Abstract
Much is known about the role of STAT3 in regulating differentiation of interleukin-17-producing Th17 cells, but its function in other lymphocyte subsets is not well understood. In this report, we reveal wide-ranging functions of STAT3 in T-cells and provide evidence that STAT3 is convergence point for mechanisms that regulate lymphocyte quiescence and those controlling T-cell activation and survival. We show here that STAT3 inhibits T-lymphocyte proliferation by up-regulating the expression of Class-O Forkhead transcription factors, which play essential roles in maintaining T-cells in quiescent state. We further show that STAT3 binds directly to FoxO1 or FoxO3a promoter and that STAT3-deficiency resulted in down-regulation of the expression of FoxO1, FoxO3a and FoxO-target genes (IκB and p27Kip1). Compared with wild-type T-cells, STAT3-deficient T-cells produced more IL-2, due in part, to marked decrease in IκB-mediated sequestration of NF-κB in the cytoplasm and resultant enhancement of NF-κB activation. However, the high level of IL-2 production by STAT3-deficient T-cells was partially restored to normal levels by overexpressing FoxO1. It is notable that their exaggerated increase in IL-2 production rendered STAT3-deficient lymphocytes more susceptible to activation-induced cell death, suggesting that STAT3 might protect T-cells from apoptosis by limiting their production of IL-2 through up-regulation of FoxO1/FoxO3a expression. Moreover, we found that STAT3 enhanced survival of activated T-cells by up-regulating OX-40 and Bcl-2 while down-regulating FasL and Bad expression, suggesting that similar to role of FoxOs in regulating the lifespan of worms, STAT3 and FoxO pathways converge to regulate lifespan of T-lymphocytes.
Keywords: Cytokine, Immunology, Interleukin, Lymphocyte, STAT Transcription Factor
Introduction
Production of Interleukin (IL)2-2 is one of the hallmarks of TCR signaling and largely responsible for T-cell proliferation and expansion (1). However, transcription of IL-2 is actively suppressed in resting T cells by T-cell quiescence factors (2, 3). Members of the FoxO (Forkhead box, class-O) subfamily of Forkhead transcription factors are important T-lymphocyte quiescence factors with broad influence on cell growth and survival. FoxO proteins maintain naïve or resting T cells in quiescent state (G0 cell cycle phase) by up-regulating the expression of cell cycle inhibitors, such as p27kip1, Gadd45, cyclin E, and p130 (4). When the resting T-cell encounters its cognate Ag in context of APC, TCR-mediated activation of Ras, PI3K/Akt, and mTOR kinases inactivate FoxOs by inducing their phosphorylation and translocation from the nucleus to the cytoplasm through 14-3-3-dependent mechanisms (5). The expulsion from the nucleus relieves the T-cell from inhibitory effects of FoxOs and allows cell cycle progression (3, 6). FoxOs also regulate T-cell quiescence by inducing expression of IκB, a protein that interacts with and sequesters NF-κB in the cytoplasm (7). Thus, by sequestrating NF-κB in the cytoplasm the naïve T-cell is deprived of a transcription factor required for transcription of IL-2 (8). IL-2 is a growth and survival factor for T cells (6). It promotes the expression of anti-apoptotic Bcl-2 members while inhibiting pro-apoptotic members (e.g. Bim) through PI3K/AKT-dependent inactivation of FoxO transcription factors (6). Paradoxically, IL-2 is required for induction of FasL and activation-induced cell death (AICD) (9), highlighting the dual role of IL-2 in regulating lymphocyte growth and survival on one hand while also orchestrating mechanisms of self-tolerance through induction of AICD.
Interleukin-17-producing T-cell (Th17) is an important T-helper subset characterized by a unique transcriptional program regulated through activation of STAT3 pathways by IL-6, IL-21, and IL-23 (10–12). Although Th17 cells constitutively express IL-2 early in their development, they rapidly lose capacity to produce IL-2 as they mature, through mechanisms mediated by STAT3/IL-23 signaling (13). Interestingly, it has also been shown that IL-2 antagonizes the differentiation and expansion of mouse Th17 cells (14, 15), suggesting that STAT3 and IL-2 signals may exert mutually antagonistic effects on Th17 differentiation and/or proliferation. Nonetheless, genome-wide analyses have revealed that STAT3 regulates many genes involved in T-cell proliferation and survival (11, 16) although it is not clear whether IL-2 and STAT3 pathways have overlapping or distinct roles in these cellular processes. It is however notable that STAT3-deficient T cells proliferate more vigorously than WT T cells in response to ionophore/PMA (mitogen that activate T cells independently of cytokines) but are severely impaired in IL-6-induced proliferation (17), indicating complex roles of STAT3 in mechanisms that regulate T-cell proliferation. Understanding the role of STAT3 in lymphocyte proliferation is further complicated by the facts that initiation of T-cell proliferation is obligatorily coupled to antigen detection and IL-2-dependent entry into S phase of cell cycle (6), while IL-6-induced proliferation does not require entry into S phase or cell cycle progression (17).
In this study, we have investigated the effects of STAT3 on T-cell proliferation, survival and IL-2 production. The data presented reveal that STAT3 promotes T-cell survival by inhibiting IL-2 production and attenuating their proliferative response through two distinct mechanisms that derive from STAT3-mediated up-regulation of class O Forkhead transcription factors. First, STAT3 inhibited T-cell proliferation by enhancing transcriptional activity of FoxO1 and FoxO3a, leading to increased expression of cell-cycle regulatory proteins. Second, STAT3 limited expansion of activated T cells by constraining their ability to produce IL-2 through FoxO1- and FoxO3a-dependent increases in IκB and IκB-mediated sequestration of NF-κB in cytoplasm (7).
MATERIALS AND METHODS
Mice
C57BL/6 mice (6–8-weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME). Mice with conditional deletion of STAT3 in CD4 T-cell compartment (STAT3KO) were derived by breeding STAT3f/f with CD4-Cre mice (Taconic Farms) as described previously (18). Littermate STAT3f/f mice, in C57BL/6 background, were used as wild-type controls. Animal care and use was in compliance with NIH guidelines.
Analysis of T-lymphocytes
Naïve CD4+ or CD8+ T cells were isolated from spleen or lymph nodes (LN) as described (18) and activated in plate-bound anti-CD3 Abs (10 μg/ml) and soluble anti-CD28 Abs (3 μg/ml) without exogenous cytokines or Abs (Th0 condition). In some experiments, the cells were activated under Th1 polarization condition (anti-CD3/CD28 Abs, anti-IL-4 Abs (10 μg/ml) and IL-12 (10 ng/ml)) or Th17 condition (anti-CD3/CD28 Abs, IL-6 (10 ng/ml), TGF-β1 (2 ng/ml), anti-IFN-γ Abs (10 μg/ml), anti-IL-4 Abs (10 μg/ml)). For propagation under Treg condition, the Th0 condition was supplemented with TGF-β1 (10 μg/ml). For intracellular cytokine detection, cells were re-stimulated for 5 h with PMA (20 ng/ml)/ionomycin (1 μm). Golgi-stop was added in the last hour and intracellular cytokine staining was performed using BD Biosciences Cytofix/Cytoperm kit as recommended (BD Pharmingen, San Diego, CA). FACS analysis was performed on a Becton-Dickinson FACSCalibur (BD Biosciences) using monoclonal antibodies specific to CD4, CD8, IL-2, CD25, IFN-γ, or OX40 mAbs and corresponding isotype control Abs (PharMingen, San Diego, CA) as described (10). In some experiments CD4+ T cells were cultured for 4 days in medium containing anti-CD3/CD28 Abs under Th1 or Th17 polarization condition.
Induction of Experimental Autoimmune Uveitis
Experimental autoimmune uveitis (EAU) is a mouse model of human uveitis and is induced by immunization with the retinal protein, interphotoreceptor-retinoid-binding protein (IRBP) (11). 6–8 week old CD4-STAT3KO or WT (littermates) mice were immunized by subcutaneous injection of 150 μg IRBP and 300 μg of human IRBP peptide (1–20) in 0.2 ml emulsion 1:1 v/v with Complete Freund Adjuvant containing Mycobacterium tuberculosis strain H37RA (2.5 mg/ml). The mice also received Bordetella pertussis toxin (0.2 μg/mouse) concurrent with immunization and clinical disease was established by histology as described previously (11). T cells were isolated from spleen and lymph nodes 21 days post-immunization.
Cell Death Analyses
Naïve CD4+ T cells were cultured for 4 days in medium containing anti-CD3/CD28 Abs under Th0, Th1, or Th17 polarization condition. The cells undergoing apoptosis were detected by FACS using PE-Annexin-V apoptosis detection kit I (BD Bioscience).
Lymphocyte Proliferation Assay
Naïve T cells were cultured for 2–5 days in quintuplet cultures containing anti-CD3/CD28 Abs. After 36 h, some cultures were pulsed with [3H]thymidine (0.5 μCi/10 μl/well) for 12 additional hours and analyzed. The presented data are mean CPM ± S.E. of responses of five replicate cultures. For CFSE dilution assay, cells were cultured for 120 h using a commercially available CFSE Cell Proliferation kit (Molecular Probes, Inc.). Graphical display showing information about each generation in the subset was obtained from FlowJo software. The threshold of cellular proliferation was determined based on analysis of unstimulated cells.
ELISA
We assayed IL-2 secreted in supernatant of the activated CD4+ T cells by Multiplex ELISA using SearchLight Arrays technology (Pierce).
Reverse Transcription (RT) PCR and Quantitative (qPCR) Analysis
All RNA samples were DNA-free. cDNA was generated as described previously (10). RT-PCR and qRT-PCR analyses were performed as described (10). Each gene-specific primer pair used for RT-PCR analysis spans at least an intron and primers and probes used for qPCR were purchased from Applied Biosystems. The mRNA expression levels were normalized to the levels of ACTB (encoding β-actin) and GAPDH housekeeping genes.
Western Blotting Analysis
Preparation of whole cell lysates and performance of Western blot analysis were as described (19). Cell extracts (20–40 μg/lane) were fractionated on 4–12% gradient SDS-PAGE, and antibodies used were: FoxO1, FoxO3a (Cell Signaling Technology); p27kip1, IκB, and β-actin (Santa Cruz Biotechnology). Preimmune serum was used in parallel as controls, and signals were detected with HRP-conjugated secondary F(ab′)2 Ab (Zymed Laboratories Inc.) using the ECL-PLUS system (Amersham Biosciences, Arlington Heights, IL).
Immunofluorescence Staining and Confocal Imaging Analysis
CD4+ T cells were stimulated with anti-CD3/anti-CD28 abs for 24 h. Cells were fixed, blocked with 5% goat serum, and then incubated with primary antibodies (p65/NF-κB) from Santa Cruz Biotechnology. Cells were washed, incubated in Alexa 568-conjugated secondary antibody (Invitrogen) containing DAPI and examined on a laser scanning confocal microscope (Olympus FV1000) as described (20).
Protein-DNA Binding Assay (Electrophoretic Mobility Shift Assay or EMSA)
Nuclear extracts were prepared using buffer containing the following protease inhibitors; 2 μm leupeptin; 2 μm pepstatin; 0.1 μm aprotinin; 1 mm AEBSF; 0.5 mm phenylmethylsulfonyl fluoride; 1 μm E-64 as described (21). Nuclear extract (5 μg) in binding buffer (20 mm HEPES, pH 7.9, 50 mm KCl, 10% glycerol, 0.5 mm dithiothreitol, 0.1 mm EDTA) containing 0.14 μg/μl poly(dI-dC) was incubated on ice for 20 min. Labeled double-stranded DNA probe was then added and incubated for an additional 15 min at room temperature. DNA-protein binding was analyzed in a gel electrophoresis DNA binding assay with a [α32P]dATP-labeled, double-stranded oligonucleotide containing the binding site for NF-κB (5′-AGCTCTGGGGACTTTCCACC-3′) (22) and for FoxO3a binding site (5′-CTAGTTTATTTTGTTTTTGTTTG-3′) (23). The double-stranded oligonucleotides were labeled by fill- in reaction using Klenow polymerase (Invitrogen) with [α-32P]dATP (3000 Ci/mmol; Amersham Biosciences). Samples were electrophoresed in 5% polyacrylamide gel in 0.25× Tris borate-EDTA buffer. For supershift assays, the indicated Abs were added to the binding buffer containing nuclear extract mixture and preincubated on ice for 10 min. 32P-labeled probe was then added and the entire mixture incubated for an additional 20 min on ice before electrophoresis. ELISA-based analysis of NF-κB or FoxO1 binding activity in the nuclear extracts was performed using TransAM NF-κB Family or FKHR kit (Active Motif, Carlsbad, CA). Specificity of the assay was monitored by use of wild-type consensus oligonucleotide that served as competitor for Fox01 or NF-κB binding while corresponding mutated consensus oligonucleotides had no effects on the binding.
Cell Transfection
CD4+ T cells were transiently transfected with plasmids expressing WT FoxO1-GFP fusion proteins (Addgene, Cambridge, MA) (5) using the Nucleofector device and corresponding kits according to the manufacturer's instructions (Amaxa, Gaitherburg, MD).
Chromatin Immunoprecipitation (CHIP) Analysis
DNA-protein complexes in naive or anti-CD3/CD28-stimulated CD4+ T cells were cross-linked for 10 min by addition of fresh formaldehyde (Sigma) to the culture medium at a final concentration of 1%, followed by quenching in 135 mm glycine. The fixed cells were lysed with lysis buffer (EZ ChipTM, Upstate Biotechnology) and sonicated (5×) for 15 s (output 5 on Sonic Dismembrator Model 1000, Fisher Scientific). Lysates were then cleared with Protein G-agarose for 1h, pelleted, and incubated overnight with control IgG or anti-STAT3 antibody (Cell Signaling, CHIP grade). Prior to antibody incubation, input samples were removed from the lysate and stored at −80 °C until extraction. Immunoprecipitation was performed according to the manufacturer's instructions (EZ ChipTM). The immunoprecipitated and input DNA were subjected to PCR and qPCR using FoxO1 (5′-CGACTTCAACACCTCATCGCTTC-3′ and 5′-AGGCGCGCAGATCCTTCGGTGA-3′) or FoxO3a (5′-CTTGCTCCCGCTGCTGCCAT-3′ and 5′-CGCGGAATCGTACGCCCTCC-3′) promoter site-specific primers. qPCR were performed with Power SYBR Green PCR master mix (Applied Biosystems, Warrington, UK). Fold enrichment relative to IgG control was calculated using the comparative CT method (ΔΔCr).
Statistical Analysis
The Student's t test was performed on the data as indicated. p values ≤ 0.05 are statistically significant and indicated by an asterisk (*).
RESULTS
STAT3 Inhibits T-cell Proliferation and IL-2 Production
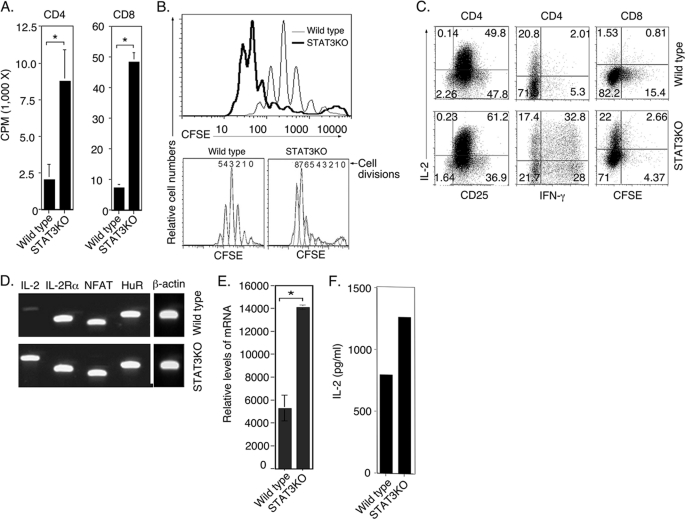
Previous studies have suggested complex roles of STAT3 in mechanisms that regulate T-cell proliferation (17) and that STAT3 and IL-2 signals may exert mutually antagonistic effects on Th17 differentiation and/or proliferation (13–15). In this study, we have used in vitro and in vivo genetic and biochemical approaches to investigate the role of STAT3 in T-cell proliferation and IL-2 production. We generated mice with targeted deletion of STAT3 in CD4+ and CD8+ T cells by Cre/Lox technology and description of these mice has previously been reported (18). Here, we isolated and purified naïve CD4+ and CD8+ T cells (>95%) from WT and STAT3-deficient mice, stimulated them with anti-CD3/CD28 for 2 or 4 days and examined their proliferative responses to TCR activation by two complementary assays. Analysis of thymidine incorporation revealed ∼4-fold and 7-fold increases in proliferative responses of CD4+ and CD8+ STAT3KO T cells, respectively (Fig. 1A). Similarly, CFSE time-series experiments (Fig. 1B) showed that the STAT3KO CD4+ T cells proliferated much faster and underwent 8-cycles of division after 4 days of stimulation compared with 5-cycles for WT counterparts (Fig. 1B, lower panels). We next examined whether loss of STAT3 had any effects on IL-2 production. Consistent with their more vigorous proliferative responses, a much higher percentage of IL-2-producing T cells were observed in TCR-activated STAT3-deficient CD4+ or CD8+ T-cell compared with their WT counterparts (Fig. 1C). To obviate the potential confounding effects of PMA/ionomycin treatment required for intracellular cytokine assay, we isolated RNA from the TCR-activated cells without treatment with PMA/ionomycin. RT-PCR and qPCR analysis performed on cDNA prepared from the RNA detected much higher levels of IL-2 expression by STAT3KO T cells compared with WT T cells (Fig. 1, D and E). The increase in IL-2 observed in STAT3KO compared with WT T cells is further confirmed by ELISA analysis of supernatants derived from the day cultures (Fig. 1F). Together these results reveal for the first time that STAT3 inhibits IL-2 production by T lymphocytes. In contrast to its role in promoting proliferation of cancer cells and most non-lymphoid cell types, our data further indicate that STAT3 may contribute to mechanisms that inhibited proliferation in lymphocytes.
FIGURE 1.
STAT3 inhibits T-cell proliferation and IL-2 production. WT or STAT3KO naïve T cells were stimulated with anti-CD3/CD28 Abs for 2 to 4 days under non-polarizing condition. Thymidine incorporation assay (A) and CFSE dilution analysis (B) were performed after 2 or 4 d stimulation, respectively. Intracellular cytokine assay (C), RT-PCR (D), or qPCR (E) analysis were performed after 3 days of stimulation. Cells for RT-PCR were not stimulated with PMA/ionomycin. F, WT or STAT3-deficient CD4+ T cells were stimulated in vitro with anti-CD3/CD28 Abs for 3 days and IL-2 secretion in the supernatants was detected by ELISA. Results are representative of three independent experiments.
STAT3 Protects T Cells from Apoptosis and Promotes Long-term Survival of T Cells
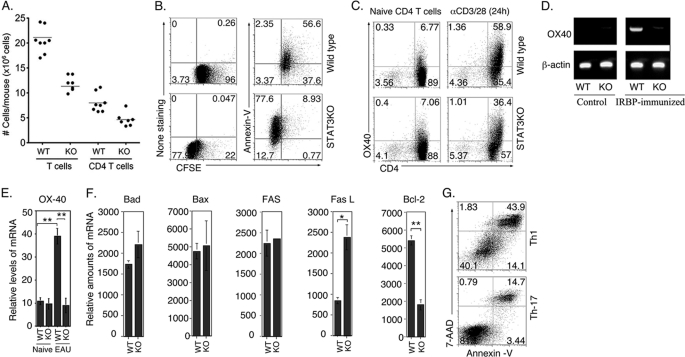
It is notable that the spleen and lymph nodes of STAT3KO mice were very small compared with WT littermates (supplemental Fig. S1). They also have significantly reduced numbers of peripheral T cells (Fig. 2A), suggesting that the increase of IL-2 production by STAT3KO lymphocytes (Fig. 1, C and D) may have rendered them more susceptible to activation-induced cell death. In view of the dual role of IL-2 in promoting lymphocyte growth and survival on one hand while also orchestrating mechanisms of self-tolerance through induction of AICD (6) (9), it was intriguing to examine how the regulation of IL-2 expression by STAT3 might affect the survival of T cells. We therefore stimulated WT or STAT3KO naïve CD4+ T cells with anti-CD3/CD28 Abs for 4 days under non-polarizing condition and determined the number of T cells undergoing apoptosis and/or proliferation by annexin-V staining assay and/or FACS. Similar to results presented in Fig. 1, the CFSE dilution assay confirmed the higher level of proliferation of the STAT3KO T cells compared with WT T-cells (Fig. 2B). Compared with WT cells, higher percentage of the STAT3KO T-cells were undergoing apoptosis after 4 days of TCR activation as revealed by annexin-V staining (compare 86.5% versus 59%) (Fig. 2B). These results thus establish a correlation between the enhanced proliferation of STAT3KO T cells and higher levels of AICD (Fig. 2, A and B), suggesting that STAT3 may enhance lymphocyte survival in vivo by constraining IL-2 production, thereby mitigating IL-2-induced AICD.
FIGURE 2.
STAT3 protects T cells from apoptosis and promotes their long-term survival. A, CD3+ and CD4+ T cells isolated from the spleen and LN from 6–8-week-old mice were quantified on a Vi-CELLTM Cell Viability Analyzer (Beckman Coulter, Inc). B, WT or STAT3KO naïve CD4+ T cells were stimulated with anti-CD3/CD28 Abs for 4 days under non-polarizing condition and levels of T cells undergoing apoptosis and/or proliferation were analyzed by Annexin-V staining or CFSE dilution assay and FACS. C, WT or STAT3KO naïve CD4+ T cells were stimulated with anti-CD3/CD28 Abs for 24 h under non-polarizing condition and numbers in quadrants indicate percentage of OX40-expressing CD4+ T cells. WT or STAT3KO mice were immunized with IRBP and after 21 days T cells were isolated and analyzed by RT-PCR (D) or qPCR (E, F) for the expression of OX-40 (D, E) or pro- and anti-apoptotic genes (F). G, naïve CD4+ T cells were activated with anti-CD3/anti-CD28 for 4 days under Th1 or Th17 polarization, and AICD was quantified by Annexin-V assay. Results are representative of at least three independent experiments.
Consistent with this notion, we observed that reduced numbers of CD4+ T cells in STAT3KO mice (Fig. 2A) correlated with marked reduction in their expression of OX40 (CD134), a TNF receptor family molecule that promotes Bcl-2 expression and survival of activated CD4+ T cells (24). As indicated, naïve WT and STAT3-deficient T-cell cultures contain comparable levels of OX40-expressing cells but after 24 h TCR activation 60% of WT expressed OX40 compared with 37% STAT3KO T cells (Fig. 2C), suggesting that STAT3 is required for optimum expression of OX40. To further characterize the role of STAT3-induced OX40 expression in long-term survival of T cells, we immunized WT and STAT3KO mice with the ocular autoantigen, IRBP and 21 days post-immunization, we harvested draining lymph node T cells and analyzed OX40 expression by RT-PCR and qPCR. The STAT3-deficient T cells expressed substantially reduced levels of OX40 compared with their WT counterparts (Fig. 2, D and E). Bcl-2 expression was also down-regulated in the STAT3KO T cells while FasL and Bad were up-regulated (Fig. 2F), further underscoring potential role of STAT3 in long-term survival of activated T cells. The Th17 developmental program is regulated by IL-6, IL-21, and IL-23 and each of these cytokines mediates its biological effects through activation of STAT3 (10–12). Because these cytokines are essential at all stages of Th17 development, STAT3 is persistently activated in this T-helper subset. We therefore examined whether the constitutive activation of STAT3 would protect Th17 cells from AICD. In comparison to the Th1 T-helper subset, we found that Th17 cells were less susceptible to AICD (Fig. 2G), suggesting that the sustained activation of STAT3 in Th17 cells may confer survival advantages to the Th17 phenotype.
STAT3 Regulates Expression and Transcriptional Activity of Forkhead Transcription Factors
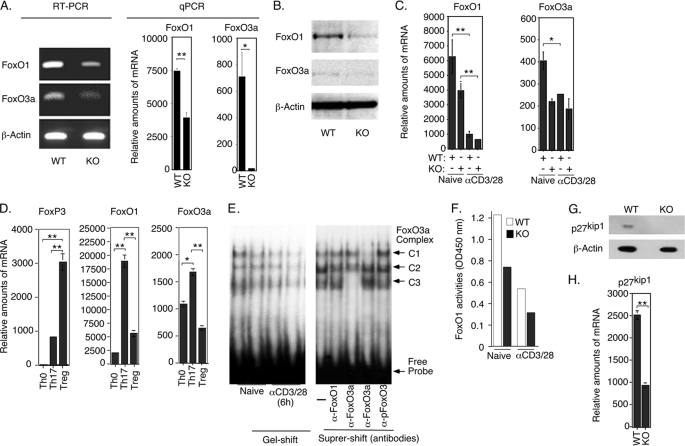
We next investigated possible molecular mechanism by which STAT3 regulates T-cell proliferation and IL-2 production in resting and activated T cells. Because class O Forkhead transcription factors maintain resting T cells in quiescent state (G0 cell cycle phase) by up-regulating expression of cell cycle inhibitors and IκB, a protein that inhibits IL-2 production by sequestering NF-κB in the cytoplasm (7), we examined whether STAT3 regulates FoxO proteins. Although FoxO1 and Fox03a are expressed at relatively high levels in resting WT T cells (2), transcription of FoxO1, FoxO3a (Fig. 3A) and expression of FoxO1 and FoxO3a proteins (Fig. 3B) are markedly reduced in naïve STAT3KO T cells, indicating that STAT3 is required for their optimal expression. It is however notable that T-cell activation induced down-regulation of FoxO1 and FoxO3a expression (Fig. 3C), indicating that STAT3- and TCR-induced pathways have diametrically opposite effects on the transcription of FoxO1 and FoxO3a genes. In contrast to other T-helper subsets, all stages of Th17 development require STAT3 activation (11). We show here that Th17 cells expressed much higher levels of FoxO1 and FoxO3a mRNA compared with Th0 or Tregs (Fig. 3D), consistent with persistent activation of STAT3 in this T-cell subset. Electrophoretic mobility shift and ELISA-based DNA binding assays further revealed a correlation between the low FoxO1 and FoxO3a levels in STAT3-deficient T cells and decrease of FoxO3a or FoxO1 DNA binding activity in these STAT3KO T cells (Fig. 3, E and F). Specificity of the FoxO3a gel-shift assay was verified by supershift assays showing complete blockade of formation of the C3 and C1 complex by N-terminal (FoxO3a Abs#1) and C-terminal (FoxO3a Abs#2) FoxO3a antibodies but not FoxO1 or pFoxO3a antibodies (Fig. 3E). Cell cycle inhibitors are direct targets of FoxO transactivation and we show here that a functional consequence of decreased FoxO1 and FoxO3a expression in T cells is marked reduction of p27Kip1 in naïve STAT3KO T cells (Fig. 3, G and H). Together these results indicate a link between loss of STAT3, decrease in p27Kip1 and increase in T-cell proliferation.
FIGURE 3.
STAT3 inhibits T-cell proliferation through up-regulation of FoxO transcription factors. Analysis of WT or STAT3KO naïve CD4+ T cells by RT-PCR or qPCR (A) and by Western blot (B). C, WT or STAT3KO CD4+ T cells were stimulated with anti-CD3/CD28 for 24 h under Th0 condition and naïve or activated CD4+ T cells were analyzed by qPCR (C). WT CD4+ T cells were polarized for 4 days under Th0, Th17, or Treg conditions and expression of FoxO1 and FoxO3a was analyzed by qPCR (D). E and F, WT or STAT3KO CD4+ T cells were stimulated with anti-CD3/CD28 for 6 h. Nuclear extracts were isolated and analyzed for FoxO3a transcriptional activity by EMSA and supershift analysis was performed with Abs specific to N (FoxO3a#1) or C (FoxO3a#2) terminus of the Foxo3a protein (E). F, FoxO1 transcriptional activity was analyzed by an ELISA-based DNA protein binding assay. G and H, we detected p27kip1 expression by naïve WT or STAT3KO T cells by Western blotting (G) or qPCR (H). Results are representative of at least three independent experiments.
STAT3 Enhances FoxO-dependent IκB Expression and NF-κB Sequestration in Cytoplasm
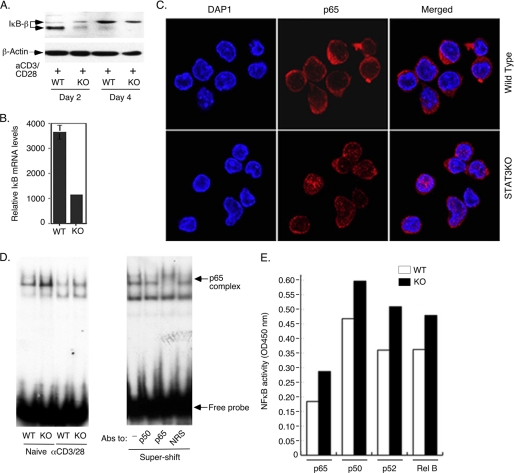
Because FoxO proteins induce expression of IκB-β, a protein that interacts with and sequesters NF-κB in the cytoplasm (7), we investigated whether STAT3 influences the level of IL-2 production through its effects on transcription of FoxO genes. We show here that the reduced expression of FoxO1 and FoxO3a in STAT3KO T cells correlates with marked decrease in IκB protein (Fig. 4A) and IκB mRNA (Fig. 4B). In addition, we observed marked increases in the localization of p65 into the nuclei of STAT3-deficient cells (Fig. 4C), suggesting that the decrease in IκB expression might have resulted in reduced capacity to sequester NF-κB in the cytoplasm of STAT3KO T cells. The STAT3KO T cells also exhibited increased binding of p65/NF-κB to target genes as revealed by EMSA/super-shift (Fig. 4D) and ELISA-based DNA-binding assay (Fig. 4E). Taken together, these results suggest that increased IL-2 production by STAT3-deficient T cells might have derived from reduced capacity to sequester NF-κB in the cytoplasm, increase of NF-κB activation and up-regulation of IL-2 transcription.
FIGURE 4.
STAT3 enhances FoxO-dependent IκB expression and NF-κB sequestration in cytoplasm. WT or STAT3KO CD4+ T cells were stimulated with anti-CD3/CD28 for varying amounts of time ranging from 6 h to 96 h. Detection of IκB expression by Western blot (A) or qRT-PCR analysis (B). Subcellular localization of NF-κB (p65) was analyzed by immuno-cytochemical staining after 24 h stimulation with anti-CD3/CD28 (C) and transcriptional activity of NF-kB in naïve or TCR-stimulated CD4+ T cells (6 h) was assessed by EMSA/supershift assay (D) or by NF-κB-specific ELISA-based DNA protein binding assay (E). Results are representative of more than three independent experiments.
STAT3 Binds to FoxO1 or FoxO3a Promoter and FoxO1 Inhibits IL-2 Production in T Cells
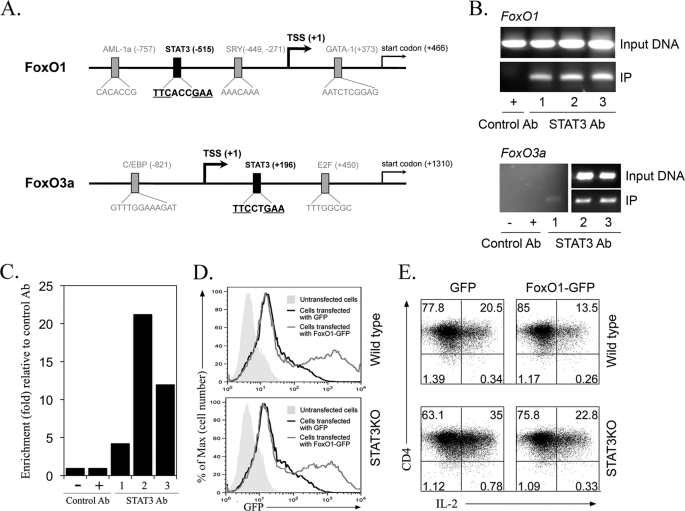
The promoter region of the FoxO1 or FoxO3a gene contains potential STAT3 DNA binding motifs (25) (Fig. 5A). We examined here whether STAT3 can bind to the FoxO1 or FoxO3a promoter by chromatin immunoprecipitation assay. Naive CD4+ T cells were stimulated with anti-CD3/CD28 in presence or absence of IL-6 and whole-cell lysates were immunoprecipitated with anti-STAT3 antibodies. PCR or qPCR analysis of the immunoprecipitated DNA indicated binding of STAT3 to the FoxO1 or FoxO3a promoter region (Fig. 5, B and C). Interestingly, binding to the STAT3 motif was detected in naïve and TCR-activated T cells, suggesting that the low level of FoxO1 and FoxO3a in STAT3-deficient naïve T cells may have derived, in part, from lack of STAT3 binding to the FoxO1 and FoxO3 promoters.
FIGURE 5.
STAT3 directly binds to FoxO1 or FoxO3a promoter and overexpression of FoxO1 inhibited IL-2 production in T cells. A, schematic structure of the FoxO1 or FoxO3a promoter region (NCBI Reference Sequence: NC_000069.5 for FoxO1 and NC_000076.5 for FoxO3a) (32). Transcription start site (TSS) is denoted by a black arrow. B and C, chromatin immunoprecipitation (CHIP) analysis was performed with naïve and anti-CD3/C28-stimulated CD4+ T cells, and STAT3 binding to the FoxO1 or FoxO3a promoter region was analyzed. Cell lysates were immunoprecipitated with anti-STAT3 Ab or control IgG. Immunoprecipitated and input DNA was analyzed by semi-quantitative PCR (B) or qPCR (C) using primers corresponding to FoxO1 or FoxO3a promoter sites. Lane 1 (mouse naïve T cell), lane 2 (T-cell activated for 4 days with anti-CD3/CD28), lane 3 (T-cell activated for 4 days with anti-CD3/CD28 and then treatment with IL-6 for 6 h). C, STAT3 binding to the FoxO3a promoter region was confirmed by qPCR. The results are presented as fold of template enrichment in immunoprecipitates of anti-STAT3 relative to those of control IgG. D and E, WT or STAT3KO T cells were transfected with plasmids expressing GFP or FoxO1-GFP. Cells were stimulated with anti-CD3/CD28 for 36 h and IL-2 expression was analyzed by the intracellular cytokine assay. Results are representative of three independent experiments.
As marked decrease of FoxO1/FoxO3a RNA and protein levels in STAT3-deficient cells correlated with their increased expression IL-2 RNA and protein (Fig. 1, C–E), it was of interest to examine whether overexpression of FoxO1 protein in T cells would inhibit IL-2 production. We transfected WT and STAT3KO T cells with control GFP or FoxO1-GFP expression vector. FACS analysis revealed similar levels of GFP or FoxO1-GFP protein (Fig. 5D), indicating equivalent transfection efficiency in WT and STAT3-deficient T cells. Analysis of IL-2 expression in the T cells transfected with the GFP control vector confirmed that the STAT3KO T cells did indeed produce higher level of IL-2 (Fig. 5E, left panels). On the other hand, transfection of WT and STAT3-deficient T cells with the FoxO1-GFP expression vector induced substantial reduction of IL-2 production (Fig. 5E, right panels). These results provide direct evidence that the level of IL-2 in T cells is inversely related to the steady state level of FoxOs and suggest that STAT3 may indirectly control the level of IL-2 production by T cells by altering FoxO protein level.
DISCUSSION
STAT3 is an evolutionarily conserved transcription factor that regulates proliferation in many mammalian cells and is considered to be an oncogene because it is constitutively activated in many types of primary tumors (26). In this report, we have uncovered a unique role played by STAT3 in promoting lymphocyte quiescence and inhibiting IL-2 production. Analysis of mice with conditional deletion of STAT3 in CD4+ and CD8+ T cells (STAT3KO) revealed that STAT3 inhibits IL-2 production and lymphocyte proliferation by enhancing the expression of FoxO3a and FoxO1 genes. We show that although Class O Forkhead transcription factors are expressed at relatively high levels in resting T cells (2), FoxO1 and FoxO3a mRNAs (Fig. 3A) and proteins (Fig. 3B) were markedly reduced in STAT3-deficient lymphocytes, indicating that STAT3 is required for their optimal expression. The requirement of STAT3 for the expression of FoxO genes in naïve T cells is particularly intriguing because STAT3 is mostly in the non-activated, unphosphorylated (U-STAT3) form and transcription of STAT3 target genes is generally attributed to tyrosine-phosphorylated STAT3 (pSTAT3). However, recent reports suggest that U-STAT3 readily enters the nucleus by carrier-independent mechanisms and also activates transcription (27, 28). Nonetheless, results of our CHIP assay showing that STAT3 binds to STAT3 consensus motif in the FoxO1 or FoxO3a promoter of naïve T cells indicates that STAT3 directly activates transcription of FoxO genes in naïve T cells.
Post-transcriptional events that lead to the phosphorylation and expulsion of FoxO proteins from the nucleus (3, 5, 6) are an important mechanism by which TCR and PI3K/AKT inhibits growth-inhibitory effects of FoxO proteins. In this study, we show that TCR activation for 1 or 2 days induced a down-regulation of FoxO1 and FoxO3a transcription (Fig. 3C), suggesting that TCR/IL-2-induced signals may also promote cell cycle progression for T-cell growth or expansion by repressing transcription of FoxO gene. However, the TCR/IL-2-mediated inhibition of FoxO1/FoxO3a transcription was not sustained because of the substantial diminution of IL-2 production that occurs during prolonged T-cell stimulation (29). Our CHIP data showing that STAT3 binds strongly to the FoxO3a promoter after 4 days of TCR activation, suggests that STAT3-mediated transcription of FoxO3a may contribute to mechanisms for restoring FoxO3a mRNA to its steady-state levels following termination of TCR activation. It is therefore of note that FoxO1 and FoxO3a bind to cognate FoxO sites in the promoter of FoxO genes and stimulate their transcription (30). Thus, the up-regulation of FoxO expression by STAT3 together with FoxO-mediated autoregulation of FoxO gene may provide a powerful positive feedback loop that potentiates FoxO expression and thus counteract physiologic effects of TCR and PI3K/AKT on FoxO expression. In this regard, it is not surprising that the loss of STAT3 in T cells resulted in defect in FoxO1 and FoxO3a expression, decreased transcriptional binding activity of FoxO1 and FoxO3a (Fig. 3, E and F), marked reduction of p27Kip1 (Fig. 3, G and H) and enhanced proliferation of STAT3KO CD4+ and CD8+ T cells (Fig. 1, A and B). Thus by enhancing the expression of FoxO1 and FoxO3a genes, STAT3 indirectly contributes to the inhibition of lymphocyte proliferation as a result of the increase of number of FoxO proteins interacting and activating growth arrest genes such as p27kip1, p21WAF1, Gadd45, cyclin E, and p130 (4). However, STAT3 has also been shown to induce IL-6-dependent T-cell proliferation by preventing apoptosis (17). In this study, we also found that STAT3 promoted stability and long-term survival of activated CD4+ T cells by up-regulating the expression of anti-apoptotic genes (Fig. 2, D and E). It is however interesting to note that despite substantial increase in AICD (Fig. 2B), the STAT3-deficient lymphocytes exhibited higher proliferative response to TCR-activation compared with WT cells. This suggests that although STAT3-mediated inhibition of apoptosis contributes to cytokine-induced proliferation, STAT3-dependent up-regulation of FoxO1, FoxO3a, and p27kip1 may exert dominant effects on T-cell proliferation induced by antigen or TCR activation.
Our results showing that STAT3 inhibits lymphocyte proliferation seem at odds with previous reports that STAT3 promotes proliferation of non-lymphoid or cancer cells (17, 26). The difference between these results and ours may derive from the fact that lymphocytes and non-lymphoid cells initiate proliferation by distinct mechanisms and highlights the unique role of STAT3 in the physiology of T-helper cells. The potential destructive power of an exuberant and unrestrained immune system necessitates stringent control over the initiation of T-cell proliferation, restricting it to T cells stimulated by cognate antigen in context of MHC molecules. Although c-Myc, pim-1, cyclin D1, and other cell cycle regulatory genes contribute to cell cycle progression in non-lymphoid and activated T cells, they have little or no effect on naïve or resting T cells that are maintained at G0.IL-2 produced following T-cell activation induces exit of the T-cell from G0 into G1 phase of the cell cycle to initiate proliferation. Understanding how STAT3 and FoxO proteins might regulate T-cell proliferation thus require appreciation of the fact that antigen-induced proliferation of CD4+ T cells is wholly dependent on the availability of IL-2, a T-cell growth factor that induces quiescent cells to enter the G1 phase of the cell cycle. And that the production of IL-2 requires NF-κB, a transcription factor that is under the stringent control of IκB. The expression of IκB is in turn regulated by FoxO proteins (7) and FoxO-dependent up-regulation of IκB promotes the sequestration of NF-κB in the cytoplasm thereby preventing NF-κB from activating transcription of IL-2 in resting T cells (31).
In this study, we have shown that STAT3-deficient T cells express barely detectable levels of IκB that correlate with reduced capacity to sequester NF-κB in the cytoplasm and marked increase of p65 nuclear localization (Fig. 4C). Thus, the observed increase in NF-κB activation (Fig. 4, D and E) and marked increase in IL-2 expression (Fig. 1, C and D) by STAT3KO T cells provides a plausible mechanistic explanation for how STAT3 and FoxO pathways might converge to constrain IL-2 production and T-cell proliferation. In a proof of principle experiment we induced substantial reduction of IL-2 in WT and STAT3-deficient T cells by overexpressing FoxO1, providing empirical evidence supporting the role of STAT3 and FoxO proteins in regulating steady state levels of IL-2 in lymphocytes. Our data thus suggest a novel role of STAT3 as regulator of T-cell quiescence, activation and proliferation by 2 distinct mechanisms that derive from its role in increasing the physiological levels and activities of FoxO1 and FoxO3a: It inhibits lymphocyte proliferation by augmenting FoxO-dependent transcriptional activation of p27kip1 while constraining IL-2 production through FoxO-dependent increase in IκB and IκB-mediated sequestration of NF-κB in the cytoplasm (7). Thus, STAT3 may have profound effect on T-cell effector functions by fine tuning TCR signaling strength. Whether the influence of STAT3 and FoxO on the level of IL-2 produced during T-cell activation determines the proliferative capacity, effector functions, and/or level of cytokines produced remains an issue of relevance to the development of effective immuno-modulation therapeutic strategies.
In summary, our data suggest STAT3 as convergence point for mechanisms that regulate cellular quiescence and T-cell activation, with STAT3 serving as a gatekeeper that influences the decision of whether the T-cell remains quiescent or proliferate. Because IL-2 production is the determinative step of the T-cell activation process, it will be important to examine in future studies whether STAT3 and FoxO proteins collaborate in: (i) establishing threshold levels of IL-2 required for T-cell activation; (ii) determining TCR signaling strength that control the balance of effector versus memory T-cell development. In terms of the broader biological significance of our data, it is of note that FoxO proteins play evolutionarily conserved roles in transcriptional control of genes that regulate proliferation, differentiation, and longevity (3). Thus, similar to their role in regulating lifespan of worms, convergence of FoxO and STAT3 pathways may serve to extend lifespan of T-lymphocytes.
Supplementary Material
Acknowledgments
We thank Dr. Pamela L. Schwartzberg (National Human Genome Research Institute, National Institutes of Health) and Dr. Igal Gery (NEI, National Institutes of Health) for critical reading of manuscript. We also thank Dr. Robert Fariss (National Eye Institute Imaging Facility) for assistance with confocal microscopy.
This work was supported, in whole or in part, by NEI, National Institutes of Health Intramural Research Programs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- IL
- interleukin
- TCR
- T-cell receptor
- AICD
- activation-induced cell death
- LN
- lymph node
- IRBP
- interphotoreceptor-retinoid-binding protein.
REFERENCES
- 1. Hoyer K. K., Dooms H., Barron L., Abbas A. K. (2008) Immunol. Rev. 226, 19–28 [DOI] [PubMed] [Google Scholar]
- 2. Liu J. O. (2005) Sci. STKE 2005, re1. [DOI] [PubMed] [Google Scholar]
- 3. Coffer P. J., Burgering B. M. (2004) Nat. Rev. Immunol. 4, 889–899 [DOI] [PubMed] [Google Scholar]
- 4. van der Horst A., Burgering B. M. (2007) Nat. Rev. Immunol. 8, 440–450 [DOI] [PubMed] [Google Scholar]
- 5. Zhao X., Gan L., Pan H., Kan D., Majeski M., Adam S. A., Unterman T. G. (2004) Biochem. J. 378, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stahl M., Dijkers P. F., Kops G. J., Lens S. M., Coffer P. J., Burgering B. M., Medema R. H. (2002) J. Immunol. 168, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 7. Lin L., Hron J. D., Peng S. L. (2004) Immunity 21, 203–213 [DOI] [PubMed] [Google Scholar]
- 8. Gaffen S. L., Liu K. D. (2004) Cytokine 28, 109–123 [DOI] [PubMed] [Google Scholar]
- 9. Refaeli Y., Van Parijs L., London C. A., Tschopp J., Abbas A. K. (1998) Immunity 8, 615–623 [DOI] [PubMed] [Google Scholar]
- 10. Amadi-Obi A., Yu C. R., Liu X., Mahdi R. M., Clarke G. L., Nussenblatt R. B., Gery I., Lee Y. S., Egwuagu C. E. (2007) Nat. Med. 13, 711–718 [DOI] [PubMed] [Google Scholar]
- 11. Dong C. (2008) Nat. Rev. Immunol. 8, 337–348 [DOI] [PubMed] [Google Scholar]
- 12. Korn T., Bettelli E., Oukka M., Kuchroo V. K. (2009) Annu. Rev. Immunol. 27, 458–517 [DOI] [PubMed] [Google Scholar]
- 13. McGeachy M. J., Chen Y., Tato C. M., Laurence A., Joyce-Shaikh B., Blumenschein W. M., McClanahan T. K., O'Shea J. J., Cua D. J. (2009) Nat. Immunol. 10, 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laurence A., O'Shea J. J. (2007) Nat. Immunol. 8, 903–905 [DOI] [PubMed] [Google Scholar]
- 15. Laurence A., Tato C. M., Davidson T. S., Kanno Y., Chen Z., Yao Z., Blank R. B., Meylan F., Siegel R., Hennighausen L., Shevach E. M., O'Shea J. J. (2007) Immunity 26, 371–381 [DOI] [PubMed] [Google Scholar]
- 16. Durant L., Watford W. T., Ramos H. L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H. W., Kanno Y., Powrie F., O'Shea J. J. (2010) Immunity 32, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeda K., Kaisho T., Yoshida N., Takeda J., Kishimoto T., Akira S. (1998) J. Immunol. 161, 4652–4660 [PubMed] [Google Scholar]
- 18. Liu X., Lee Y. S., Yu C. R., Egwuagu C. E. (2008) J. Immunol. 180, 6070–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egwuagu C. E., Yu C. R., Zhang M., Mahdi R. M., Kim S. J., Gery I. (2002) J. Immunol. 168, 3181–3187 [DOI] [PubMed] [Google Scholar]
- 20. Yu C. R., Mahdi R. M., Ebong S., Vistica B. P., Chen J., Guo Y., Gery I., Egwuagu C. E. (2004) J. Immunol. 173, 737–746 [DOI] [PubMed] [Google Scholar]
- 21. Li W., Nagineni C. N., Efiok B., Chepelinsky A. B., Egwuagu C. E. (1999) Dev. Biol. 210, 44–55 [DOI] [PubMed] [Google Scholar]
- 22. Arenzana-Seisdedos F., Thompson J., Rodriguez M. S., Bachelerie F., Thomas D., Hay R. T. (1995) Mol. Cell. Biol. 15, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J., Yusuf I., Andersen H. M., Fruman D. A. (2006) J. Immunol. 176, 2711–2721 [DOI] [PubMed] [Google Scholar]
- 24. Rogers P. R., Song J., Gramaglia I., Killeen N., Croft M. (2001) Immunity 15, 445–455 [DOI] [PubMed] [Google Scholar]
- 25. Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. (2001) J. Biol. Chem. 276, 6675–6688 [DOI] [PubMed] [Google Scholar]
- 26. Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T., Akira S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3801–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mertens C., Darnell J. E., Jr. (2007) Cell 131, 612. [DOI] [PubMed] [Google Scholar]
- 28. Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. (2007) Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Villarino A. V., Tato C. M., Stumhofer J. S., Yao Z., Cui Y. K., Hennighausen L., O'Shea J. J., Hunter C. A. (2007) J. Exp. Med. 204, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Essaghir A., Dif N., Marbehant C. Y., Coffer P. J., Demoulin J. B. (2009) J. Biol. Chem. 284, 10334–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henkel T., Machleidt T., Alkalay I., Krönke M., Ben-Neriah Y., Baeuerle P. A. (1993) Nature 365, 182–185 [DOI] [PubMed] [Google Scholar]
- 32. Church D. M., Goodstadt L., Hillier L. W., Zody M. C., Goldstein S., She X., Bult C. J., Agarwala R., Cherry J. L., DiCuccio M., Hlavina W., Kapustin Y., Meric P., Maglott D., Birtle Z., Marques A. C., Graves T., Zhou S., Teague B., Potamousis K., Churas C., Place M., Herschleb J., Runnheim R., Forrest D., Amos-Landgraf J., Schwartz D. C., Cheng Z., Lindblad-Toh K., Eichler E. E., Ponting C. P. (2009) PLoS Biol. 7, e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.