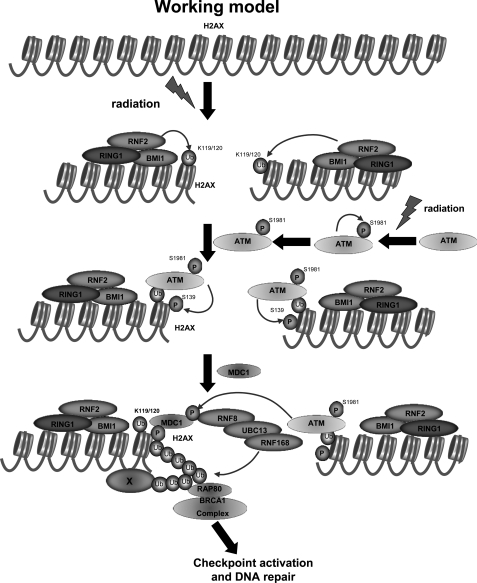
On the basis of our finding, we propose a working model by which monoubiquitination of H2AX regulates DDR (Fig. 8). Upon IR treatment, the BMI1-RNF2-RING1 E3 ligase complex is recruited to DNA damage foci and subsequently induced monoubiquitination of H2AX, which in turn facilitates the recruitment of active ATM to DNA damage foci, followed by facilitating the formation of γ-H2AX and the recruitment of MDC1 and other DNA repair proteins to DNA damage sites. Our study therefore reveals an important cross-talk between H2AX monoubiquitination and H2AX phosphorylation, providing novel paradigms for DDR and potentially therapeutic strategies for the treatment of human cancers.
FIGURE 8.
Working model for the role of monoubiquitination of H2AX in DDR. Upon DSBs, the RNF2-RING1-BMI1 E3 ligase complex is recruited to DNA damage foci and induces monoubiquitination of H2AX at Lys-119/Lys-120. Monoubiquitination of H2AX recruits active ATM to sites of DNA breaks, in turn facilitating the formation of γ-H2AX and the recruitment of MDC1 to DNA damage foci. MDC1 is then phosphorylated by ATM and recruits RNF8 and RNF168 E3 ligases to DNA damage foci. RNF8 and RNF168 cooperate to promote the polyubiquitination of H2AX, H2A or unidentified proteins, which may recruit the BRCA1-RAP80 complex to DNA damage foci to facilitate DNA repair.
Supplementary Material
Acknowledgments
We thank Drs. J. Chen, S. Y. Lin, M. C. Hung, and B. Wang for reagents. We are grateful to D. Sarbassov and M. G. Lee for insightful comments and suggestions. We extend special thanks to S. Zhang for technical support. We also thank Dr. Z. Han for assistance with the confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants 1RO1CA149321-01 and 1R01CA136787-01A2. This work was also supported by the Trust Scholar Fund from M.D. Anderson Cancer Center and New Investigator Award PC081292 from the Department of Defense (to H. K. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- DDR
- DNA damage response
- ATM
- ataxia telangiectasia mutated
- p-ATM
- ATM phosphorylation
- IR
- ionizing radiation
- MEF
- mouse embryonic fibroblast
- DSB
- DNA double strand break
- IP
- immunoprecipitation
- IB
- immunoblotting
- IFA
- immunofluorescence assay
- Gy
- gray.
REFERENCES
- 1. Huen M. S., Sy S. M., Chen J. (2010) Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim H., Chen J. (2008) Mol. Cells 25, 457–461 [PMC free article] [PubMed] [Google Scholar]
- 3. Ayoub N., Jeyasekharan A. D., Bernal J. A., Venkitaraman A. R. (2009) Cell Cycle 8, 1494–1500 [DOI] [PubMed] [Google Scholar]
- 4. van Attikum H., Gasser S. M. (2009) Trends Cell Biol. 19, 207–217 [DOI] [PubMed] [Google Scholar]
- 5. Bassing C. H., Alt F. W. (2004) Cell Cycle 3, 149–153 [DOI] [PubMed] [Google Scholar]
- 6. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. (2008) Nat. Rev. Cancer 8, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook P. J., Ju B. G., Telese F., Wang X., Glass C. K., Rosenfeld M. G. (2009) Nature 458, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao A., Li H., Shechter D., Ahn S. H., Fabrizio L. A., Erdjument-Bromage H., Ishibe-Murakami S., Wang B., Tempst P., Hofmann K., Patel D. J., Elledge S. J., Allis C. D. (2009) Nature 457, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 10. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 12. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 13. Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., Kimura H., Ikura M., Nishikubo S., Ito T., Muto A., Miyagawa K., Takeda S., Fishel R., Igarashi K., Kamiya K. (2007) Mol. Cell. Biol. 27, 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhoj V. G., Chen Z. J. (2009) Nature 458, 430–437 [DOI] [PubMed] [Google Scholar]
- 15. Hoeller D., Dikic I. (2009) Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 16. Yang W. L., Wu C. Y., Wu J., Lin H. K. (2010) Cell Cycle 9, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin H. K., Bergmann S., Pandolfi P. P. (2004) Nature 431, 205–211 [DOI] [PubMed] [Google Scholar]
- 18. Lin H. K., Wang G., Chen Z., Teruya-Feldstein J., Liu Y., Chan C. H., Yang W. L., Erdjument-Bromage H., Nakayama K. I., Nimer S., Tempst P., Pandolfi P. P. (2009) Nat. Cell Biol. 11, 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., Lin H. K. (2009) Science 325, 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lukas C., Melander F., Stucki M., Falck J., Bekker-Jensen S., Goldberg M., Lerenthal Y., Jackson S. P., Bartek J., Lukas J. (2004) EMBO J. 23, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leatherbarrow E. L., Harper J. V., Cucinotta F. A., O'Neill P. (2006) Int. J. Radiat. Biol. 82, 111–118 [DOI] [PubMed] [Google Scholar]
- 22. Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 23. Cao R., Tsukada Y., Zhang Y. (2005) Mol. Cell 20, 845–854 [DOI] [PubMed] [Google Scholar]
- 24. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 25. Ismail I. H., Andrin C., McDonald D., Hendzel M. J. (2010) J. Cell Biol. 191, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu J., Huen M. S., Lu L. Y., Ye L., Dou Y., Ljungman M., Chen J., Yu X. (2009) Mol. Cell. Biol. 29, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kinner A., Wu W., Staudt C., Iliakis G. (2008) Nucleic Acids Res. 36, 5678–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kühne M., Riballo E., Rief N., Rothkamm K., Jeggo P. A., Löbrich M. (2004) Cancer Res. 64, 500–508 [DOI] [PubMed] [Google Scholar]
- 30. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 31. Wang H., Wang M., Wang H., Böcker W., Iliakis G. (2005) J. Cell. Physiol. 202, 492–502 [DOI] [PubMed] [Google Scholar]
- 32. Pan M. R., Peng G., Hung W. C., Lin S. Y. (2011) J. Biol. Chem. 286, 28599–28607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.