Abstract
Background
Lactobacillus reuteri harbors the genes responsible for glycerol utilization and vitamin B12 synthesis within a genetic island phylogenetically related to gamma-Proteobacteria. Within this island, resides a gene (lreu_1750) that based on its genomic context has been suggested to encode the regulatory protein PocR and presumably control the expression of the neighboring loci. However, this functional assignment is not fully supported by sequence homology, and hitherto, completely lacks experimental confirmation.
Results
In this contribution, we have overexpressed and inactivated the gene encoding the putative PocR in L. reuteri. The comparison of these strains provided metabolic and transcriptional evidence that this regulatory protein controls the expression of the operons encoding glycerol utilization and vitamin B12 synthesis.
Conclusions
We provide clear experimental evidence for assigning Lreu_1750 as PocR in Lactobacillus reuteri. Our genome-wide transcriptional analysis further identifies the loci contained in the PocR regulon. The findings reported here could be used to improve the production-yield of vitamin B12, 1,3-propanediol and reuterin, all industrially relevant compounds.
Background
Lactobacillus reuteri is a heterofermentative lactic acid bacterium colonizing the gastrointestinal tract (GI tract) of various mammals, including humans [1]. It is able to convert glycerol to 1,3-propanediol in a two-step enzymatic conversion, yielding NAD+ [2]. In the first reaction, glycerol dehydratase (EC 4.1.2.30), converts glycerol to 3-hydroxypropionaldehyde requiring the presence of vitamin B12 as a coenzyme [3]. Reuterin, a mixture of 3-hydroxypropionaldehyde isomers [4], is a potent antimicrobial, bestowing L. reuteri with an important growth advantage over other residents of the GI tract, such as Gram-negative enteric bacteria [5,6].
We have shown previously that L. reuteri CRL1098 encodes the complete machinery necessary for de novo synthesis of vitamin B12 in a single chromosomal gene cluster [7]. This cluster was shown to be very similar to that present in various representatives of γ-Proteobacteria, standing out against canonical phylogeny. Complete genome sequence analysis of the type strain of L. reuteri revealed that the region immediately upstream of the vitamin B12 biosynthesis cluster maintains a gene order similar to that of Salmonella [8]. The functionality of this upstream region was demonstrated to also match Salmonella where the pdu gene cluster is located. The latter encodes the assembly machinery of metabolosomes and the several subunits of a large diol dehydratase that can metabolize both glycerol and 1,2-propanediol [9].
Also within this cluster resides a gene (lreu_1750) predicted to encode a 359 amino acid long putative transcription factor of the AraC type family, containing a typical helix-turn-helix domain. Based strictly on its conserved genomic context, this gene has been suggested to encode PocR, a regulatory protein that modulates propanediol utilization (pdu) and vitamin B12 biosynthesis in enteric bacteria [8-10]. This functional annotation, however, does not seem to be fully supported by sequence homology. And more importantly, to the best of our knowledge, it completely lacks experimental confirmation.
Here we provide the first experimental evidence to support the functional assignment of Lreu_1750. This was achieved by overexpression and inactivation of lreu_1750, assessing its impact on central carbon and energy metabolism, and on reuterin and vitamin B12 synthesis. In addition, we characterized the genome-wide transcriptional response of both constructs in comparison to their parent strains leading to the identification of the genes comprised in the PocR regulon of Lactobacillus reuteri.
Results and Discussion
Phylogenetic analysis of Lreu_1750
Phylogenetic comparisons between Lreu_1750 and other PocR sequences raise serious doubts about its functional annotation (Figure 1). When compared to the PocR found in enteric bacteria, Lreu_1750 reveals limited amino acid sequence identity (19.1%) and a large percentage of gaps (40.1%). The sequence identity and percentage of gaps (20.5% and 38.8%, respectively) of the PocR-like regulatory proteins of other vitamin B12-producing Firmicutes, such as Listeria monocytogenes, suggest that it is slightly more related. The closest homolog of Lreu_1750 present in the complete genomes available is found in L. brevis ATCC 367 (GI:116334199) with 36.1% identity and only 1.4% gaps. L. brevis is also able to produce 1,3-propanediol [3], but it cannot synthesize vitamin B12 [7]. The predicted products of lreu_1750 and its homolog in L. brevis are approximately 60 amino acid residues longer in the C-terminus in comparison to PocR from S. typhimurium LT2. This could additionally affect its functionality and further urges the experimental confirmation of its tentative annotation. The putative PocR of two different wild type L. reuteri strains, JCM1112 and ATCC PTA 6475, have also been aligned and found to display 100% sequence identity and 0% gaps (Figure 1). Subsequent experiments have been carried out using derivatives of both strains predominantly for technical convenience. Nevertheless, this choice is also important to further substantiate the generality of our findings regarding the role of this PocR-like protein in L. reuteri strains.
Figure 1.
Amino acid sequence alignment of Lreu_1750 and putatively related sequences. Alignments were obtained using ClustalW with default settings [20] and visualized in CLC Sequence Viewer 6.5. Abbreviations stand for: LRE, GI:148544956 (Lactobacillus reuteri JCM1112); LAR, GI:325683301 (Lactobacillus reuteri ATCC PTA 6475); LBR, GI:116334199 (Lactobacillus brevis ATCC367); LMO, GI:16410566 (Listeria monocytogenes EGDe); LIN, GI:16413573 (Listeria innocua Clip11262); STL, GI:16420566 (Salmonella typhimurium LT2); SEN, GI:29136962 (Salmonella enterica Ty2); KPN, GI:152971720 (Klebsiella pneumoniae MGH 78578); YEN, GI:123442942 (Yersinia enterocolitica 8081). Darker blue background stands for higher percentage conservation of respective residue.
Physiological effects
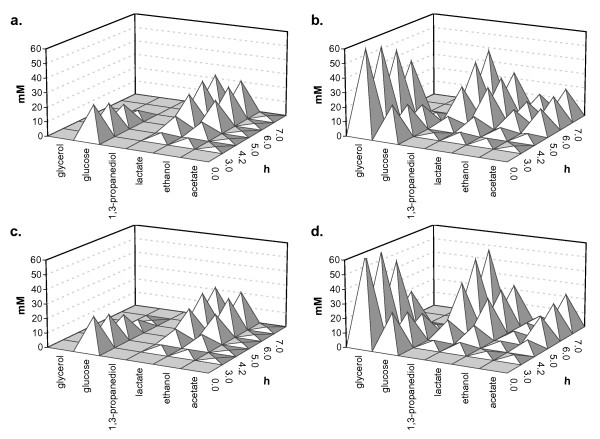
The functional assignment of lreu_1750 was initiated by characterizing the impacts of its overexpression on central carbon and energy metabolism. Since functionally active glycerol metabolizing enzymes are encoded in the vicinity of lreu_1750, the experimental focus was on glycerol metabolism. In the absence of glycerol, except for a slight impairment (< 10%) of μmax (Figure 2), no metabolic effects were observed related to the overexpression of lreu_1750 (Figure 3, panels a. and c.). In the presence of glycerol, however, the overexpressing strain in comparison with JCM1112 transformed with pNZ7021 (empty plasmid), displays a drop in the final ethanol concentration from 13.7 to 6.2 mM while acetate increased approximately 4 mM. This enhancement of the shift from ethanol to acetate formation (p-value < 0.025, paired two-tailed t-test) is accompanied by a 22.5% increase of 1,3-propanediol production, which is produced on a 2:1 molar ratio with acetate, assuring the regeneration of reducing equivalents (Figure 3, panels b. and d.).
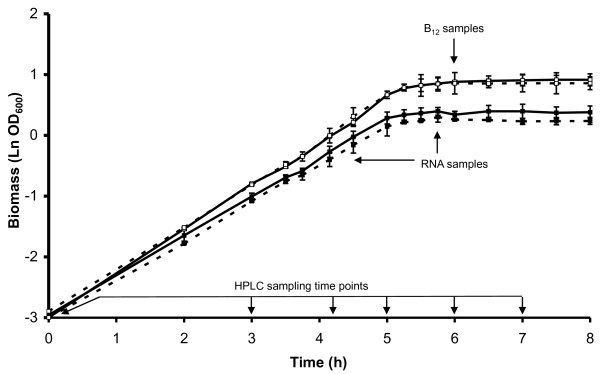
Figure 2.
Biomass formation and sampling scheme in pH-controlled batch fermentations. Experiments were carried out in CDM in the presence (white squares) or absence (black square) of glycerol by L. reuteri transformed with pNZ7021 (empty plasmid, solid lines) or pNZ7748 (harboring lreu_1750, dashed lines).
Figure 3.
Substrate consumption and product formation in pH-controlled batch fermentations. Substrate consumption and product formation by different constructs of L. reuteri in CDM and in CDM with 0.5% glycerol (v/v). a. L. reuteri pNZ7021 (empty plasmid) in CDM; b. L. reuteri pNZ7021 in CDM with glycerol; c. L. reuteri pNZ7748 (harboring lreu_1750) in CDM; d. L. reuteri pNZ7748 in CDM with glycerol.
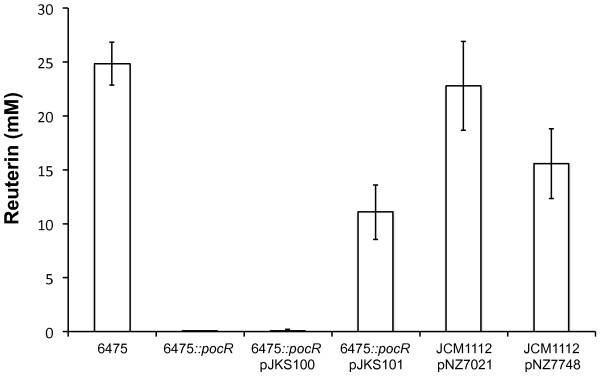
Overexpressing Lreu_1750 does not lead to significant changes (p-value = 0.07, paired two-tailed t-test) in reuterin production (Figure 4). This is not totally unexpected, since changes in the level of enzymes involved in central carbon metabolism often do not result in drastic changes in fluxes [11]. In contrast, the disruption of the lreu_1750 gene leads to an abrupt decrease in reuterin production from 25 mM in the parent strain to undetectable levels (< 0.1 mM) in the mutant. When this strain is complemented with a plasmid harboring lreu_1750 under control of its native promoter (pJKS101), reuterin production is restored to levels in the same order of magnitude as L. reuteri 6475 (11 mM). Mostly human-derived L. reuteri strains can produce reuterin, and therefore, it is thought that this may be important for their survival in the human GI tract [1]. The observed reduction by more than 250-fold in reuterin production most likely debilitates the probiotic functionality of the PocR mutant strain [1,12]. Furthermore, it will condition the potential utilization of glycerol for the regeneration of NAD+, limiting its biomass yield on carbon substrate [10].
Figure 4.
Reuterin production by L. reuteri strains in MRS media. Bars represent average values along with standard deviation (error bars) of at least three independent biological replicates using strains 6475 (parent strain), 6475::pocR (PocR deficient mutant), 6475::pocR pJKS100 (PocR deficient mutant transformed with empty plasmid as negative control), 6475::pocR pJKS101 (PocR deficient mutant complemented with putative pocR of 6475 under control of its native promoter), JCM1112 pNZ7021 (type strain transformed with empty plasmid) and JCM1112 pNZ7748 (type strain carrying the lreu_1750 overexpression). Experiment was performed at least twice with similar results.
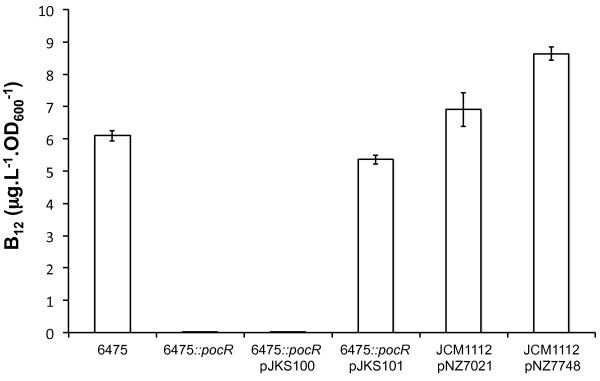
The regulatory role of Lreu_1750 on vitamin B12 synthesis is clearly illustrated by the drastic inhibitory effect that its inactivation exerts over vitamin B12 production (Figure 5). In contrast to the parent strain, the deletion mutant of the putative PocR did not produce detectable levels of B12 (6.09 and less than 0.01 μg.L-1.OD600-1, respectively). Furthermore, the complementation of the mutant with pJKS101 (harboring the putative pocR) leads to the reestablishment of B12 production (5.36 μg.L-1.OD600-1). Additionally, in the strain overexpressing Lreu_1750 (JCM1112 pNZ7748) we observe a significant increase (p-value < 0.016, paired two-tailed t-test) of more than 25% in vitamin B12 production in comparison to JCM1112 transformed with the empty plasmid (pNZ7021). This increase was obtained regardless of the addition of glycerol, and was observed in all the media and conditions tested including the pH-controlled batch fermentations using CDM (data not shown).
Figure 5.
Vitamin B12 production by L. reuteri strains in commercial Vitamin B12-assay medium supplemented with glycerol. Bars represent average values along with standard deviation (error bars) of at least two independent biological replicates measured in triplicate using strains 6475 (parent strain), 6475::pocR (PocR deficient mutant), 6475::pocR pJKS100 (PocR deficient mutant transformed with empty plasmid), 6475::pocR pJKS101 (PocR deficient mutant complemented with putative pocR of 6475 under control of its native promoter), JCM1112 pNZ7021 (type strain transformed with empty plasmid) and JCM1112 pNZ7748 (type strain carrying the lreu_1750 overexpression).
The physiological effects observed for the overexpression and inactivation of Lreu_1750 are all in agreement with its functional assignment as the regulatory protein PocR.
Transcriptomic response
In order to probe the global regulatory role of the putative PocR of L. reuteri, we compared the transcriptomes of the deficient and overexpressing strains relative to their parent strains. Considering that (i) glycerol has been shown to induce the expression of lreu_1750 [10], masking the effect of its overexpression; (ii) consequently the differentiating phenotype of the PocR deficient strain can be best observed under conditions in which its growth kinetics are hampered (such as in the presence of glycerol - Figure 2); and (iii) there is a large redundancy between the different transcriptome analyses carried out; most emphasis in this report has been put on the data related to the lreu_1750 overexpression in the absence of glycerol. The complete list of differentially regulated genes under all conditions assayed is available in Additional file 1: Transcriptome analysis data.
Upon overexpressing lreu_1750 only 120 genes (approximately 6% of the genome) are differentially regulated, of which, all but two are up-regulated. Three functional classes were represented with 10% or more of its members in the list of differentially expressed loci, namely the ones related to coenzymes, secondary metabolites and energy production (Additional file 1, Table S2). A closer inspection of the list of differentially regulated genes shows that lreu_1750 is clearly involved in the regulation of the same processes that have been linked to PocR in Salmonella [13]. These genes include the pdu cluster flanking lreu_1750, encoding the several subunits of the diol dehydratase and the metabolosome-assembly proteins [9], along with the two operons of the B12 biosynthesis cluster [7] (Figure 6). The lack of statistical significance observed for some of the expression data of the B12 synthesis cluster is easily explained. This cluster is divided into two multicistronic operons with a relatively low abundance and a remarkably large size. This raises great technical difficulties during mRNA purification as reported in the past [7,14].
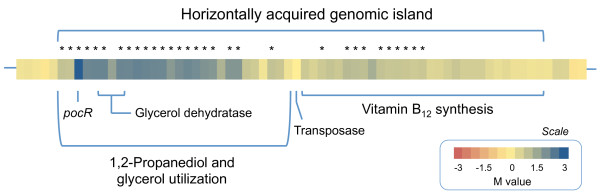
Figure 6.
Relative expression levels of the loci situated within the genetic island that harbors PocR. Color-scale plot of the relative expression levels of the loci situated between lreu_1690 and lreu_1757 (not to scale). All genes situated within the genetic island are up-regulated when lreu_1750 is overexpressed with the exception of the transposase. The lack of statistical significance observed for some of the loci is discussed in the text. Abbreviations stand for: M, log2(intensity of signal of L. reuteri pNZ7748/intensity of signal of L. reuteri pNZ7021); "*", p-value ≤ 0.05.
Besides these genes stretching from lreu_1695 to lreu_1752, which are expected to be regulated by PocR by homology with Salmonella [2], we found two genes (lreu_0429 and lreu_0430) predicted to be co-transcribed and annotated with unknown function in GenBank, that are up-regulated ~3.5-fold (Table 1). A closer look at their sequence indicates that these are most likely two subunits of an ATPase transporter [15], which have been tentatively associated with copper transport in ERGO [16]. We speculate that these are cobalt- rather than copper-transporters, based on the fact that cobalt availability is essential for the synthesis of vitamin B12. We also found regulated a few genes related to sugar uptake and carbon metabolism. These are presumably related to the up-regulation of lreu_0088 (transcription factor of LacI family), which is most likely a consequence of a slight drop in growth rate caused by the extra burden of the lreu_1750 overexpression (Figure 2). A considerable number of enzymes involved in recombination and DNA repair were also up-regulated. This is probably a consequence of the homologous region to the genome of L. reuteri present in plasmid pNZ7748 (harboring lreu_1750) and not in pNZ7021 (empty vector), as previously observed [17,18].
Table 1.
Relative expression levels of loci associated to PocR and not within its flanking regiona.
| Locus | Function | Mb. | pc. | Accession number |
|---|---|---|---|---|
| Lreu_0088 | Transcriptional regulator, LacI family | 1.11 | 0.03 | gi|148543330 |
| Lreu_0103 | 3-hydroxybutyryl-CoA dehydrogenase (EC 1.1.1.157) | 0.89 | 0.03 | gi|148543342 |
| Lreu_0429 | Putative cobalt-transporting ATPased. | 1.85 | 0.00 | gi|148543665 |
| Lreu_0430 | Putative cobalt-transporting ATPased. | 1.76 | 0.00 | gi|148543666 |
| Lreu_0479 | Arabinose-proton symporter | 1.67 | 0.00 | gi|148543714 |
| Lreu_0631 | Pyruvate dehydrogenase alpha subunit (EC 1.2.4.1) | 0.88 | 0.02 | gi|148543863 |
| Lreu_0632 | Pyruvate dehydrogenase beta subunit (EC 1.2.4.1) | 0.86 | 0.04 | gi|148543864 |
| Lreu_0633 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (EC 2.3.1.12) | 0.81 | 0.03 | gi|148543865 |
| Lreu_0910 | Alpha-galactosidase (EC 3.2.1.22) | 0.90 | 0.03 | gi|148544139 |
| Lreu_1007 | Transcription regulator, Crp family | -0.82 | 0.04 | gi|148544234 |
| Lreu_1531 | Fumarate hydratase (EC 4.2.1.2) | 1.09 | 0.05 | gi|148544743 |
| Lreu_1768 | Lactose permease | 0.97 | 0.05 | gi|148544974 |
| Lreu_1832 | Histidine decarboxylase (EC 4.1.1.22) | -1.67 | 0.03 | gi|148545038 |
a. Genes predicted to encode phage-related proteins, recombinases, mobile elements, DNA repair and general or unknown functions were omitted (for full list, please see Additional file 1, Table S1)
b. M, log2(intensity of signal of L. reuteri pNZ7748/intensity of signal of L. reuteri pNZ7021).
c. p, p-value.
d. Annotated as hypothetical protein in GenBank and as copper-transporting ATPase (EC 3.6.3.10) in ERGO database [16].
The transcriptome studies carried out for the PocR insertion mutant were consistent with the results obtained through the overexpression of lreu_1750. We mainly observed in the PocR mutant compared to the wild-type strain, a down-regulation of the genes located within the genetic island that comprises the pdu and vitamin B12 operons (Additional file 1, Table S3). Again due to the rarity and fragility of these transcripts [7,14] only 30 out of 58 loci are differentially expressed significantly (p-value ≤ 0.05) even though the whole region, excluding the transposase, appears co-regulated.
There is strong phylogenetic evidence supporting that the pdu and vitamin B12 synthesis gene clusters have been acquired by L. reuteri through distant horizontal gene transfer [7,8]. The confinement of the PocR regulon to mostly one continuous stretch of the chromosome (Figure 6), with exception of the putative cobalt transporter, further substantiates this hypothesis.
Conclusions
In this study, we have provided experimental evidence that lreu_1750 encodes a PocR-like regulatory protein, despite its lack of sequence homology to PocR from enteric bacteria. This was achieved by overexpression and inactivation of lreu_1750, and assessment of its impact on central carbon and energy metabolism, and on reuterin, 1,3-propanediol and vitamin B12 biosynthesis. In addition, we characterized the genome-wide transcriptional response of both constructs in comparison to the wild-type leading to the identification of the genes encompassed in the PocR-like regulon of L. reuteri. The latter were found to be similar to the ones present in some representatives of γ-Proteobacteria. Ultimately, the demonstrated stimulatory effects of PocR on vitamin B12, 1,3-propanediol and reuterin synthesis could be applied to improving the production yield of these industrially relevant compounds.
Methods
Phylogenetic analysis of Lreu_1750
The sequence of Lreu_1750 (GI:148544956) was entered as a string to search for closely related homologs within available microbial genomes using the protein-protein BLAST algorithm [19]. Relevant sequences were retrieved and aligned using ClustalW with default settings [20] and visualized in CLC Sequence Viewer 6.5.
Strains, plasmids, primers and cultivation conditions
The bacterial strains, plasmids and primers used in this study are listed in Table 2. L. reuteri strains were cultivated at 37°C in undefined MRS broth [21], in Vitamin B12 assay medium (Sigma-Aldrich, Zwijndrecht, The Netherlands) enriched with 0.5% glycerol (v/v), in the semi-defined medium LDMIIIG [12] and in a chemically defined medium (CDM) previously used to study vitamin B12 production in L. reuteri [14]. When appropriate, erythromycin and/or chloroamphenicol were added to a final concentration of 10 μg/mL.
Table 2.
Strains, plasmids and primers used in this study
| Materials | Relevant features | Source or reference |
|---|---|---|
| Strains | ||
|
L. reuteri JCM1112 |
Type strain, synonymous to ATCC 23272, DSM 20016 and F275. Human isolate. | Japanese Collection of Microorganisms (Riken, Japan) |
|
L. reuteri ATCC PTA 6475 |
Synonymous to MM4-1A. Finnish mother's milk isolate. | Biogaia AB (Stockholm, Sweden) |
| 6475::pocR | EmR, pocR insertion mutant derivative of L. reuteri ATCC PTA 6475 | This study |
|
Lc. lactis NZ9000 |
MG1363 pepN:nisRK, cloning host. | NIZO culture collection (Ede, The Netherlands) |
|
L. delbrueckii NIZO235 |
L. delbrueckii subsp. lactis ATCC 7830. Vitamin B12 assay indicator strain. | NIZO culture collection (Ede, The Netherlands) |
| Plasmids | ||
| pCR®2.1 | Used in routine cloning and to construct pJKS100 | Invitrogen (Carlsbad, CA) |
| pLEM5 | L. reuteri replication origin used to construct pJKS100 | [28] |
| pNZ7021 | CmR, pNZ8148 derivative with the nisin promoter replaced by the pepN promoter | [23] |
| pNZ7748 | CmR, pNZ7021 derivative harboring lreu_1750 downstream of the pepN promoter. | This study |
| pVE6007 | CmR, repA-positive temperature-sensitive derivative of pWV01 | [27] |
| pORI28 | EmR, repA-negative derivative of pWV01 | [35] |
| pORIpocR | EmR, pORI28 derivative harboring internal fragment of gene encoding putative PocR | This study |
| pJKS100 | CmR, E. coli-L. reuteri shuttle vector | This study |
| pJKS101 | CmR, pJKS100 derivative expressing 6475 pocR gene under control of its natural promoter | This study |
| Primers | 5' - 3' | Application |
| P180 | AAAAGGTACCGTAGGCGAAATTCAAATGTACG | Amplification of lreu_1750 and addition of KpnI site |
| P181 | GAATAAATAAGAGGCTGGGCAC | Amplification of lreu_1750 |
| P182 | ATGAACTCTATTCAGGAATTG | Control of pNZ7748 |
| LR0062F-BHI | TGACGGATCCTAACACAAGCATTACCGGAGCAATTG | Amplification of internal fragment of putative pocR, addition of BamHI site and translational stop codon |
| LR0062R-ERI | TGACGAATTCGCGTCTGATTCTATATGTGATTC | Amplification of internal fragment of putative pocR and addition of EcoRI site |
| LR0062 FL F | CGCTTTATCCTCAATTTGTTACG | Amplification of wild-type pocR gene and natural promoter |
| LR0062 FL R | GCTTTTACCATTGCATCAGCAG | Amplification of wild-type pocR gene and natural promoter |
Construction of putative pocR overexpression and deletion mutants
Gene lreu_1750, encoding the putative PocR in JCM1112T, was overexpressed constitutively under control of the pepN promoter in a similar fashion as previously described [22]. A fragment containing lreu_1750 was amplified from chromosomal DNA of L. reuteri using Herculase II DNA polymerase (Stratagene, La Jolla, USA), and primers P180 and P181 (Table 2). After digestion with KpnI, the modified amplicon was purified and cloned in pNZ7021 making use of the KpnI and PmlI restriction sites directly downstream of the pepN promoter. The resulting plasmid, termed pNZ7748, was used directly from the ligation reactions to transform Lactococcus lactis NZ9000 by electroporation [23]. Subsequently, pNZ7748 was purified from Lc. lactis as previously described [24] and, after confirming the sequence of the insert using both P181 and P182, it was used to transform L. reuteri also by electroporation [25].
The disruption of the putative pocR gene was carried out in L. reuteri ATCC PTA 6475, which shares an identical sequence with the type strain (JCM1112) for this region of the chromosome [1]. This was achieved by site-specific integration of plasmid pORIpocR as described previously [26] using the temperature-sensitive plasmid pVE6007 [27] as the helper plasmid. The internal fragment of the target gene was amplified by PCR using primers LR0062F-BHI and LR0062R-ERI (Table 2), and inserted into pORI28 by directional cloning using standard techniques [24]. The resulting insertion mutant was designated 6475::pocR.
Complementation of L. reuteri 6475::pocR
An E. coli-L. reuteri shuttle vector (pJKS100) was constructed by combining an L. reuteri replicon from pLEM5 [28], the chloramphenicol resistance gene (CmR) from pVE6007 [27], the L. lactis promoter (P23) [29], and the pUC origin and multiple cloning site (MCS) from pCR®2.1 (Invitrogen, Carlsbad, CA). Each fragment was PCR amplified from their respective template, restriction enzyme digested and subsequently ligated to generate the final shuttle-vector, pJKS100. To create the complementation vector for 6475::pocR, the L. reuteri 6475 pocR gene with its natural promoter was PCR-amplified from genomic DNA using LR0062 FL F and LR0062 FL R primers and cloned into pJKS100 using standard techniques [24]. Both constructs, pJKS100 and pJKS101, were electroporated seperately into L. reuteri 6475::pocR as previously described [25].
Fermentation conditions and substrate and product analysis
The physiological effects of the overexpression of lreu_1750 were studied in pH-controlled batch cultivations of L. reuteri pNZ7748 (lreu_1750 overexpression) and L. reuteri pNZ7021 (empty plasmid) in CDM in the presence or absence of glycerol carried out as described previously [14]. At different time points, samples were taken for transcriptome, supernatant and vitamin B12 analysis (Figure 2). We determined the extracellular concentration of main fermentation substrates and products by HPLC, as described elsewhere [6,30].
The comparison between the insertion mutant, 6475::pocR, and its parent strain was established in batch fermentations of LDMIIIG or MRS carried out in an anaerobic chamber (80% N2, 10% H2, and 10% CO2; Microbiology International). Transcriptome comparisons were carried out at the end of the fermentation when biomass concentration became stable.
Vitamin B12 and reuterin determination
Vitamin B12 levels were determined as described in the Official methods of analysis of AOAC International [31], using a bioassay with L. delbrueckii subsp. lactis ATCC 7830 as the indicator strain. Reuterin production was measured with a bioassay and carried out as previously described [5].
Transcriptome analyses
- Transcriptional analysis of the putative PocR overexpression mutant
The transcriptome of cells transformed with pNZ7748 (lreu_1750 overexpression) and pNZ7021 (empty plasmid) were compared using cDNA microarrays as previously detailed [14] using a hybridization scheme comprising 17 arrays in a loop-design. The following samples were hybridized per array labeled with cyanine3 and cyanine5, respectively: sta-F6 and sta-F5, sta-F7 and sta-F8, sta-F5 and sta-F7, sta-F8 and sta-F6, sta-F3 and sta-F4, sta-F1 and sta-F3, sta-F2 and sta-F1, sta-F4 and sta-F2, exp-F3 and exp-F4, exp-F1 and exp-F3, exp-F2 and exp-F1, exp-F4 and exp-F2, exp-F4 and sta-F4, sta-F3 and exp-F3, sta-F2 and sta-F8, sta-F4 and sta-F6, exp-F2 and sta-F2. Here, F1 and F5 represent completely independent biological duplicates of L. reuteri pNZ7021 cultured in the absence of glycerol; F2 and F6 represent completely independent biological duplicates of L. reuteri pNZ7748 cultured in the absence of glycerol; F3 and F7 represent completely independent biological duplicates of L. reuteri pNZ7021 cultured in the presence of glycerol; and F4 and F8 represent completely independent biological duplicates of L. reuteri pNZ7748 cultured in the presence of glycerol. The prefix exp- and sta- stand for cells harvested at mid-logarithmic and early-stationary growth phases, respectively. The custom probe design of the Agilent 11 K microarray platform (Agilent Technologies, Santa Clara, CA, USA) used is available at the Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo under accession number GPL6856, and the data obtained were deposited in the same repository under accession number GSE13289.
- Transcriptional analysis of the putative PocR insertion mutant
The transcriptome of the insertion mutant, 6475::pocR, and its parent strain were compared using two-color microarrays as previously detailed [32]. Briefly, oligonucleotides (60-mers) were designed and synthesized for 1,966 open reading frames from a draft genome sequence of L. reuteri ATCC PTA 6475 [1]. For expression analyses, three biological replicates of the insertion mutant and parent strain were compared. Moreover, dye-swap hybridization was performed for each comparison. Following mRNA isolation [32], cDNA synthesis, labeling, and hybridization were performed as previously described [32]. Information regarding the microarray platform and data obtained is deposited at NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under GPL7541 and GSE22926, respectively.
GenePix Pro 4.0.12 software was utilized for image analysis of the 6475 microarrays. Normalization within arrays and between arrays was performed by applying the Loess algorithm [33] using the Limma package [34] in R http://www.r-project.org. Normalized intensities were used for further analysis. The average signal intensities of three biological replicates were calculated in order to compare the relative gene expression of mutant and wild type strains. The statistical significance of differences was calculated based on variation in biological duplicates, using the eBayes function in Limma (cross-probe variance estimation) and false discovery rate (FDR) adjustment of the p-values. Only genes that were differentially expressed by least 1.5-fold with FDR-adjusted p-values lower than 0.05 were considered significant.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FS performed the phylogenetic analysis and all experiments related to the overexpression of PocR and measurement of vitamin B12 production, and wrote the first draft of the manuscript. JS constructed the PocR mutant and measured reuterin production and helped to draft the manuscript. DS performed the fermentations and cDNA microarrays related to the PocR mutant and helped to draft the manuscript. DM supervised the statistical analysis and interpretation of the cDNA microarrays related to the PocR overexpression. BT supervised the interpretation of the fermentation data. WV helped in the design and supervision of the PocR overexpression experiments and helped to draft the manuscript. JV helped in the design and supervision of the PocR deletion experiments and helped to draft the manuscript. JH conceived the study, participated in its coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Transcriptome analysis data. Single PDF containing three tables with additional transcriptome analysis data: Additional file 1, Table S1, Complete list of transcripts from L. reuteri that are differentially expressed by the overexpression of lreu_1750; Additional file 1, Table S2, Distribution of transcripts listed in Additional file 1, Table S1 throughout categories of clusters of orthologuous groups (COG, [35]); Additional file 1, Table S3, Complete list of loci from L. reuteri ATCC PTA 6475 that are differentially expressed by the disruption of the putative PocR.
Contributor Information
Filipe Santos, Email: filipe.santos@falw.vu.nl.
Jennifer K Spinler, Email: spinler@bcm.tmc.edu.
Delphine MA Saulnier, Email: Delphine.Saulnier@nizo.nl.
Douwe Molenaar, Email: douwe.molenaar@falw.vu.nl.
Bas Teusink, Email: bas.teusink@falw.vu.nl.
Willem M de Vos, Email: willem.devos@wur.nl.
James Versalovic, Email: jamesv@bcm.tmc.edu.
Jeroen Hugenholtz, Email: jhugenholtz@eur.ko.com.
Acknowledgements
This work was supported by the Department of Defense through the Defense Advanced Research Projects Agency (DARPA), National Institutes of Health, National Institute of Diabetes, Digestive and Kidney Diseases (R01 DK065075), and National Center for Complementary and Alternative Medecine (R01 AT004326). We thank Eammon Connoly for providing the L. reuteri 6475 strain, P. Hemarajata and M. Balderas for their technical efforts, and TA Misttetta for assistance in the statistical analyses of the 6475 microarray data.
References
- Walter J, Britton RA, Roos S. Microbes and Health Sackler Colloquium: Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A. 2010. pp. 4645–4652. [DOI] [PMC free article] [PubMed]
- Chen P, Ailion M, Bobik T, Stormo G, Roth J. Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium. Journal of bacteriology. 1995;177:5401–10. doi: 10.1128/jb.177.19.5401-5410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Bobik TA, Gottschalk G. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS microbiology reviews. 1998;22:553–566. doi: 10.1111/j.1574-6976.1998.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Vollenweider S, Grassi G, König I, Puhan Z. Purification and structural characterization of 3-hydroxypropionaldehyde and its derivatives. Journal of agricultural and food chemistry. 2003;51:3287–93. doi: 10.1021/jf021086d. [DOI] [PubMed] [Google Scholar]
- Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14:166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleusix V, Lacroix C, Vollenweider S, Duboux M, Le Blay G. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC microbiology. 2007;7:101. doi: 10.1186/1471-2180-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Vera JL, van der Heijden RTJM, Valdez GF, de Vos WM, Sesma F, Hugenholtz J. The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiology. 2008;154:81. doi: 10.1099/mic.0.2007/011569-0. [DOI] [PubMed] [Google Scholar]
- Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T, Murakami M, Hisamatsu S, Kato Y, Takizawa T. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA res. 2008;15(3):151–161. doi: 10.1093/dnares/dsn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, Warren MJ, Prentice MB. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. Journal of bacteriology. 2008;190:4559–67. doi: 10.1128/JB.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F. Vitamin B12 synthesis in Lactobacillus reuteri. 2008. p. 274.
- Rossell S, Solem C, Jensen PR, Heijnen JJ. Towards a quantitative prediction of the fluxome from the proteome. Metabolic engineering. 2011;13:253–62. doi: 10.1016/j.ymben.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Jones SE, Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC microbiology. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Ailion M, Roth JR. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Teusink B, Molenaar D, van Heck M, Wels M, Sieuwerts S, de Vos WM, Hugenholtz J. Effect of amino acid availability on vitamin B12 production in Lactobacillus reuteri. Applied and Environmental Microbiology. 2009;75:3930–3936. doi: 10.1128/AEM.02487-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic acids research. 2007;35:D237–40. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Larsen N, Walunas T, D'Souza M, Pusch G, Selkov E, Liolios K, Joukov V, Kaznadzey D, Anderson I, Bhattacharyya A, Burd H, Gardner W, Hanke P, Kapatral V, Mikhailova N, Vasieva O, Osterman A, Vonstein V, Fonstein M, Ivanova N, Kyrpides N. The ERGO genome analysis and discovery system. Nucleic acids research. 2003;31:164–71. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegkamp A, Mars AE, Faijes M, Molenaar D, de Vos RC, Klaus SMJ, Hanson AD, de Vos WM, Smid EJ. Physiological responses to folate overproduction in Lactobacillus plantarum WCFS1. Microbial cell factories. 2010;9:100. doi: 10.1186/1475-2859-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegkamp A. Modulation of folate production in lactic acid bacteria. 2008.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix. Nucleic acids research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Man JD, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Bact. 1960;23:130–135. [Google Scholar]
- Santos F, Wegkamp A, Vos WM de, Smid EJ, Hugenholtz J. High-level folate production in fermented foods by the B12 producer Lactobacillus reuteri JCM1112. Applied and environmental microbiology. 2008;74:3291. doi: 10.1128/AEM.02719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegkamp A, Van Oorschot W, De Vos WM, Smid EJ. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Applied and environmental microbiology. 2007;73:2673. doi: 10.1128/AEM.02174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Walter J, Heng NCK, Hammes WP, Loach DM, Tannock GW, Hertel C. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Applied and environmental microbiology. 2003;69:2044. doi: 10.1128/AEM.69.4.2044-2051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Chagnaud P, Tannock GW, Loach DM, Dal Bello F, Jenkinson HF, Hammes WP, Hertel C. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Applied and environmental microbiology. 2005;71:979. doi: 10.1128/AEM.71.2.979-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. Journal of bacteriology. 1992;174:5633–8. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fons M, Hégé T, Ladiré M, Raibaud P, Ducluzeau R, Maguin E. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid. 1997;37:199–203. doi: 10.1006/plas.1997.1290. [DOI] [PubMed] [Google Scholar]
- van der Vossen JM, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Applied and environmental microbiology. 1987;53:2452–7. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starrenburg MJC, Hugenholtz J. Citrate Fermentation by Lactococcus and Leuconostoc spp. Applied and environmental microbiology. 1991;57:3535. doi: 10.1128/aem.57.12.3535-3540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz W, Latimer GW. Official methods of analysis of AOAC International. 18. Gaithersburg, Md. AOAC International; 2006. [Google Scholar]
- Saulnier DM, Santos F, Roos S, Mistretta T, Spinler JK, Molenaar D, Teusink B, Versalovic J. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS ONE. 2011;6:e18783. doi: 10.1371/journal.pone.0018783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall T, Bath K, Britton RA, Jonsson H, Versalovic J, Roos S. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Applied and environmental microbiology. 2007;73:3924. doi: 10.1128/AEM.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic acids research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic acids research. 2000;28:33–6. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptome analysis data. Single PDF containing three tables with additional transcriptome analysis data: Additional file 1, Table S1, Complete list of transcripts from L. reuteri that are differentially expressed by the overexpression of lreu_1750; Additional file 1, Table S2, Distribution of transcripts listed in Additional file 1, Table S1 throughout categories of clusters of orthologuous groups (COG, [35]); Additional file 1, Table S3, Complete list of loci from L. reuteri ATCC PTA 6475 that are differentially expressed by the disruption of the putative PocR.