Abstract
Podoviridae are double-stranded DNA bacteriophages that use short, non-contractile tails to adsorb to the host cell surface. Within the tail apparatus of P22-like phages, a dedicated fiber known as the “tail needle” likely functions as a cell envelope-penetrating device to promote ejection of viral DNA inside the host. In Sf6, a P22-like phage that infects Shigella flexneri, the tail needle presents a C-terminal globular knob. This knob, absent in phage P22 but shared in other members of the P22-like genus, represents the outermost exposed tip of the virion that contacts the host cell surface. Here, we report a crystal structure of the Sf6 tail needle knob determined at 1.0 Å resolution. The structure reveals a trimeric globular domain of the TNF fold structurally superimposable with that of the tail-less phage PRD1 spike protein P5 and the adenovirus knob, domains that in both viruses function in receptor binding. However, P22-like phages are not known to utilize a protein receptor and are thought to directly penetrate the host surface. At 1.0 Å resolution, we identified three equivalents of l-glutamic acid (l-Glu) bound to each subunit interface. Although intimately bound to the protein, l-Glu does not increase the structural stability of the trimer nor it affects its ability to self-trimerize in vitro. In analogy to P22 gp26, we suggest the tail needle of phage Sf6 is ejected through the bacterial cell envelope during infection and its C-terminal knob is threaded through peptidoglycan pores formed by glycan strands.
Keywords: Cell Cycle, Chromatin, Cyclins, Microarray, Transcription, Androgen Receptor, Prostate Cancer
Introduction
Tailed bacteriophages are grouped in three broad families based on tail morphology: Myoviridae, Siphoviridae, and Podoviridae (1). The first two are formed by bacteriophages that present long contractile and long non-contractile tails, respectively (1). In contrast, the Podoviridae family is characterized by a short tail apparatus attached to a unique vertex of the icosahedral capsid. Many Podoviridae bind the host surface via dedicated tailspike proteins that provide adsorption specificity and initiate the subsequent infection process. Much is known about the tail apparatus of P22, the type species of the genus (2, 3). In P22, the tail apparatus is ∼2.8 MDa in molecular mass and contains 51 polypeptide chains (4). This includes a dodecamer of gene 1 product (gp1)2 portal protein (5), six gp9 tailspike trimers (6), 12 copies of gp4 (7), a hexamer of gp10 (8), and a trimer of gp26 (9). The tailspike possesses endorhamnosidase activity that hydrolyzes the O antigen on the outer surface of Salmonella enterica LT2 (10). Like the other component proteins of the tail, the tail needle gp26 (9) is essential for viability (11). Gp26 functions as a molecular “plug” that seals the portal protein channel after DNA packaging to stabilize viral DNA in the capsid (11, 12). Accordingly, disrupting the gene encoding gp26 results in non-infective particles that have lost their DNA (11). The tail needle may also be involved in cell wall penetration, an idea supported by the observation that gp26 is injected into the host during infection, followed by viral DNA ejection (10, 13).
Like tailspikes, tail needles are well conserved among P22-like phages. All tail needles characterized so far consist of trimeric fibers ∼220–320 Å in length3 (12). Trimerization is mediated by a long trimeric coiled-coil core that contains 11–16 trimerization heptads (12). As revealed by the crystal structure of P22 gp26, the tail needle is only 20–25 Å in diameter (14, 15) and possesses remarkably high structural stability (16). The tail needle N-terminal ∼60 residues are highly conserved in all P22-like phages, providing an attachment point for gp10 and serving to plug the portal channel (12, 14, 16). In contrast, the tail needle C terminus is variable both in sequence and morphology; it represents the most exposed moiety of the virion distal to the head (6) and is thought to be involved in penetrating the host. The C-terminal domain is helical in P22 (14, 15) and HK620 (12) but forms a globular “knob” in other phages such as Sf6 and HS1 (12), which infect the Gram-negative bacteria Shigella flexneri and Escherichia coli, respectively.
The process by which tailed phages eject their genome across the cell envelope is poorly understood. In Myoviridae such as T4, infection involves complex rearrangements of tail proteins, together with a lysozyme-like activity that creates a hole in the cell envelope, which allows the tail tube to interact with the cytoplasmic membrane and hence to initiate phage DNA transfer (17). In Siphoviridae such as λ or SPP1, the long (∼1000Å) non-contractile tail usually binds a specific host receptor to initiate genome ejection. This process is well understood in SPP1, where infection is initiated by specific interaction of the virus tail with its receptor Bacillus subtilis YueB (18). YueB is a membrane protein whose extracellular domain spans the thick Gram-positive cell wall. The interaction between SPP1 tail and YueB triggers a cascade of conformational changes transmitted along the tail tube, which leads to genome ejection (19). Despite these differences, recently observed similarities among the tail proteins of Myoviridae and Siphoviridae suggest that there may be mechanistic similarities that are not yet understood (20, 21).
In Podoviridae, the mechanisms of cell envelope penetration and genome delivery are less well understood. The P22-like phages do not appear to recognize a proteinaceous receptor, but use the combined action of an adhesion protein (tailspike) and a tail needle (gp26) to bind and penetrate the host cell envelope. The O antigen chain of the LPS functions as the primary receptor site for specific adsorption of P22-like phages (10). Tailspike hydrolyzes the O antigen, thereby leading to the delivery of the phage particle to the outer surface of the host. Although the molecular details of subsequent events are largely unknown, the tail needle is released from the virion to open a passage for the ejection proteins and DNA (10, 13). Andres et al. (22) recently developed an in vitro assay to study P22 genome ejection, which is specifically triggered by aggregates of purified Salmonella LPS. This suggests that the signal for tail needle ejection and genome release lays in the tailspike, an idea supported by observations that LPS with different O antigen structure, lipid A, phospholipids, or soluble O antigen polysaccharide fail to trigger P22 gp26 ejection and genome release (22). Thus, P22 and likely all P22-like phages do not appear to use a secondary protein receptor located in the bacterial outer membrane.
In this work, we have determined the 1.0 Å crystal structure of Sf6 tail needle knob and examined some of its biological features. The knob folds into a trimeric jellyroll characteristic of viral proteins of the PRD1-adenovirus lineage.
EXPERIMENTAL PROCEDURES
Cloning and Protein Expression of Needle Protein Knob
The gene-encoding phage Sf6 tail needle was PCR-amplified from genomic DNA (23) and ligated between the XbaI and HindIII restriction sites of the vector pMal-c2e (New England Biolabs), fusing the Sf6 tail needle to an N-terminal maltose binding protein (MBP) (12). A PreScission Protease cleavage site was engineered between the MBP and the tail needle gene. A construct the tail needle knob was constructed by long PCR by removing residues 1–131 from the Sf6 tail needle gene. The final knob construct includes residues 132–282 plus additional four amino acids from the plasmid polylinker (sequence FGSS). A mutant of Sf6 tail needle knob that fails to bind phosphate was generated by introducing two alanine substitutions at Asp233 and Ser231 using the QuikChange site-directed mutagenesis kit (Stratagene). All constructs generated in this study were entirely sequenced to ensure the correctness of the DNA sequence. All expression plasmids were induced in E. coli BL21(DE3) pLysE for 22 h at 22 °C. The recombinant MBP-fused Sf6 knob was purified over amylose-agarose beads (New England Biolabs). Fusion proteins were digested with PreScission Protease in Buffer A (50 mm NaCl, 20 mm Tris-HCl, pH 8.0, and 5 mm β-mercaptoethanol) and purified by Superdex 200 gel filtration (GE Healthcare) pre-equilibrated in Buffer A, followed by passage over a 5-ml DEAE column (Sigma) that captured contaminating MBP, whereas pure knobs are recovered in the flow-through. The final purified knob (residues 132–282) was analyzed by SDS-PAGE, revealing a purity of >95%. Denaturant-treated samples were prepared by dialyzing purified knob against 1.5 m guanidine hydrochloride (GdnHCl) and 500 mm NaCl for 24 h, followed by a second dialysis against Buffer A for 12 h.
Negative Stain Transmission Electron Microscopy
Electron microscopic examination used a Joel JEM-2100 transmission electron microscope at 200 kV. Sf6 tail needles (∼1 μg/ml) were applied to glow-discharged carbon-coated copper grids and were stained with 1% uranyl formate. Grids were imaged on a TVIPS F415MP CCD camera at an optical magnification of 100,000×. Two-dimensional projection averages of negatively stained needles were computed using EMAN software (24) as described previously (12), using an average of 100 particles manually selected from a single micrograph.
Crystallization and Structure Determination
Sf6 tail needle knob was crystallized under 22% PEG 3350, 50 mm Tris, at pH 8.0. Crystals were analyzed at beamline X6A at the Brookhaven National Synchrotron Light Source on an Area Detector System Corporation Q-270 CCD detector. The best crystals diffracted to ∼1.0 Å resolution (see Table 1). Diffraction data were reduced to intensities using the programs of the HKL-2000 package (25) and further analyzed using software of the CCP4 package (26). For phasing, a single Sf6 tail needle knob crystal was soaked in cryoprotectant (26% PEG 3350, 50 mm Tris, at pH 8.0, 27% ethylene glycol) supplemented with ∼1 m sodium bromide for ∼60 s. An ultracomplete and redundant data set to 1.8 Å was collected at beamline F1 at the Cornell High Energy Synchrotron Source, on an Area Detector System Corporation Q-270 CCD detector, at the bromine edge (∼0.92 Å) (see Table 1). Twenty five bromine sites were located by single-wavelength anomalous diffraction phasing protocol in PHENIX (27), which also built an initial main chain model of the tail needle knob. This model was then refined to the highest resolution available (1.0 Å) with iterative restrained refinement using PHENIX (27) and manual model building with COOT (28). At 1.0 Å resolution, the quality of the electron density maps was excellent. A phosphate, three l-Glu and 1017 ordered water molecules were modeled into the Fo − Fc electron density using the program COOT. The complete protein model with l-Glu and solvent was then further subjected to several cycles of anisotropic B-factor and individual occupancy refinement. Riding hydrogen atoms were added in the final round of refinement. The final model includes C-terminal residues 132–282, in addition to four linker residues (FGSS) and has an Rwork/Rfree of 13.28/14.18%, respectively. The model has exceptional geometry, with 93% of the residues in the most favored regions of the Ramachandran plot (see Table 1). Strands in the jellyroll motif were labeled according to convention (29). The GdnHCl-treated knob was crystallized under the same conditions as the wild type knob. Crystals were diffracted on an Xcalibur PX Ultra manufactured by Oxford at the Kimmel Cancer Center X-ray Crystallography and Molecular Characterization shared resource facility at Thomas Jefferson University. Diffraction data to 2.8 Å resolution were collected on a 165 mm Onyx CCD detector using a single crystal cryo-protected with 27% ethylene glycol. The structure was solved by molecular replacement and refined using PHENIX (27) to an Rwork/Rfree of 26.99/30.84% (see Table 1). Inspection of electron density maps using COOT (28) revealed that GdnHCl-treated knob is free of l-Glu. Ribbon diagrams in this work were generated using PyMOL (30). The coordinates and structure factors for the Sf6 tail needle knob have been deposited in the Protein Data Bank with accession code 3RWN.
TABLE 1.
Data collection, phasing, and refinement statistics for the Sf6-knob domain
| Native | NaBr | Dialyzed | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 58.4, 88.3, 89.1 | 58.6, 88.5, 88.9 | 182.74, 53.24, 101.72 |
| α, β, γ | 90, 90, and 90° | 90, 90, and 90° | 90.00, 111.51, and 90.00° |
| Resolution (Å) | 15.0-1.0 (1.04-1.0)a | 15-1.8 (1.86-1.80)a | 30-2.80 (2.95-2.80)a |
| Measured reflections | 932,344 | 656,305 | 51,084 |
| Unique reflections | 222,826 | 43,724 | 18,253 |
| Rsymb | 5.9 (57.3) | 10.0 (37.1) | 13.6 (33.7) |
| I/σI | 25.9 (1.9) | 49.2 (10.2) | 6.6 (2.8) |
| Completeness (%) | 90.1 (71.5) | 99.9 (99.4) | 80.0 (84.4) |
| Redundancy | 4.2 (3.3) | 15 (14.7) | 2.8 (2.5) |
| Molecules per asymmetric unit | 1 | 1 | 2 |
| No. of bromine sites | 25 | ||
| FOMd | 0.326 | ||
| Refinement | |||
| Resolution (Å) | 15.0-1.0 | 15.0-2.8 | |
| No. of reflections | 393,623 | 18,043 | |
| Rwork/R—-freec | 13.28/14.18 | 26.99/30.84 | |
| No. of atoms | |||
| Protein | 3499 | 6938 | |
| l-Glu/ion | 3/1 | ||
| Water | 1,017 | 6 | |
| B-factors | |||
| Protein overall | 8.04 | 22.86 | |
| l-Glu/ion | 7.66/7.0 | ||
| Water | 20 | 25.00 | |
| R.m.s.d. | |||
| Bond lengths (Å) | 0.005 | 0.008 | |
| Bond angles | 1.163° | 0.96° | |
| Ramachandran plot (%) | |||
| Core region | 93 | 91.2 | |
| Allowed region | 7 | 8.8 | |
| Generously allowed region | 0 | 0 | |
| Disallowed region | 0 | 0 | |
a The highest resolution shell is shown in parentheses.
b Rsym = Σi,h|I(i,h) − 〈I(h)〉|/Σi,h|I(i,h)|, where I(i,h) and 〈I(h)〉 are the ith and mean measurement of intensity of reflection h.
c The Rfree value was calculated using 2000 reflections.
d Figure of Merit.
SDS Resistance Assay and Thermal Stability
For the SDS resistance assay, 20 μl of Sf6 tail needle fiber or the tail needle knob at ∼1 mg/ml were incubated either at 22 or 95 °C for 5 min in Laemmli sample buffer (31), followed by electrophoretic separation on 12.5% SDS-PAGE. Thermal stabilities were analyzed by circular dichroism using a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control system. All measurements were carried out at 6 μm protein in 20 mm sodium phosphate, pH 8.0, and 50 mm NaCl. Temperature-induced unfolding was measured by monitoring changes in the ellipticity at 218 nm in 1.0 °C intervals from 20 to 80 °C. Measurements were done using a heating rate of 20 °C/h in a 0.1-cm path length cuvette. Slow cooling to 20 °C followed by a second run to 80 °C was carried out to probe the reversibility of unfolding (32).
RESULTS
Tail Needle Fiber of Bacteriophage Sf6 Contains a C-terminal Knob
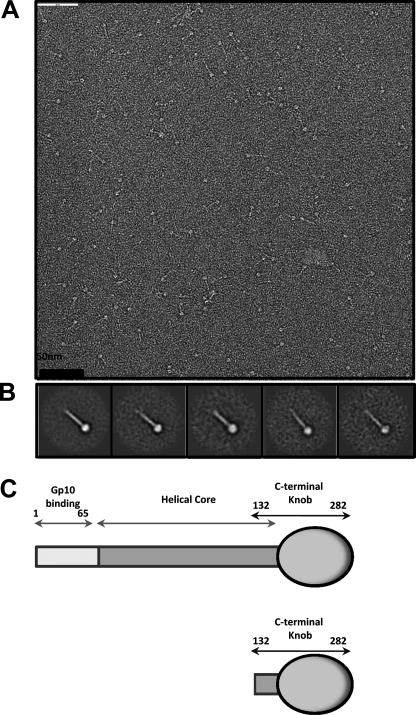
The tail needle of bacteriophage Sf6 consists of 282 amino acid residues and assembles into a triple stranded coiled-coil (12). Similar to P22 gp26, the Sf6 tail needle is characterized by an exceedingly high structural stability, consistent with a putative role in cell envelope penetration (14). To determine its morphology, the purified recombinant Sf6 tail needle was spotted on a carbon-coated copper grid, negatively stained with 1% uranyl formate and visualized by transmission electron microscopy. Micrographs showed a characteristic rod-like elongated fiber, characterized by a terminal globular knob (Fig. 1A). To improve the signal-to-noise ratio, ∼350 individually selected particles were subjected to single particle averaging. The resultant two-dimensional averaged projections revealed a ∼220 Å long straight fiber with a discrete ∼50 Å wide globular knob at one end, thought to be the C terminus (Fig. 1B) (12). Based on this topology and previous biochemical analyses (12), we define the architecture of the Sf6 tail needle as being composed of three linear domains: (i) an N-terminal gp10-binding domain (residues 1–60) that is ∼92% identical in all gp26-like fibers (12), (ii) a trimeric helical coiled-coil core (residues 61–140), which in Sf6 consists of ∼80 residues and 11 trimerization heptads (12); (iii) a C-terminal globular domain, which represents the outermost exposed moiety of the virion (residues 141–282) (Fig. 1C). The N-terminal domain and middle coiled-coil core domain are highly conserved throughout the gp26 superfamily, whereas the C-terminal knob (referred to as the “tail needle knob”) is found only in certain phages such as Sf6 and HS1 (12). To shed light on the three-dimensional architecture of Sf6 C-terminal domain, we generated an N-terminal deletion construct of the tail needle spanning residues 132–282. This fragment includes the last trimerization heptad found in the helical core followed by the C-terminal knob (Fig. 1C) (12). The Sf6 tail needle knob was overexpressed in E. coli and purified to homogeneity in its native trimeric conformation (molecular mass ∼ 51.4 kDa).
FIGURE 1.
Bacteriophage Sf6 tail needle forms an elongated fiber with a C-terminal globular domain. A, micrographs of negatively stained Sf6 tail needle fibers. B, five class averages resulting from several iterations of multireference alignment (24). The approximate length of the Sf6 tail needle estimated from the projection average is ∼220 Å. C, schematic diagram of the Sf6 tail needle (residues 1–282) and the knob construct used in this study (residues 132–282).
Structure of Sf6 Tail Needle Knob at 1.0 Å Resolution
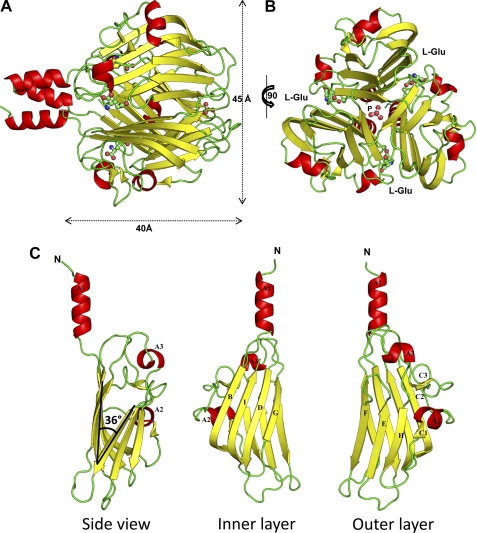
We crystallized the Sf6 tail needle knob in an orthorhombic space group with one trimeric knob per asymmetric unit. Crystals of the tail needle knob diffracted to 1.0 Å using synchrotron radiation (Table 1). The structure was phased by single-wavelength anomalous diffraction using a crystal transiently soaked in ∼1 m sodium bromide. The positions of 25 bromine atoms were determined using a highly redundant dataset collected at the bromine K-absorption edge (Table 1). Initial single-wavelength anomalous diffraction phases to 1.8 Å resolution were extended to the highest resolution available (1.0 Å) using 3-fold non-crystallographic averaging and used to autobuild a complete atomic model, one that also includes hydrogen atoms. Analysis of electron density difference maps defined the position of 1017 water molecules, one phosphate ion, and three molecules of l-Glu bound to the knob. The final model was refined to an Rwork/Rfree of 13.28/14.18% (Table 1).
In agreement with the averaged two-dimensional projections, the knob has a globular shape with diameter of ∼45 Å (Fig. 2A). The proximal end of the knob is attached to the helical core, largely missing in the construct, with the exception of heptad 16 (residues 133–139) (12). The 51.4-kDa knob folds as a homotrimer that displays a characteristic TNF-like fold (Fig. 2, A and B). Each of the three identical subunits adopts a jellyroll topology, characterized by two layers of β-sheets crossing at an angle of ∼36° and three α-helices. The inner layer contains four antiparallel β-strands (B, I, D, and G) (Fig. 2C). In contrast, the outer layer has three β-strands (H, E, and F) in addition to a short β-strand C1 hydrogen bonded to strand H and a short β-hairpin C2-C3 as part of an extended loop connecting to inner layer strand I (Fig. 2C). α-Helix A1 forms the N-terminal helical core and is rigidly connected to strand B of the inner layer. α-Helix A2 links strand C1 to β-hairpin C2-C3, and α-helix A3 connects strand G and H, respectively (Fig. 2C). The residues between the helical core end and the TNF fold do not contain flexible side chains, suggesting the knob is rigidly connected to the tail needle helical core. This is distinct from the C-terminal helical domain of P22 gp26, which is connected to the helical core by a flexible hinge, severely tilted in different crystal forms (8, 15, 16).
FIGURE 2.
Atomic structure of bacteriophage Sf6 tail needle knob bound to l-Glu visualized crystallographically at 1.0 Å resolution. A and B, ribbon diagram of the trimeric Sf6 tail needle knob shown in side and top views, respectively. The structure is colored by secondary structure elements with β-strands and α-helices in yellow and red, respectively. l-Glu and phosphate ion are shown as ball-and-stick diagrams. C, ribbon diagram of the tail needle knob monomer reveals the characteristic architecture of a TNF jellyroll fold consisting of eight β-strands organized in two β-sheets with a tilt angle of 36°.
Overall, the β-sheets from adjacent knob subunits form ∼60 hydrogen bonds to build a tightly packed trimer interface that buries a total surface area of ∼3366 Å2. Such a topology creates a solvent-filled closed cavity located along the central 3-fold axis of symmetry occupying a volume of 1080 Å3. A phosphate ion is tightly bound at the distal tip of the knob trimeric interface, in a shallow cleft formed by three pairs of Asn233/Ser231 projecting from each of the subunits (Fig. 2B). Replacing these two residues with alanine does not reduce the knob structural stability (data not shown), suggesting that the phosphate ion is only trapped at the 3-fold interface and does not play a major structural role.
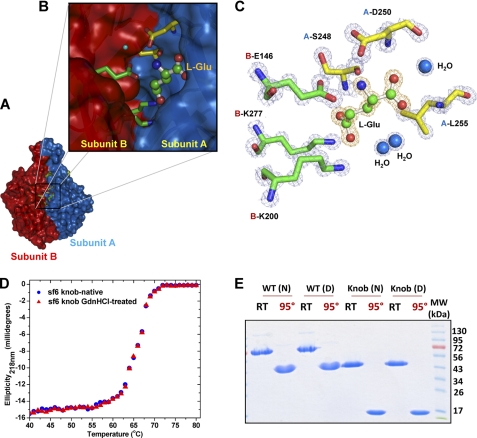
l-Glu Is Bound at Tail Needle Knob Subunit Interface
Perhaps the most intriguing feature of the Sf6 tail needle knob structure lies in the solvent-accessible cleft between monomers (Fig. 3, A and B). At 1.0 Å resolution, we identified an l-Glu bound to each monomer. l-Glu is visible as a ∼3.8σ peak of positive electron density in a difference map calculated with the coefficient Fo − Fc and phases from the refined knob structure (Fig. 3C). Each l-Glu is wedged into a narrow cleft of approximate 1082 Å3 formed by two neighboring subunits, where it forms close contacts with six protein side chains and three water molecules (Fig. 3C). Notably, two sets of intermolecular salt bridge interactions are observed between l-Glu and residues emanating from both subunits of a dimeric interface (referred herein as subunit A and B) (Fig. 3C). First, the cationic amine (−NH3+) of l-Glu engages in salt bridges with the carboxyls of A-Asp250 (Oδ2 atom) and B-Glu146 (Oϵ1 and Oϵ2 atoms), which both project into the cleft. Second, the anionic carboxylates (Oϵ1 and Oϵ2 atoms) of l-Glu form salt bridges with the ϵ-amino groups of B-Lys200 and B-Lys277. In addition, l-Glu also hydrogen bonds with A-Ser248 and with main chain amino groups of A-Asp250 and A-Leu255. Finally, three water molecules within 3.0 Å distance are coordinated to the carboxylate C-terminal oxygen atom (OXT), and Oϵ2 of l-Glu. The refined B-factor of l-Glu is lower than that of average protein atoms in the structure (∼7.66 Å2 versus ∼8.04 Å2), suggesting a full occupancy in the crystal structure. This is quite remarkable, as l-Glu was not added to the tail needle knob before or during crystallization but is likely captured in the expression host.
FIGURE 3.
l-Glu binding cleft in Sf6 tail needle knob. A, surface representation of Sf6 knob highlighting the interface between subunits A and B (colored in blue and red, respectively). B, zoom-in view of the subunit interface where l-Glu is bound. Subunit A and B side chains from the final refined model are shown in sticks, and l-Glu is shown in a stick-and-ball representation. C, the final refined model for l-Glu and the interacting residues from both subunits are overlaid to the final 2Fo − Fc electron density map calculated at 1.0 Å resolution and contoured at 1.5σ above background (in blue). The l-Glu is bound in a cleft formed at the interface of two knob monomers, closely surrounded by B-Glu146, B-Lys277, A-Leu255, B-Lys200, A-Ser248, and A-Asp250. D, thermal stability of the native and GdnHCl-treated Sf6 knob. Both samples unfold irreversibly with a melting temperature (Tm) of ∼67 °C. E, SDS resistance assay of wild type Sf6 tail needle and Sf6 knob that were previously treated under native conditions (N) or dialyzed against 1.5 m GdnHCl, 0.5 m NaCl (D). RT and 95° denote samples that were either incubated at 22 °C or boiled for 5 min prior to being electrophoresed.
l-Glu Is Not Major Structural Determinant for Knob Stability and Trimerization in Vitro
To explore the functional role of l-Glu, we first hypothesized that it had a structural role. For example, l-Glu may increase knob conformational stability by “filling” an empty volume between monomers. To test this hypothesis, we subjected the Sf6 knob to extensive dialysis followed by recrystallization under conditions identical to those used for the native protein. Several crystal structures of dialyzed knob were determined at resolutions between 1.7 and 3 Å; in all cases, the cleft was fully occupied by l-Glu (data not shown), suggesting that l-Glu has a low rate of dissociation from the protein, which, as seen in the structure, buries the small molecule in a deep cleft. In a second set of experiments, we dialyzed purified knob against 1.5 m GdnHCl and 0.5 m NaCl. This concentration of GdnHCl is not sufficient to unfold the trimer (which denatures at >4.5 m) (12) but should enhance any protein breathing and allow a faster release of l-Glu. After crystallization of the GdnHCl-treated knob, a 2.8 Å data set was collected, and the structure was solved by molecular replacement (Table 1). This structure had two knob molecules in the asymmetric unit and revealed empty l-Glu-binding sites. Instead, a water molecule was seen hydrogen bonding Ser248, in a similar position occupied by the carboxyl group of l-Glu in the native structure (data not shown). Adding l-Glu instead of water followed by crystallographic refinement resulted in a 0.5% increase in Rfree, confirming that GdnHCl-treated knob is free of l-Glu. With the exception of l-Glu, structural models for both native and GdnHCl-treated knob superimposed within experimental uncertainty (root mean square deviation (r.m.s.d.) 0.194 Å). Thus, dialysis of purified tail knob against moderate concentrations of GdnHCl removes l-Glu from the knob without affecting the rest of the structure.
To determine whether the apostructure had reduced structural stability relative to the native knob, and therefore whether l-Glu enhances the knob structural stability, we carried out thermal melting experiments, monitoring changes in ellipticity at 218 nm. Both GdnHCl-treated and native samples displayed irreversible thermal unfolding with an apparent melting temperature (Tm) of 67 °C (Fig. 3D). Likewise, both the full-length tail needle/tail needle knob and equivalent samples previously dialyzed against 1.5 m GdnHCl, 0.5 m NaCl migrated as a SDS-resistant trimeric species on 12.5% SDS-PAGE (Fig. 3E). Based on these data, l-Glu does not appear to be a major structural determinant for knob stability and trimerization in vitro. However, because the knob thermal denaturation is strictly irreversible (Fig. 3D), this experiment measures only the kinetics of denaturation and provides limited information about the knob folding, which, in vivo (inside the bacterial expression host), occurs in the presence of l-Glu. Therefore, although not essential in vitro, l-Glu may still play a role in the assembly of Sf6 tail needle knob in vivo.
Comparison of Sf6 Tail Needle Knob to TNF Jellyroll β-Structures of PRD1-adenovirus Lineage
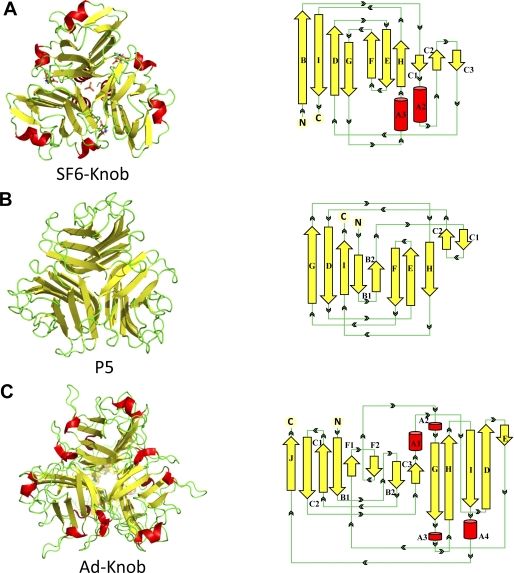
A comprehensive three-dimensional comparison of the Sf6 tail needle knob structure against structures in the Protein Data Bank was carried out using the DALI server (33). DALI identified 23 structures with a TNF-like fold that superpose to Sf6 tail needle knob with an r.m.s.d. between 2.9–5.0 Å. Interestingly, the viral protein with the greatest structural similarity (r.m.s.d. 3.2 Å) to the Sf6 knob is PRD1 vertex protein P5 (PDB code 1YQ5) (Fig. 4B). PRD1 P5 and the adenovirus knob receptor-binding domains have been shown to be evolutionarily related (34). Although the adenovirus knob (PDB code 1NOB) was not identified by DALI, alignment using secondary structure matching (35) revealed an overall significant structural similarity with the Sf6 knob (r.m.s.d. ∼ 4.5 Å) (Fig. 4C).
FIGURE 4.
Comparison of the Sf6 tail needle knob with the receptor-binding domains of PRD1 and adenovirus. Left panel, Sf6 knob (A); PRD1 P5 (PDB code 1YQ5) (B); and adenovirus knob (PDB code 1NOB) (C). Right panel, topological diagrams of each protomer (generated using PDBsum (50)). Red and yellow are α-helices and β-strands, respectively.
Despite less than 10% amino acid sequence similarity, many structural similarities exist among the Sf6 tail needle knob, PRD1 vertex protein P5, and adenovirus knob (34, 36, 37). The three proteins are topologically similar with ∼48% β-strands. The β-sheet angle for Sf6 knob is 36° compared with 39° and 42° for PRD1 P5 and the adenovirus knob, respectively. This contributes to the slightly tighter packing of the Sf6 tail needle knob relative to the PRD1/adenovirus knobs, which is also exemplified by its slightly narrower external width (45, 47, and 60 Å for Sf6, PRD1, and adenovirus knobs, respectively) (Fig. 4, A–C). Noticeable differences among the three structures exist in their secondary structure content: PRD1 P5 lacks α-helices and presents a high percentage of loops, whereas the Sf6 and adenovirus knobs are only 7 and 9.2% α-helical, respectively (Fig. 4, A–C). However, the position of these helices varies greatly between Sf6 and adenovirus. For instance, helix A2 in Sf6 contains six amino acids (163LVSLLK168) and folds into a β-strand in PRD1 P5 (Fig. 4, A and B) or Sf6 helix A3 (254FLATN258) is localized in a loop in PRD1 P5 (Fig. 4, A and B). Interestingly, Sf6 helix A3 extends to the lid of the l-Glu binding cleft and adopts in this knob a conformation drastically different as compared with PRD1 vertex protein P5 (local r.m.s.d. ∼ 8 Å) and adenovirus knob (local r.m.s.d. ∼ 7 Å) (Fig. 4, A–C). This is noteworthy, in particular at the level of l-Glu that is located at the subunit interface of the Sf6 knob, analogous to the adenovirus knob recognition pockets where the coxsackie adenovirus receptor interacts with the knob (36).
Three-dimensional Model for Full-length Sf6 Tail Needle
The N-terminal 59 residues of the Sf6 tail needle contain a gp10-binding domain, whose sequence is ∼92% identical in the P22-like gp26 family. Residues 60–140 of Sf6 helical core reveal 56% sequence similarity to P22 gp26 and contain 11 predicted adjacent heptads with conserved hydrophobic residues at positions a and d (12). The only region of the Sf6 helical core present in our structure, helix A1 (Fig. 2C) is superimposable to the equivalent N-terminal heptad region of P22 gp26 (PDB code 2POH) (Fig. 5A) (14). We used homology modeling to generate a three-dimensional model for the Sf6 helical core. The model output by the Phyre server (38) showed 100% confidence and is essentially identical to the helical core of P22 gp26 (Fig. 5B). Inspection of non-identical residues in the two fibers revealed that these residues are solvent exposed and do not change to overall trimer structure. The Sf6 helical core is very slender, with an average diameter of 25 Å and a length of 180 Å, which is followed by the ∼45 Å wide knob. Interestingly, the receptor-binding protein of the lactococcal phage TP901-1 base plate (39) and the receptor-binding protein from p2 phage (20) also share a TNF-like structure. This phage infects the Gram-positive bacterium Lactococcus lactis, and although the composition of Gram-negative and -positive cell envelopes are drastically different, the TP901-1 receptor-binding protein exhibits strong structural similarities to gp26 tail needles (Fig. 5C). The N-terminal region of the TP901-1 protein forms a short trimeric helical hairpin similar to the gp10-binding domain of gp26 (14, 15); the helical core is also connected to a triple β-helix similar to that in P22 gp26. In addition, a domain resembling the Sf6 knob is attached at the distal end of the TP901-1 helical core. Notably, the lactococcal phages p2 and TP901-1 receptor-binding proteins bind saccharides in between subunits, at a topologically similar position of the jellyroll motif as the Sf6 tail needle knob binds l-Glu (40–42). Thus, the tail needle fold appears to be conserved in phages that infect very different hosts; individual phages have likely optimized a fiber length best suited to attach and penetrate the host cell envelope.
FIGURE 5.
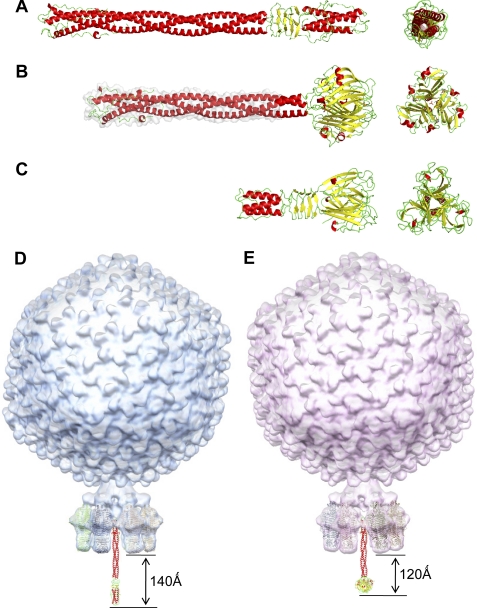
Modeling bacteriophage tail needles. A, crystal structure of P22 tail needle gp26 (PDB code 3C9I). B, homology model of the full-length Sf6 tail needle. The portion of the tail needle helical core missing (residues 1–132) in the construct used for crystallography was modeled using Phyre (38) and is highlighted in gray. C, crystal structure of the receptor binding protein from lactococcal phage TP901-1 base plate (PDB code 3EJC) (20, 39). D, a composite model of bacteriophage P22 and Sf6 (E) mature virions generated by fitting the atomic structure of gp26 tail needles and tailspikes into the asymmetric cryo-EM reconstruction of mature P22 (EMDB ID 1220) (6).
DISCUSSION
Phages have evolved different mechanisms to eject their genome through the host cell envelope. The structure of the Sf6 tail needle knob presented in this paper expands our understanding of cell wall penetrating devices. A number of Sf6 tail needle features are particularly interesting. First, the knob adopts a TNF-fold similar to that observed in classical TNF receptor-binding domain shared by both phages (e.g. PRD1) and animal viruses (e.g. adenovirus) (Fig. 4). This is surprising as a protein receptor is neither known to be required for P22 infection in vivo nor necessary for P22 genome ejection in vitro (22). Second, a molecule of l-Glu is tightly bound at each subunit interface of the Sf6 tail needle knob. Although in vitro l-Glu does not appear to play a major structural role for tail needle assembly, this small molecule is intimately bound to what appears to be a highly dedicated binding cleft. Because we have no convincing evidence for proteolytic activity with Sf6 knob,4 l-Glu found in the knob is likely picked up from the soluble pool of l-Glu present in the expression host. Third, an identical knob as that found in phage Sf6 (which infects S. flexneri) was also identified and characterized in phage HS1 that infects E. coli (12). However, the tail needle length varies considerably in P22-like phages, likely to match the host cell envelope, which in Gram-negative bacteria is a complex multilayer compartment of 150 Å in thickness (43).
The actual function of a tail needle in cell envelope penetration and viral genome ejection remains uncertain. Morphologically, the tail needle is the most exposed feature protruding from the tail, which makes it likely to interact first with the outer membrane. Structural studies by cryo-EM (44, 45) and x-ray crystallography (14, 15) have determined that in the mature P22 virion the tail needle projects ∼140 Å beyond the plane formed by the tips of the tail spikes (Fig. 5D). In Sf6, which is similar to P22, this distance is expected to be ∼120 Å (Fig. 5E). Furthermore, recent in vitro work has demonstrated that P22 tail needle can be ejected from virions by treatment with lipopolysaccharide (22), suggesting that gp26 does not interact with a proteinaceous receptor and is mechanically ejected into the cell during infection. Enzymatic studies using either isolated tailspike or mature P22 virions have determined that O antigen cleavage by tailspike stops about five repeat units from the core LPS (22). The tail needle was suggested to function as a simple “pressure sensor” that triggers genome ejection after contacting the LPS inner core or lipid A. A similar role can be hypothesized for the Sf6 tail needle because the P22 and Sf6 tailspikes are remarkably similar (46, 47), and despite the differences between the distal ends of the Sf6 and P22 tail needle, the length of their helical core is also very similar (Fig. 5, A, B, D, and E). Thus, the mechanisms by which the Sf6 and P22 tail needles are ejected during infection may be the same.
Model for Tail Needle-mediated Penetration of Gram-negative PG
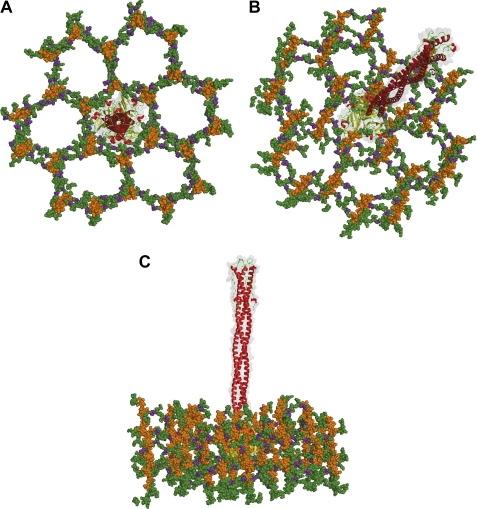
In Gram-negative bacteria, the cell wall consists of glycan strands, alternating residues of N-acetylmuramic acid and N-acetylglucosamine, periodically cross-linked by a tetrapeptide: l-Ala, d-Glu, meso-diaminopimelic acid, and d-Ala (48). NMR studies on synthetic fragments of the cell wall have revealed that the PG backbone forms a right-handed helix, with a periodicity of three N-acetylglucosamine-N-acetylmuramic acid repeats (49). The tetrapeptide cross-links 3-fold symmetric units of N-acetylglucosamine-N-acetylmuramic acid generating pores with a predicted diameter of ∼70 Å, which is large enough to accommodate enzymes involved in its biosynthesis (49). To determine how the Sf6 knob would fit inside the Gram-negative PG, we modeled the structure of Sf6 tail needle inside the proposed structure of PG. Notably, the knob diameter (∼45 Å) fits well inside a “PG pore” (Fig. 6, A–C), and the slender conformation of the tail needle can be threaded through a 6-fold symmetric PG pore formed by glycan strands (49). This is compatible with a model whereby the Sf6 tail needle is ejected through the host cell envelope without interfering or potentially clashing with PG. As a corollary, the host cell wall does not represent a physical barrier that prevents tail needle penetration.
FIGURE 6.
A structural model for Sf6 tail needle penetration of Shigella cell wall. Top (A), tilted (B), and side (C) view of the Sf6 tail needle embedded into a pore of the Gram-negative cell wall, according to the structure of PG determined by Meroueh et al. (49). Glycan strands are colored orange, and the cross-linking stem peptides are colored green, with d-Glu in purple. The Sf6 tail needle is shown as a transparent solvent-accessible gray surface overlaid to the ribbon structure.
A potential multistep mechanism that describes the infection of Shigella by bacteriophage Sf6 can be hypothesized: (i) the phage interacts with the host cell surface via tailspikes (at least three (10)) that have endorhamnosidase activity (46) and provide the driving force to adhere onto the host surface and remove repeated units of O antigen repeats. (ii) Upon contact of the tail needle knob with the outer lipid A leaflet, a series of conformational changes are triggered, which propel the tail needle through the outer cell envelope membrane. Tail needle ejection is energized by the pressure created by the highly condensed DNA in the head. (iii) In the periplasm, the tail needle knob is threaded through a PG pore. This interaction facilitates passage of the tail needle through the PG layer and avoids curving or bending of the virion, which would result in ejection of DNA into the periplasm compartment. (iv) In analogy to that proposed for P22 (13), the tail needle is ejected inside the host followed by ejection proteins and phage genome. The molecular details of this fascinating cell envelope-penetrating molecular fiber need further investigation, but the structure of the tail needle knob described in this paper is a step forward in understanding how P22-like bacteriophages penetrate the host cell envelope to eject their genome.
Acknowledgments
We thank Stephan Wilkens (The State University of New York Upstate) for assistance with electron microscopy and Dr. Shahriar Mobashery (University of Notre Dame) for sharing the coordinates of the cross-linked cell wall PG. We are grateful to Vivian Stojanoff and staff at the the Brookhaven National Synchrotron Light Source beamline X6A and at macCHESS for beam time and beamline assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R56AI076509-01A1 (to G. C.) and AI074825 (to S. R. C.).
The atomic coordinates and structure factors (code 3RWN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
A. Bhardwaj, unpublished data.
I. J. Molineux, unpublished data.
- gp
- gene product
- l-Glu
- l-glutamic acid
- MBP
- maltose-binding protein
- GdnHCl
- guanidine hydrochloride
- r.m.s.d.
- root mean square deviation
- Tm
- melting temperature
- PDB
- Protein Data Bank
- PG
- peptidoglycan.
REFERENCES
- 1. Ackermann H. W. (2003) Res. Microbiol. 154, 245–251 [DOI] [PubMed] [Google Scholar]
- 2. Teschke C. M., Parent K. N. (2010) Virology 401, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casjens S. R., Thuman-Commike P. A. (2011) Virology 411, 393–415 [DOI] [PubMed] [Google Scholar]
- 4. Lander G. C., Khayat R., Li R., Prevelige P. E., Potter C. S., Carragher B., Johnson J. E. (2009) Structure 17, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olia A. S., Prevelige P. E., Jr., Johnson J. E., Cingolani G. (2011) Nat. Struct. Mol. Biol. 18, 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lander G. C., Tang L., Casjens S. R., Gilcrease E. B., Prevelige P., Poliakov A., Potter C. S., Carragher B., Johnson J. E. (2006) Science 312, 1791–1795 [DOI] [PubMed] [Google Scholar]
- 7. Olia A. S., Al-Bassam J., Winn-Stapley D. A., Joss L., Casjens S. R., Cingolani G. (2006) J. Mol. Biol. 363, 558–576 [DOI] [PubMed] [Google Scholar]
- 8. Olia A. S., Bhardwaj A., Joss L., Casjens S., Cingolani G. (2007) Biochemistry 46, 8776–8784 [DOI] [PubMed] [Google Scholar]
- 9. Andrews D., Butler J. S., Al-Bassam J., Joss L., Winn-Stapley D. A., Casjens S., Cingolani G. (2005) J. Biol. Chem. 280, 5929–5933 [DOI] [PubMed] [Google Scholar]
- 10. Israel V. (1978) J. Gen. Virol. 40, 669–673 [DOI] [PubMed] [Google Scholar]
- 11. Strauss H., King J. (1984) J. Mol. Biol. 172, 523–543 [DOI] [PubMed] [Google Scholar]
- 12. Bhardwaj A., Walker-Kopp N., Casjens S. R., Cingolani G. (2009) J. Mol. Biol. 391, 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Israel V. (1977) J. Virol. 23, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olia A. S., Casjens S., Cingolani G. (2007) Nat. Struct. Mol. Biol. 14, 1221–1226 [DOI] [PubMed] [Google Scholar]
- 15. Olia A. S., Casjens S., Cingolani G. (2009) Protein Sci. 18, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhardwaj A., Olia A. S., Walker-Kopp N., Cingolani G. (2007) J. Mol. Biol. 371, 374–387 [DOI] [PubMed] [Google Scholar]
- 17. Leiman P. G., Chipman P. R., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2004) Cell 118, 419–429 [DOI] [PubMed] [Google Scholar]
- 18. Baptista C., Santos M. A., São-José C. (2008) J. Bacteriol. 190, 4989–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plisson C., White H. E., Auzat I., Zafarani A., São-José C., Lhuillier S., Tavares P., Orlova E. V. (2007) EMBO J. 26, 3720–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sciara G., Bebeacua C., Bron P., Tremblay D., Ortiz-Lombardia M., Lichière J., van Heel M., Campanacci V., Moineau S., Cambillau C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andres D., Hanke C., Baxa U., Seul A., Barbirz S., Seckler R. (2010) J. Biol. Chem. 285, 36768–36775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Casjens S., Winn-Stapley D. A., Gilcrease E. B., Morona R., Kühlewein C., Chua J. E., Manning P. A., Inwood W., Clark A. J. (2004) J. Mol. Biol. 339, 379–394 [DOI] [PubMed] [Google Scholar]
- 24. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) J. Struct Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Collaborative Computational Project, N (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 27. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Chelvanayagam G., Heringa J., Argos P. (1992) J. Mol. Biol. 228, 220–242 [DOI] [PubMed] [Google Scholar]
- 30. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 1.3, Schrodinger, LLC, San Carlos, CA [Google Scholar]
- 31. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Bhardwaj A., Walker-Kopp N., Wilkens S., Cingolani G. (2008) Protein Sci. 17, 1475–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 34. Merckel M. C., Huiskonen J. T., Bamford D. H., Goldman A., Tuma R. (2005) Mol. Cell 18, 161–170 [DOI] [PubMed] [Google Scholar]
- 35. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 36. Bewley M. C., Springer K., Zhang Y. B., Freimuth P., Flanagan J. M. (1999) Science 286, 1579–1583 [DOI] [PubMed] [Google Scholar]
- 37. van Raaij M. J., Mitraki A., Lavigne G., Cusack S. (1999) Nature 401, 935–938 [DOI] [PubMed] [Google Scholar]
- 38. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 39. Bebeacua C., Bron P., Lai L., Vegge C. S., Brøndsted L., Spinelli S., Campanacci V., Veesler D., van Heel M., Cambillau C. (2010) J. Biol. Chem. 285, 39079–39086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spinelli S., Campanacci V., Blangy S., Moineau S., Tegoni M., Cambillau C. (2006) J. Biol. Chem. 281, 14256–14262 [DOI] [PubMed] [Google Scholar]
- 41. Tremblay D. M., Tegoni M., Spinelli S., Campanacci V., Blangy S., Huyghe C., Desmyter A., Labrie S., Moineau S., Cambillau C. (2006) J. Bacteriol. 188, 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spinelli S., Desmyter A., Verrips C. T., de Haard H. J., Moineau S., Cambillau C. (2006) Nat. Struct. Mol. Biol. 13, 85–89 [DOI] [PubMed] [Google Scholar]
- 43. Doerrler W. T. (2006) Mol. Microbiol. 60, 542–552 [DOI] [PubMed] [Google Scholar]
- 44. Tang J., Lander G. C., Olia A., Li R., Casjens S., Prevelige P., Jr., Cingolani G., Baker T. S., Johnson J. E. (2011) Structure 19, 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen D. H., Baker M. L., Hryc C. F., DiMaio F., Jakana J., Wu W., Dougherty M., Haase-Pettingell C., Schmid M. F., Jiang W., Baker D., King J. A., Chiu W. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chua J. E., Manning P. A., Morona R. (1999) Microbiology 145, 1649–1659 [DOI] [PubMed] [Google Scholar]
- 47. Müller J. J., Barbirz S., Heinle K., Freiberg A., Seckler R., Heinemann U. (2008) Structure 16, 766–775 [DOI] [PubMed] [Google Scholar]
- 48. Vollmer W., Blanot D., de Pedro M. A. (2008) FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 49. Meroueh S. O., Bencze K. Z., Hesek D., Lee M., Fisher J. F., Stemmler T. L., Mobashery S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4404–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laskowski R. A. (2009) Nucleic Acids Res. 37, D355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]