Abstract
The fish swimbladder is a unique organ in vertebrate evolution and it functions for regulating buoyancy in most teleost species. It has long been postulated as a homolog of the tetrapod lung, but the molecular evidence is scarce. In order to understand the molecular function of swimbladder as well as its relationship with lungs in tetrapods, transcriptomic analyses of zebrafish swimbladder were carried out by RNA-seq. Gene ontology classification showed that genes in cytoskeleton and endoplasmic reticulum were enriched in the swimbladder. Further analyses depicted gene sets and pathways closely related to cytoskeleton constitution and regulation, cell adhesion, and extracellular matrix. Several prominent transcription factor genes in the swimbladder including hoxc4a, hoxc6a, hoxc8a and foxf1 were identified and their expressions in developing swimbladder during embryogenesis were confirmed. By comparison of enriched transcripts in the swimbladder with those in human and mouse lungs, we established the resemblance of transcriptome of the zebrafish swimbladder and mammalian lungs. Based on the transcriptomic data of zebrafish swimbladder, the predominant functions of swimbladder are in its epithelial and muscular tissues. Our comparative analyses also provide molecular evidence of the relatedness of the fish swimbladder and mammalian lung.
Introduction
The swimbladder is a specialized organ in teleosts that regulates buoyancy. It is a sac filled by several types of gas, mainly oxygen and carbon dioxide [1], [2], and is located between the peritoneum and the vertebral column in the dorsal part of the body. The volume of gas in the swimbladder can be actively regulated to maintain neutral buoyancy as fish ascend or descend in the water column. The long-term maintenance of internal gas pressure and also compensatory inflation and deflation are under reflex autonomic control. The homology of the vertebrate lung and swimbladder was noted by the British comparative anatomist Richard Owen as early as in 1846 [3]. It has been noted that both the swimbladder and lung are originated from the same ancestral organ, namely the respiratory pharynx, which is the posterior region of the pharynx [4], [5]. The diversified morphologies and functions of the swimbladder in different fish species illustrate its evolutionary relationship with tetrapod lungs. All ray-finned fish except the Polypteriformes develop the dorsal part directly from the ancestral respiratory pharynx as a pulmonoid swimbladder, which has a homologous blood supply with the lung. Although the homology of the lung and swimbladder has been well recognized based on morphological and embryological evidence, molecular evidence is still lacking [6], [7].
Despite the publication of a few papers recently on zebrafish swimbladder development, the swimbladder is still an organ understudied [8], [9], [10], [11]. In particular, we have characterized in detail the early development of zebrafish swimbladder with three distinct tissue layers [9]. Our study has also illustrated some conserved gene expression and regulatory mechanisms during early swimbladder and lung development, including the Hedgehog signaling pathway [9]. The study provides evidence that the budding and initial growth of the two organs is conserved, and that the Hedgehog signaling pathway is involved in the early development of the two organs. Thus, the difference of the two organs is likely to lie in the branching morphogenesis in lung, which is absent in the swimbladder.
Transcriptomic analyses, both descriptive and quantitative, are important for interpreting the functional elements of the genome and revealing the molecular constituents of cells and tissues. The transcriptome of zebrafish tissues have been characterized based on expressed sequence tag (EST) or microarray techniques [12], [13], [14], [15]. With the rapid advance of DNA sequencing technology, here we used Illumina next generation sequencing (NGS) platform for high content analysis of the zebrafish swimbladder transcriptome. We first described the molecular constitution of this organ, and then focused on the unique features, including the enriched genes, transcription factors and biological pathways. We also established the relatedness between fish swimbladder and mammalian lung by transcriptome comparison.
Results
General features of the zebrafish swimbladder transcriptome
The swimbladders were isolated from 90 adult zebrafish and pooled to make representatives for deep sequencing analysis. One cDNA library was constructed and sequenced for the swimbladder. A total of 34 million of read pairs was generated (Table S1), which is comparable to several recently published data using the Illumina Genome Analyzer [16], [17]. All sequence tags were mapped to known transcripts in ZGC (Zebrafish Gene Collection) in order to reveal the molecular characteristics of the swimbladder transcriptome. A total of 9,315 transcript entries were identified with as few as one mapped read pairs, constituting 55.6% of total known zebrafish transcript entries in the ZGC database. As indicated in Figure 1, the swimbladder transcriptome showed a relatively continuous distribution of gene expression levels. Similar to previous RNA-seq studies in other tissues [17], there were only a few transcripts which had high expression levels, while most transcripts were expressed at very low levels. More than 60% of the transcript body consisted of the highest expressed transcripts which accounted for less than 10% of the transcript entries, while the lowest expressed 60% of the transcript entries only contributed for 10% of the total transcript counts. It has been documented in previous studies that RNA-seq can readily detect gene expression level across a broad dynamic range [18], [19]. The expression level of genes in the swimbladders ranged from 0.54 to 11,178 RPKM, showing a dynamic range of more than five orders of magnitude in RNA concentration. Real-time PCR was carried out to verify relative abundance of several selected transcripts determined by RNA-seq and the result indicated a good correlation of the two methods (Figure 2). Since transcripts with marginal expression levels could be due to leaky expression, we implemented a general cutoff at 10 RPKM (∼3 transcripts per cell) for analyzing physiologically more relevant transcripts [18]. Finally, 5,758 transcript entries above the cutoff were used to represent the total swimbladder transcriptome. The list was subsequently mapped to 5,506 zebrafish Unigene clusters.
Figure 1. Distribution of transcript entries and total transcript counts over different tag abundance categories.
Categories of transcript abundance were assigned by setting the lower limit of the count number that includes the transcript as a category member. The percentages of total transcript counts and number of different transcript entries per category are plotted on a logarithmic scale (base 10).
Figure 2. Real-time validation of RNA-seq data.
The relative expression level of the genes selected was shown in log2 fold change as compared with a housekeeping gene, ef1a (10680.1 RKPM).
Functional implications of the swimbladder transcriptome
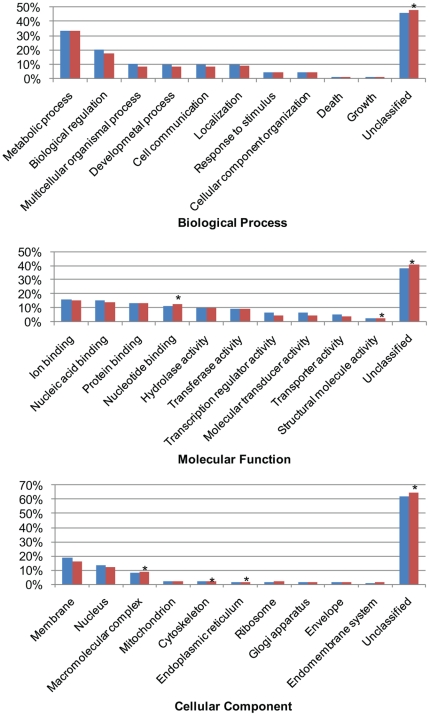
The 5,506 zebrafish Unigene clusters identified in the swimbladder were classified based on Gene Ontology (GO). Comparing to the distribution of GO categories of the total ZGC database (9,631 Unigene clusters), the swimbladder had significantly more expressed genes with unknown function in all the three classifications: Biological Process, Molecular Function, and Cellular Component, indicating the fact that the swimbladder is a less studied organ (Figure 3, Table S2). Under the Biological Process classifications, large proportions of genes were involved in housekeeping functions such as metabolic process and biological regulation. In Molecular Function classification, the categories of nucleotide binding and structural molecular activity were significantly enriched in the swimbladder, whereas in Cellular Component classification genes functioning in the endoplasmic reticulum was enriched in the swimbladder, suggesting the active synthesis and transportation of proteins. In particular, enriched categories under Molecular Function and Cellular Component together implicated the abundance of cytoskeleton genes in the swimbladder.
Figure 3. Gene ontology slim classification for the entire swimbladder transcriptome under Biological process, Molecular function and Cellular component classifications.
Slim classifications of the total ZGC database entries and swimbladder transcriptome are represented by blue and red bars, respectively. Astrid is used to label significantly enriched categories in the swimbladder (FDR<0.01).
Next we compared the Gene Ontology and energy distribution of the swimbladder under Molecular Function category. Energy distribution describes how a given tissue distributes its transcriptional energy based on relative abundance of total transcripts in different GO groups, thus yielding information on the main function of the tissue [13]. As shown in Figure 4, genes with nucleotide binding function are the second most diversified group in the swimbladder, and this group occupies a much heavier proportion in the energy distribution, indicating these transcripts tend to have higher expression levels. At the same time, there were a few categories which showed high diversity but have low expression levels, including genes with hydrolase, transferase, transcription regulator, molecular transducer, and enzyme regulator activities. These categories are crucial for maintaining basic metabolisms and performing specific functions for the swimbladder, although they are expressed at relatively lower levels.
Figure 4. Gene ontology classification (a) and energy distribution (b) of the swimbladder transcriptome.
Gene ontology classification and energy distribution are based on GO Slim classification of molecular functions. Genes without gene ontology information constitute 40% of the total swimbladder transcriptome and they were not included in the pie chart.
The original swimbladder transcriptome list contains many ribosomal protein genes and other housekeeping genes (see Table S3). To extract a more specific swimbladder transcriptome, a list of 888 enriched genes in the swimbladder was generated (bold-labeled in Table S3) using t-test by comparing with three other sets of zebrafish transcriptome data from the heart, brain and head kidney. The list efficiently excluded commonly expressed housekeeping genes and retained rarely expressed genes coding for transcription factor and signaling activity if they were enriched in the swimbladder.
A detailed enrichment analysis of GO terms was performed to examine the functional distribution of the 888 enriched genes (Table S4). Genes located in the endoplastic reticulum and extracellular region were enriched in the list, which implicated the active protein modification and transportation in the swimbladder. The results of enriched functional group in terms of Biological Process and Molecular Function together support the enrichment of signaling molecules in the list. We furthered examined the composition of these signaling molecules (Table S5). Among the 201 zebrafish Unigenes identified in the KEGG pathway database, 31 of them were involved in focal adhesion or extracellular matrix (ECM)-receptor interaction, suggesting the critical role of ECM in the swimbladder. Genes involved in adherens junction and tight junction were also enriched, which is essential for epithelial morphology and function. Particularly, genes involved in Hedgehog and TGF beta signaling pathways were enriched. Previous research in the lab has shown that Hedgehog signaling is critical for swimbladder specification and organization during embryogenesis [9]. The current transcriptome data correlates with the early developmental mechanism, suggesting that Hedgehog pathway remains active in the adulthood stage and may be important to maintain swimbladder regular function. Furthermore, GSEA (gene set enrichment analysis) pre-ranked analysis produced similar results in a quantitative manner (Table S6).
Top enriched genes in the swimbladder
In the list of top 50 transcribed genes (Table 1), the most abundant category was extracellular matrix (13 zebrafish Unigenes). Among them, three different glycoprotein genes were present: sparc, dcn and chad.
Table 1. Top 50 enriched Unigenes in the swimbladder with annotation.
| UGCluster | Gene Symbol | Gene Name | RPKM | P value | |
| 1 | Dr.20277 | acta2 | Actin, alpha 2, smooth muscle, aorta | 75742.5 | 1.17E-06 |
| 2 | Dr.82256 | tagln | Transgelin | 7296.7 | 2.43E-05 |
| 3 | Dr.75554 | sparc | Secreted acidic cysteine rich glycoprotein | 3751.4 | 3.04E-06 |
| 4 | Dr.75575 | col1a2 | Collagen, type I, alpha 2 | 2351.1 | 1.94E-04 |
| 5 | Dr.76397 | mmp2 | Matrix metalloproteinase 2 | 2038.5 | 6.96E-05 |
| 6 | Dr.105413 | tpm1 | Tropomyosin 1 (alpha) | 1903.5 | 1.90E-05 |
| 7 | Dr.76093 | s100a10b | S100 calcium binding protein A10b | 1821.1 | 8.08E-03 |
| 8 | Dr.67796 | icn | Ictacalcin | 1584.0 | 4.87E-03 |
| 9 | Dr.42794 | ctgf | Connective tissue growth factor | 1537.3 | 6.25E-05 |
| 10 | Dr.24504 | pabpc1a | Poly A binding protein, cytoplasmic 1 a | 1248.1 | 1.14E-03 |
| 11 | Dr.114623 | cald1 | Caldesmon 1 | 1226.2 | 5.84E-04 |
| 12 | Dr.79279 | lum | Lumican | 1175.7 | 3.22E-05 |
| 13 | Dr.34240 | fbp2 | Fructose-1,6-bisphosphatase 2 | 1053.9 | 3.96E-06 |
| 14 | Dr.79127 | stm | Starmaker | 1023.5 | 9.47E-03 |
| 15 | Dr.89765 | zgc:103467 | Myosin, light chain 9, regulatory | 1014.0 | 1.90E-03 |
| 16 | Dr.122523 | dap1b | Death associated protein 1b | 944.6 | 1.09E-02 |
| 17 | Dr.76351 | dcn | Decorin | 839.7 | 2.12E-04 |
| 18 | Dr.77427 | b2m | Beta-2 microglobulin | 798.2 | 1.41E-04 |
| 19 | Dr.7877 | mylka | Myosin, light chain kinase a | 718.8 | 3.27E-04 |
| 20 | Dr.76950 | tpm4 | Topomyosin alpha-4 chain | 699.1 | 7.02E-04 |
| 21 | Dr.88679 | fhl2a | Four and a half LIM domains 2a | 693.6 | 5.34E-03 |
| 22 | Dr.43046 | c1qtnf1 | C1q and tumor necrosis factor related protein 1 | 682.7 | 1.05E-06 |
| 23 | Dr.155448 | bactin2 | Bactin2 | 661.0 | 2.56E-04 |
| 24 | Dr.86222 | rergl | RERG/RAS-like | 651.3 | 1.82E-04 |
| 25 | Dr.80811 | si:ch211-237l4.5 | Si:ch211-237l4.5 | 628.6 | 2.24E-03 |
| 26 | Dr.80402 | chad | Chondroadherin | 558.6 | 5.77E-06 |
| 27 | Dr.142266 | itgb1b | Integrin, beta 1b | 536.1 | 7.00E-03 |
| 28 | Dr.105090 | ckba | Creatine kinase, brain a | 480.9 | 3.89E-03 |
| 29 | Dr.110680 | mxra8a | Matrix-remodelling associated 8a | 476.5 | 4.97E-05 |
| 30 | Dr.150732 | sdc2 | Syndecan 2 | 465.2 | 7.39E-04 |
| 31 | Dr.104772 | LOC100332090 | Thymosin, beta 4-like | 453.5 | 1.15E-02 |
| 32 | Dr.78058 | myh11 | Myosin, heavy polypeptide 11 | 448.5 | 2.10E-03 |
| 33 | Dr.33255 | tpm3 | Tropomyosin 3 | 428.3 | 1.37E-02 |
| 34 | Dr.81804 | dkk3 | Dickkopf homolog 3 (Xenopus laevis) | 427.7 | 1.86E-04 |
| 35 | Dr.76952 | actn4 | Actinin, alpha 4 | 407.0 | 2.55E-03 |
| 36 | Dr.29018 | cygb1 | Cytoglobin 1 | 391.4 | 6.09E-05 |
| 37 | Dr.30646 | nt5c2b | 5'-nucleotidase, cytosolic IIb | 389.2 | 2.50E-06 |
| 38 | Dr.560 | fbln1 | Fibulin 1 | 387.4 | 4.79E-05 |
| 39 | Dr.15501 | cyr61 | cystein-rich, angiogenic inducer, 61 | 365.6 | 2.04E-03 |
| 40 | Dr.75720 | myl6 | Myosin, light chain 6 | 356.6 | 1.38E-03 |
| 41 | Dr.80990 | htra1b | HtrA serine peptidase 1b | 353.3 | 3.41E-04 |
| 42 | Dr.76054 | tpm4 | Tropomyosin 4 | 350.0 | 1.82E-03 |
| 43 | Dr.83404 | csrp1a | Cysteine and glycine-rich protein 1a | 349.3 | 3.08E-06 |
| 44 | Dr.118039 | tnmd | tenomodulin | 341.5 | 3.31E-06 |
| 45 | Dr.75641 | cnn2 | Calponin 2 | 339.9 | 3.35E-03 |
| 46 | Dr.104797 | cdc42 | Cell division cycle 42 | 319.7 | 1.00E-02 |
| 47 | Dr.26461 | gpc1 | Glypican 1 | 309.2 | 2.72E-03 |
| 48 | Dr.76307 | cav1 | Caveolin 1 | 292.7 | 1.51E-02 |
| 49 | Dr.94036 | pthlh | Parathyroid hormone-like hormone | 270.1 | 1.15E-04 |
| 50 | Dr.11532 | fbxo32 | F-box protein 32 | 264.6 | 1.10E-02 |
Sparc encodes a prototypic matricellular protein, which is conserved in a wide variety of evolutionarily diverse organisms [20], [21]. Sparc can bind calcium, hydroxyapatite, and multiple types of collagens [22]. In mammals, Sparc is highly expressed in many developing tissues, including heart, thymus, lung, and gut [23], [24]. However, upon organ maturation, levels of Sparc decrease and remain relatively low in most adult tissues with the exception of those undergoing high rates of matrix production and proliferation such as bone, skin and gut epithelia. Moreover, there is robust elevation of Sparc expression upon injury, particularly those associated with excessive deposition of collagen [25]. Hence, expression patterns of Sparc are consistent with a critical role of this protein in collagen production and deposition, as collagen is also highly expressed in the swimbladder.
Dcn and chad belong to another glycoprotein family, the small leucine-rich repeat proteoglycan (SLRP) family. The SLRP family is found in a variety of extracellular matrix tissues, including bone, cartilage and tendon. Dcn is known to bind to different types of collagens [26]. It can be located in the ECM or in the cell membrane interacting with cell surface receptors. In muscles, Dcn located in the ECM function as components of it, regulating the matrix structure as well as modulating the bioavailability of several growth factors, including BMP-4 and TGF-b [27], [28], [29]. Overexpression of Dcn can induce migration of fibroblasts. A number of the intracellular regulators and effectors involved in cell migration can be up-regulated, including the focal adhesion proteins, and some of the small Rho GTPase such as RhoA, Rac1 and Cdc42 [30].
The second most abundant category in the top 50 transcribed genes list was cytoskeleton genes, especially those important for muscle contraction. The third most abundant category was membrane protein genes, including immune-related genes. Bacterial and fungal infections of the swimbladder are occasionally reported in various fish species [31], [32]. Having an open swimbladder that connects to the gastro-intestinal tract, the zebrafish swimbladder is more vulnerable to infection than physoclistous fishes. Our observation indicated that the swimbladder had its own defensive mechanism by expressing high levels of surface recognition molecules.
Top enriched transcription factors in the swimbladder
Next, we compiled the list of top transcribed transcription factors in the swimbladder enriched gene list based on Gene Ontology (Table 2).
Table 2. Top 20 enriched transcription factors in the swimbladder.
| UGCluster | Gene Symbol | Gene Name | RPKM | P value* | |
| 1 | Dr.132201 | hoxc8a | Homeo box C8a | 185.8 | 8.06E-05 |
| 2 | Dr.132203 | hoxc6a | Homeo box C6a | 143.2 | NA |
| 3 | Dr.15390 | foxl1 | Forkhead box L1 | 108.0 | 1.45E-03 |
| 4 | Dr.89399 | foxf1 | Forkhead box F1 | 85.8 | 4.48E-04 |
| 5 | Dr.12437 | tsc22d3 | TSC22 domain family, member 3 | 74.2 | 4.84E-03 |
| 6 | Dr.155563 | atf1 | Activating transcription factor 1 | 63.0 | 4.85E-04 |
| 7 | Dr.81025 | foxk1 | Forkhead box K1 | 61.5 | 2.41E-02 |
| 8 | Dr.152531 | foxq1l | Forkhead box Q, like | 46.6 | NA |
| 9 | Dr.139 | tgif1 | TGFB-induced factor homeobox 1 | 45.9 | 1.96E-02 |
| 10 | Dr.80310 | stat6 | Signal transducer and activator of transcription 6, interleukin-4 induced | 44.6 | 1.33E-02 |
| 11 | Dr.82149 | zhx3 | Zinc fingers and homeoboxes 3 | 33.3 | 2.45E-02 |
| 12 | Dr.20916 | isl2b | Islet2b | 28.0 | NA |
| 13 | Dr.510 | hoxc4a | Homeo box C4a | 27.5 | 2.47E-03 |
| 14 | Dr.15663 | cebpg | CCAAT/enhancer binding protein (C/EBP), gamma | 26.8 | 1.28E-02 |
| 15 | Dr.32618 | hoxa3a | Homeo box A3a | 26.1 | 1.71E-03 |
| 16 | Dr.91917 | mnx1 | Motor neuron and pancreas homeobox 1 | 25.9 | 1.63E-03 |
| 17 | Dr.80606 | creb3l2 | cAMP responsive element binding protein 3-like 2 | 22.4 | 3.49E-03 |
| 18 | Dr.8233 | tbx2b | T-box 2b | 22.0 | 1.03E-02 |
| 19 | Dr.83529 | hsf5 | heat shock transcription factor family member 5 | 20.6 | 1.46E-03 |
| 20 | Dr.114796 | foxm1l | Forkhead box M1-like | 17.0 | 7.81E-03 |
*NA, P value not available because the transcripts were detected only in the swimbladder.ummary of sequencing results.f Gene Ontology terms in the swimbladder enriched gene list.
One unique observation is that three genes from the hoxC cluster are enriched in the swimbladder, including hoxc8a, hoxc6a and hoxc4a. In order to confirm the expression of hoxC genes in swimbladder, we examined the expression of hoxc4a/6a/8a during zebrafish embryogenesis (Figure 5). The early expression pattern was consistent with previously reported results [33]. Expression of these genes in the notochord all had clear anterior boundaries, following the colinearity rule. Hoxc4a started to express in the notochord at the position of hindbrain, while hoxc6a and hoxc8a have the anterior expression boundary at approximately somite 2 and 4 respectively. None of their expression domain had a clear posterior boundary. Expression of all three genes in the swimbladder primordium could be detected at 36 hpf. The expression of hoxc8a became very prominent in the swimbladder starting from 48 hpf and was persistent at least until 72 hpf. Cross-section confirmed that hoxc8a was expressed strongly in the mesenchyme and relatively weakly in the mesothelium. Hoxc6a was expressed at a slightly lower level from 48 hpf to 72 hpf, and it was also expressed in the swimbladder mesenchyme and mesothelium. Hoxc4a was expressed at a barely visible level in the swimbladder and likely also in the mesoderm, though the exact expression domain could not be confirmed by cross-section because of its weak expression.
Figure 5. Expression of hoxc4a, hoxc6a and hoxc8a in developing zebrafish swimbladder. (a, e, i, m).
Expression of hoxc4a in the swimbladder at 36 hpf (a), 60 hpf (e) and 72 hpf (i, m). (b, f, j, n) Expression of hoxc6a in the swimbladder at 36 hpf (b), 60 hpf (f) and 72 hpf (j, n). (c, g, k, o) Expression of hoxc8a in the swimbladder at 36 hpf (c), 60 hpf (g) and 72 hpf (k, o). (d, h, l, p) Expression of foxf1 in the swimbladder at 36 hpf (d), 60 hpf (h), and 72 hpf (l, p). Panels (a–l) are lateral view of embryos after whole mount in situ hybridization and panels (m–p) are cross-sections of in situ hybridized embryos. Swimbladder is indicated by red dashed-line circles or red arrows. Numbers are used to mark the position of somite 1–4.
Two closely related Forkhead homeobox genes, foxl1 and foxf1 were also on the top of the list of enriched transcription factors [34]. Foxf1 was expressed in the swimbladder primordium as early as 36 hpf (Figure 5d) and the expression was persistent in the swimbladder until at least 72 hpf (Figure 5i). At the same time, prominent expression was also observed along the alimentary tract. Cross-section confirmed that the expression of foxf1 was restricted to the mesenchyme layer in both the swimbladder and the alimentary tract (Figure 5p). However, although the expression level of foxl1 is higher than foxf1 in the adult swimbladder as revealed by the RNA-seq data, expression of foxl1 was not detected in the developing swimbladder by in situ hybridization (data not shown).
Resemblance of swimbladder transcriptome to mammalian lung
In order to gain insight into molecular resemblance of fish swimbladder and mammalian lung, our swimbladder transcriptomic data were compared with the transcriptomic data from various human and mouse tissues based on microarray studies. The enriched gene list of each zebrafish tissue was used to represent its transcriptome. As shown in Figure 6, based on normalized enrichment scores (NES), the zebrafish brain show high resemblance to the human fetal and adult brains as well as the cerebellum and hippocampus of mouse. Meanwhile, the zebrafish heart closely resembles the mammalian heart and skeletal muscle, indicating similar cellular constitutions of the two tissues and thus validating the methodology. Among all the endodermal organs compared, it is interesting to note from Figure 6 that the zebrafish swimbladder has the highest and significant NES to both human and mouse lung, indicating that indeed the fish swimbladder has the highest resemblance with lung at the transcriptome level.
Figure 6. Comparison of zebrafish and human (a) or mouse (b) transcriptome tissues by GSEA.
Each intersection of the two zebrafish and human or mouse tissues was split into two cells. Upper cell shows NES, and lower cell shows the corresponding FDR. **: very significant (p<0.001), *: significant (p<0.05).
To further analyze the molecular resemblance of swimbladder and lung, GSEA leading edge genes, i.e., zebrafish swimbladder enriched genes appearing in the ranked list of human lung transcriptome at or before the point at which the running sum score reaches its maximum deviation from zero [35], were examined and presented in Table S7. These leading edge genes contains both constitution of the ECM (LUM, FN1, COL1A2, CYR61 and SPARC) and regulators of the ECM (TFPI, MMP2, RNPEP and HPSE), indicating that the zebrafish swimbladder and human lung may have some similar ECM characteristics. A few molecules belong to the small GTPase signaling pathway are identified (TNFAIP1, RND3, ARHGAP29, RASL12 and MX1), suggesting that the small GTPase signaling pathway may play an important role in both organs. Besides, genes involved in MAPK (DUSP1), TGF (TGFBI), and BMP (BMP5) signaling pathways are also identified. Several transcription factors are present in the list, including TGIF1, FOXF2, FOXF1, AATF and PFDN1. Examination of the leading edge gene list between zebrafish swimbladder and mouse lung showed a similar profile (data not shown).
Discussion
Epithelial tight junctions allow selective permeability of the swimbladder
The epithelium is the inner most layer of the swimbladder and is in direct contact with the gas inside. It has been shown by transmitted electron microscopy that the swimbladder epithelial cells are polarized even prior to inflation [36]. Tight junctions serve to form seals between epithelial cells, creating a selectively permeable barrier to intercellular diffusion. Consistent with this, our KEGG pathway analysis indicated that the tight junction pathway genes were indeed enriched in the swimbladder. Among the swimbladder enriched gene, the zebrafish homologs of cldn4/5/6/7/9 were identified, together with members of the Rho small GTPase subfamily including cdc42, rhoA and rab13. Claudins are transmembrane proteins which act in concert with other transmembrane and peripheral proteins to form the physical basis for tight junction. There are roughly two dozens of different claudins. In human airways, both bronchi and bronchioles express Claudin 1, 3, 4, 5 and 7. Particularly, CLDN3/4/5 have been found to be co-expressed by type II alveolar epithelial cells [37]. It has been revealed by immunofluorescence staining that CLDN4 is increasingly localized to the apical tight junction region, but with lower expression at the lateral region [38]. In contrast, CLDN3 and 5 are localized exclusively in the apical-most region of the tight junctions. Altered Claudin expression pattern can change the paracellular permeability characteristics of the epithelium. For example, CLDN3 overexpression decreases solute permeability, whereas CLDN5 increases permeability [39]. In summary, the expression of CLDN/cldn 4, 5 and 7 is conserved between the human lung and the zebrafish swimbladder.
However, cldn9, which is the one of the highest expressed in the swimbladder, is not identified in the human lung. Interestingly, Cldn9 is the most highly expressed in the inner ear of all the Claudin family members [40], and it is present in all of the major epithelial cell types that line the endolymphatic space. Analysis of Cldn9 mutant mice shows that Cldn9 is a paracellular ion permeability barrier for Na+ and K+, and loss of Cldn9 expression in the inner ear disrupts the Na+/K+ barrier and causes deafness. In contrast, a mutant zebrafish line with K+ channel defect shows both hearing defect and swimbladder over-inflation [10], suggesting that K+ channel plays a very important role in regulating swimbladder volume. In the zebrafish, the larvae surface and swallow a bolus of air, which is passed down through the esophagus and into the swimbladder via the pneumatic duct, to inflate their swimbladders [41]. However, how the larvae and adult fish maintain and regulate the swimbladder volume is unclear and seems to be independent of surface contact. Based on these findings, we speculate that cldn9 is likely to be involved in forming a Na+/K+ barrier in the swimbladder and to regulate swimbladder volume. It is also interesting to note that swimbladder has long been recognized to function for sound production and hearing [42].
Smooth muscle regulation and the ECM
It has been previously revealed by phalloidin labeling of muscle fibers revealed that smooth muscles are the major muscle constitution in the swimbladder and myocytes form thick bands along the ventral surface of the anterior chamber and bilaterally along the posterior chamber. In contrast, striated muscle fibers constitute a sphincter at the junction of the esophagus with the pneumatic duct [43]. The abundance of muscle-related genes identified in the swimbladder transcriptome correlates with this feature. Besides, KEGG pathway and GSEA analysis showed critical role of interaction between the cells and surrounding extracellular matrix.
The viscoelasticity of smooth muscle is contributed by a complex extracellular matrix. The ECM is not only a supporting structure of the smooth muscles, but also a dynamic structure constantly turning over its contents. This explains the abundant ECM-relating transcripts and the active protein transportation process. The major protein constituting ECM are collagens, glycoproteins and proteoglycans. In our transcriptome data, we also observed these transcripts expressing at high levels in the swimbladder. Collagen I is the only type of collagen identified in the swimbladder transcriptome, and it is also the most abundant collagen in the human body. In mammalian tissues, type I collagen shows the highest expression in the cardiomyocytes and smooth muscles [44].
Previously, it has been reported that human airway smooth muscle cells in culture can secrete various ECM proteins [45], [46]. The ECM can store inflammatory mediators and growth factors, which can be released via the action of MMPs (matrix metalloproteinases) to modulate smooth muscle proliferative and synthetic capacity. The composition of the ECM can be regulated by the synthesis of new proteins, and by the action of MMPs and TIMPs (tissue inhibitor of metalloproteinases). In the swimbladder, mmp2 and timp2 are the only MMP and TIMP identified. Mmp2 functions to degrade type IV collagen, which is a major structural component of the basement membranes. The activity of Mmp2 is often associated with excessive extracellular turnover, which is consistent with our observations that sparc is the most abundant transcript in the swimbladder. Interestingly, TIMP2 has been shown to be able to directly bind and inhibit MMP2 activity [47]. Therefore, mmp2 and timp2 may function to balance the extracellular turnover rate in the swimbladder.
Possible roles of hoxC family genes in the swimbladder
Hox genes are one of the master regulators of pattern formation during embryogenesis. They regulate pattern formation by coordinating cell proliferation, migration, adhesion and differentiation. Our data on the embryonic expression pattern of hoxC family members and the adult transcriptome data together suggest that the expression of embryonic hox genes is persistent until adult stage. This is in consistent with the previous findings that hox genes might have an enduring role in maintaining positional identity throughout the lifetime of an organism [48], [49]. As the expression of hoxc4a/6a/8a in developing swimbladder was identified, the function of these genes remains an open question; thus, it is worth further exploring their regulatory mechanisms in future studies.
In humans, HOXC6 mRNA is detected in both fetal and normal adult lung. On contrary, HOXC8 mRNA is present in the fetal lung, but absent from normal adult lung. Interestingly, HOXC8 is consistently up-regulated in emphysematous lungs, a disease in which the alveolar septum is disintegrated and the alveoli gradually lose the elasticity. However, the human lung has a different expression profile of Hox genes. In both human fetal and adult lungs, the most abundant expressed Hox genes are HOXA5, HOXB2 and HOXB5. Among these genes, only the homolog of HOXB5 is expressed in the zebrafish swimbladder at a relatively low level. It is mostly accepted that the swimbladder and lung were evolved from the same ancestral organ, namely the respiratory pharynx. The swimbladder arises from the dorsal part, while the lung originated from the ventral part. The different expression profiles of HOX/hox genes in the swimbladder and lung are consistent with this double origin theory.
In recent years, it becomes increasingly clear that hox genes have regulatory roles in the adult, likely involved in cell renewal and in the normal physiological changes that occur in the adult life [50]. The deregulated expression of hox genes in adulthood is associated with cancer development and malignant progression such as invasion and metastasis [51], [52]. Noticeably, HOXC cluster genes have been shown to be selectively overexpressed in prostate carcinoma and may play key roles in the acquisition of invasive and metastatic phenotypes of prostate cancer cells [53], [54]. Both of Hoxc6 and Hoxc8 have been shown to be able to regulate the cross-talk between Wnt, BMP, and FGF signaling pathways by directly targeting a few important regulators in the pathways [55], [56], [57]. Thus, the expression of HoxC cluster genes in the swimbladder may not only serve to memorize the positional identity of epithelial cells, but also act as master regulator for adult swimbladder function, likely in cellular adhesion and mobility.
Evolutionary insights between the fish swimbladder and mammalian lung
Epithelial cells of air-breathing organs of vertebrates are covered with a thin layer surfactant, which reduces and modifies surface tension at the air-liquid interphase. Surfactant consists of mixtures of lipids and surfactant proteins (SPs). In humans, four surfactant proteins have been identified: SP-A, SP-B, SP-C, and SP-D. These four proteins belong to three different superfamilies. Both SP-A and SP-D are collectins, and they are known to play a role in innate immune defense of the lungs by binding a wide array of pathogens, including viruses, bacteria, and fungi, and facilitating their uptake by immune cells. Both of them are rooted by the MBL (Mannose binding lectin) sequence [58]. Homologs of SP-A has been identified in the swimbladder of goldfish by western and northern blot analyses [59]. We also identified a zebrafish homolog in this family, lman2 (lectin,manose binding2), expressed in the swimbladder, which was confirmed by real-time qRT-PCR (Figure 2). SP-B, which is highly hydrophobic, belongs to the superfamily of saposin-like proteins, a diverse group of lipid-interacting proteins. We identified prosaposin (Dr.75922) transcripts in the zebrafish swimbladder at intermediate abundance (78.9 RPKM), and it is also enriched in the swimbladder. SP-C belongs to the chondromodulin I (CHM1) family. One of the zebrafish homolog from the gene family, tenomodulin (Dr.118039, 341.47 RPKM) is highly transcribed and enriched in the swimbladder. Taken together, the homologs of all four human SPs have been identified in the zebrafish swimbladder transcriptome, further supporting the evolutionary relationship of the fish swimbladder and mammalian lung. In human lung, SP-A is the most dominant surfactant protein expressing [60]. However, in the zebrafish swimbladder transcriptome, homologs of SP-B and SP-C are highly expressed surfactant-related genes. Since both of them are hydrophobic, the higher expressing level may due to the fact that the fraction of lipid (mainly cholesterol) in the swimbladder is higher than in lung surfactant of mammals [61]. In contrast to lung surfactant, swimbladder surfactant mainly acts as an antiglue to facilitate reopening of the swimbladder after a collapse or partial collapse, and it may prevent edema [62].
Gas gland cells of physostome have been shown to produce surfactant in vivo and in culture [63]. Lamellar bodies are also observed in the apical region of these cells. No anatomical evidence for a gas gland was found in the zebrafish swimbladder in previous study [43]. However, many species of physostomes that are known to secrete gas into their swimbladders do not have a morphologically identifiable gas gland, and it has been proposed that the gas-secreting cells may be scattered singly or in small groups in the wall of the swimbladder in these species [64], [65].Immunohistochemistry staining suggested the presence of gas-secreting cells in the zebrafish swimbladder by showing nerve terminal concentration of autonomic nerve terminals [43].
Another clue of the evolutionary homology is the parathyroid hormone-related protein (PTHrP). Ligand-receptor signaling involving PTHrP is crucial for the development and proper functioning of lungs in all vertebrates studied. Its expression correlates with lung maturation, homeostasis, and repair as well as alveolar size, septal thickness and composition of the matrix [66]. It is expressed throughout vertebrate phylogeny, beginning with its expression in the fish swimbladder as an adaption to gravity. The zebrafish swimbladder transcriptome provides supporting evidence by showing the high expression of parathyroid hormone (pth, Dr.94036).
Materials and Methods
Ethics statement
All experimental protocols were approved by Institutional Animal Care and Use Committee (IACUC) of National University of Singapore (Protocol 079/07).
RNA sample preparation and library sequencing
Healthy Singapore wildtype adult zebrafish (around 6 months old) were purchased from a local fish farm. The swimbladders including the attached pneumatic ducts were isolated from 45 female and 45 male fish and pooled. Brains, hearts and head kidneys were also collected from the same batch of fish for comparative studies. Total RNA was extracted using TRIzol® Reagent (Invitrogen). mRNA (polyA+) was purified using DynaBeads® Oligo(dT)25 (Invitrogen) according to the manufacture’s protocol and treated with DNaseI (Ambion)to remove DNA contamination. The resulted mRNA sample was quantified on NanoDrop® ND-100 Spectrophotometer (Thermo Scientific). Prior to cDNA synthesis, mRNAs were hydrolyzed by RNA Fragmentation Reagent (Ambion). Paired-ends sequencing was performed using Sanger-modified Illumina protocol [67], [68].
We used MAQ (Mapping and Assembly with Qualities) to align the sequence tags to transcriptome database [69]. MAQ assign each alignment a phred-scaled quality score (Qs), which measures the probability that the true alignment is not the one found by MAQ. The data have been submitted to the European Bioinformatics Institute (EBI) database (Accession number: ERP000447). ZGC database (retrieved on Jan 28, 2011) was used in this study, which contains 16,739 ORFs (Open Reading Frames). The sequencing results were summarized in Table S1. The mapped sequence tags for each transcript entry were normalized into RPKM as previously described [18].
Annotation
To facilitate functional implications of zebrafish transcriptome, all zebrafish genes were mapped to annotated human and mouse genes in order to use existing online software developed in human genes. Thus, Unigene annotation of zebrafish transcript entries (GenBank accession ID) and human and mouse homology mapping of zebrafish Unigene clusters were retrieved from the Genome Institute of Singapore Zebrafish Annotation Database (http://giscompute.gis.a-star.edu.sg/~govind/unigene_db/) as previously described [70]. For Unigene clusters mapped by more than one transcript entries, the highest RPKM was used to represent the expression level of the Unigene cluster [71]. In this study, the transcript entries of the ZGC database were mapped to 6392 unique human Unigene clusters and 6793 unique mouse Unigene clusters. Some zebrafish Unigene clusters were mapped to more than one human or mouse Unigene clusters, which usually came from the same gene family. To remove redundancy and avoid causing bias in functional analyses, only the first human or mouse Unigene cluster in the list was selected to represent the zebrafish Unigene clusters. Functional characterization of human and mouse Unigenes clusters was based on Gene Ontology and can be obtained from Stanford’s SOURCE database [72].
Swimbladder-enriched gene selection by t-test
While Gene Ontology analysis can provide a general picture of the swimbladder transcriptome, the unique features of the swimbladder may only be unmasked by removing those housekeeping genes which are commonly expressed in all tissues. Therefore, one-sample t-test was conducted to select enriched Unigenes in the swimbladder against other zebrafish tissues. One sample t-test was performed according to the standard method implemented in MATLAB. The p value is the probability, under the null hypothesis, of observing a value as extreme or more extreme of the test statistic
where is the sample mean or RPKM values of a transcript in the swimbladder, µ is the population mean or mean RKPM values of the same transcript in the other three comparing tissues, s is the sample standard deviation calculated from population means in the three comparing tissues, and n is the sample size and the value is 3 here. Unigene clusters with p value smaller than 0.025 are defined as enriched genes. At the same time, a second threshold of RPKM>10 and RPKM>average RPKM of the four comparing zebrafish tissues (swimbladder, brain, heart and head kidney) is added to ensure that the selected genes are relatively abundant and physiologically relevant. The enriched gene lists contain 888, 1,732 and 535 zebrafish Unigene clusters for the swimbladder, brain and heart, respectively. The lists were subsequently converted into 491, 967 and 323 homologous human Unigene clusters and 483, 963 and 311homologous mouse Unigene clusters.
Gene Ontology slim classification and enrichment analysis
Gene ontology slim classification was performed using WebGestalt against the total ZGC database (containing 9,631 zebrafish Unigene clusters) and the total zebrafish swimbladder transcriptome (containing 5,506 zebrafish Unigene clusters). The significance level of enrichment was indicated by false discovery rate (FDR)-corrected p-value from hypergeometric test. The cutoff is FDR<0.01.
Gene ontology enrichment analysis was performed using DAVID (The Database for Annotation, Visualization and Integrated Discovery) with the total zebrafish genome information as the background and p-values representing a modified Fisher’s exact t-test. Gene Ontology Fat categories were used for this analysis. GO Fat is a term that the DAVID team used to describe a subset of the GO term set. It is coined after GO slim which serves as a subset of the broadest GO terms. In contrast, the GO Fat attempts to filter out the broadest terms so that they will not overshadow the more specific terms. FDR score was also provided as a multiple testing correction method. Unless specifically indicated, the cut-off of p-value is <0.01. KEGG pathway analysis was also performed similarly using DAVID.
Analysis of the tissue-specific enriched gene list using GSEA pre-ranked analysis
GSEA Pre-ranked option was used to analyze the entire swimbladder enriched gene list. Briefly, the gene symbols of human homologs of the enriched zebrafish Unigene clusters were ranked using logarithm transformed p-value (base 10). The number of permutation used was 1000. Pathways with nominal p-value (NP) <0.05 were considered statistically significant.
Cross-species and cross-platforms analysis
Two sets of transcriptome data for healthy human and mouse tissues (GSE2361 and GSE97) were obtained from GEO (Gene Expression Omnibus). Annotation information was retrieved from the Genome Institute of Singapore Annotation Database (http://giscompute.gis.a-star.edu.sg/~govind/unigene_db/). For multiple probes which can be mapped to one Unigene cluster, the maximum signal intensity was selected to represent the expression level of the Unigene cluster.
We used GSEA to establish the relatedness between zebrafish and mammalian tissues. GSEA is a computational method that determines whether a priori defined set of genes shows statistically significant, concordant differences between two biological samples; it calculates an enrichment score using a running-sum statistic through a ranked list of gene expression data set [35]. The zebrafish swimbladder, brain and heart transcriptome lists were converted into human and mouse homolog Unigene clusters. The enriched gene list of each tissue was used to represent its transcriptome. The statistical significance of the enrichment score was estimated by using an empirical phenotype-based permutation test procedure. An FDR value was provided by introducing adjustment of multiple hypothesis testing.
Real-time PCR
Real-time PCR was performed using the LightCycler system (Roche Applied Science) with LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science) according to the manufacturer’s instructions. cDNA was synthesized from the same RNA sample which were used for the RNA-seq. For comparison between real-time PCR and RNA-seq results, Cp and RPKM values for each gene were normalized against Cp and RPKM of ef1a (Dr.31797).
Whole mount in situ hybridization
In situ hybridization probes were generated from available sequences in the public databases. The plasmids were linearized to synthesize both sense and antisense probes with T7 or SP6 RNA polymerase by using digoxigenin (DIG) RNA labeling mix (Roche Applied Science). Whole mount in situ hybridization (WISH) was performed using standard protocols as described previously [76].
Supporting Information
Summary of sequencing results for the zebrafish swimbladder, brain, heart and head kidney.
(DOC)
Detailed results of Gene Ontology slim classification of the entire swimbladder transcriptome.
(DOC)
Complete list of zebrafish genes expressed in the swimbladder. The genes are annotated based on Unigene cluster ID and ranked by RPKM. Swimbladder enriched genes are indicated in bold.
(XLS)
Enrichment of Gene Ontology terms in the swimbladder enriched gene list. The counts are presented in Unigene cluster counts. The percentage for each GO term represents the percentage of Unigene clusters in the GO term in the total transcript entries identified in the DAVID database. P-values represent a modified Fisher’s exact t-test. Only GO terms with p-value<0.01 were shown in the table.
(DOC)
KEGG pathway analysis of the swimbladder enriched gene list. The counts are presented in Unigene cluster counts. The percentage for each GO term represents the percentage of Unigene clusters in the GO term in the total transcript entries identified in the DAVID database. P-values represent a modified Fisher’s exact t-test. Only GO terms with p-value<0.05 were shown in the table.
(DOC)
GSEA analysis of the swimbladder enriched gene list. Gene sets that are statistically enriched with nominal p-value (NP) are shown. The sizes of the gene sets mean the number of the genes from the pre-defined canonical pathway database which are identified from the swimbladder enriched gene list. Values of normalized enrichment score (NES) indicate the activities of the enriched gene sets.
(DOC)
GSEA leading edge genes between zebrafish swimbladder and human lung.
(DOCX)
Acknowledgments
The authors are grateful to personnel in Gong’s lab for their suggestions and support during the process of this project. The authors are also thankful to supporting staff from the Sanger Institute for their excellent services.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The project was supported by a grant from the Ministry of Education (MOE), Singapore. JEC, RMA and DS were supported by the Wellcome Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fange R. Gas exchange in fish swim bladder. Rev Physiol Biochem Pharmacol. 1983;97:111–158. doi: 10.1007/BFb0035347. [DOI] [PubMed] [Google Scholar]
- 2.Pelster B. pH regulation and swimbladder function in fish. Respir Physiol Neurobiol. 2004;144:179–190. doi: 10.1016/j.resp.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Owen R. London: Longman, Brown, Green, and Longmans; 1846. Lectures on Comparative Anatomy and Physiology of the Vertebrate Animals. [Google Scholar]
- 4.Neumayer L. Die Entwicklung des Darms von Acipenser. Acta Zoologica. 1930;39:1–151. [Google Scholar]
- 5.Wassnetzov W. Über die Morphologie der Schwimmblase. Zoologische Jahrbücher Abteilung Anatomie und Ontogenie der Tiere. 1932;56:1–36. [Google Scholar]
- 6.Perry SF, Wilson RJ, Straus C, Harris MB, Remmers JE. Which came first, the lung or the breath? Comp Biochem Physiol A Mol Integr Physiol. 2001;129:37–47. doi: 10.1016/s1095-6433(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 7.Perry SF, Sander M. Reconstructing the evolution of the respiratory apparatus in tetrapods. Respir Physiol Neurobiol. 2004;144:125–139. doi: 10.1016/j.resp.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Winata CL, Korzh S, Kondrychyn I, Korzh V, Gong Z. The role of vasculature and blood circulation in zebrafish swimbladder development. BMC Dev Biol. 2010;10:3. doi: 10.1186/1471-213X-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winata CL, Korzh S, Kondrychyn I, Zheng W, Korzh V, et al. Development of zebrafish swimbladder: The requirement of Hedgehog signaling in specification and organization of the three tissue layers. Dev Biol. 2009;331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 10.Abbas L, Whitfield TT. Nkcc1 (Slc12a2) is required for the regulation of endolymph volume in the otic vesicle and swim bladder volume in the zebrafish larva. Development. 2009;136:2837–2848. doi: 10.1242/dev.034215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 12.Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A, et al. Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One. 2008;3:e1791. doi: 10.1371/journal.pone.0001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng S, Gong Z. Expressed sequence tag analysis of expression profiles of zebrafish testis and ovary. Gene. 2002;294:45–53. doi: 10.1016/s0378-1119(02)00791-6. [DOI] [PubMed] [Google Scholar]
- 14.Wen C, Zhang Z, Ma W, Xu M, Wen Z, et al. Genome-wide identification of female-enriched genes in zebrafish. Dev Dyn. 2005;232:171–179. doi: 10.1002/dvdy.20210. [DOI] [PubMed] [Google Scholar]
- 15.Lo J, Lee S, Xu M, Liu F, Ruan H, et al. 15000 unique zebrafish EST clusters and their future use in microarray for profiling gene expression patterns during embryogenesis. Genome Res. 2003;13:455–466. doi: 10.1101/gr.885403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H. Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics. 2008;92:187–194. doi: 10.1016/j.ygeno.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Hegedus Z, Zakrzewska A, Agoston VC, Ordas A, Racz P, et al. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol. 2009;46:2918–2930. doi: 10.1016/j.molimm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki K, Suzuki T, Weiss KM. Genetic basis for the evolution of vertebrate mineralized tissue. Proc Natl Acad Sci U S A. 2004;101:11356–11361. doi: 10.1073/pnas.0404279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka S, Nambu F, Nambu Z. Isolation of a cDNA encoding a putative SPARC from the brine shrimp, Artemia franciscana. Gene. 2001;268:53–58. doi: 10.1016/s0378-1119(01)00419-x. [DOI] [PubMed] [Google Scholar]
- 22.Giudici C, Raynal N, Wiedemann H, Cabral WA, Marini JC, et al. Mapping of SPARC/BM-40/osteonectin-binding sites on fibrillar collagens. J Biol Chem. 2008;283:19551–19560. doi: 10.1074/jbc.M710001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sage H, Vernon RB, Decker J, Funk S, Iruela-Arispe ML. Distribution of the calcium-binding protein SPARC in tissues of embryonic and adult mice. J Histochem Cytochem. 1989;37:819–829. doi: 10.1177/37.6.2723400. [DOI] [PubMed] [Google Scholar]
- 24.Mundlos S, Schwahn B, Reichert T, Zabel B. Distribution of osteonectin mRNA and protein during human embryonic and fetal development. J Histochem Cytochem. 1992;40:283–291. doi: 10.1177/40.2.1552170. [DOI] [PubMed] [Google Scholar]
- 25.Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kresse H, Hausser H, Schonherr E, Bittner K. Biosynthesis and interactions of small chondroitin/dermatan sulphate proteoglycans. Eur J Clin Chem Clin Biochem. 1994;32:259–264. [PubMed] [Google Scholar]
- 27.Chen XD, Fisher LW, Robey PG, Young MF. The small leucine-rich proteoglycan biglycan modulates BMP-4-induced osteoblast differentiation. FASEB J. 2004;18:948–958. doi: 10.1096/fj.03-0899com. [DOI] [PubMed] [Google Scholar]
- 28.Brandan E, Cabello-Verrugio C, Vial C. Novel regulatory mechanisms for the proteoglycans decorin and biglycan during muscle formation and muscular dystrophy. Matrix Biol. 2008;27:700–708. doi: 10.1016/j.matbio.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Cabello-Verrugio C, Brandan E. A novel modulatory mechanism of transforming growth factor-beta signaling through decorin and LRP-1. J Biol Chem. 2007;282:18842–18850. doi: 10.1074/jbc.M700243200. [DOI] [PubMed] [Google Scholar]
- 30.Tufvesson E, Westergren-Thorsson G. Biglycan and decorin induce morphological and cytoskeletal changes involving signalling by the small GTPases RhoA and Rac1 resulting in lung fibroblast migration. J Cell Sci. 2003;116:4857–4864. doi: 10.1242/jcs.00808. [DOI] [PubMed] [Google Scholar]
- 31.Wada S, Hatai K, Tanaka E, Kitahara T. Mixed infection of an acid-fast bacterium and an imperfect fungus in a Napoleon fish (Cheilinus undulatus). J Wildl Dis. 1993;29:591–595. doi: 10.7589/0090-3558-29.4.591. [DOI] [PubMed] [Google Scholar]
- 32.Aho R, Koski P, Salonen A, Rintamaki P. Fungal swimbladder infection in farmed Baltic salmon (Salmo salar L.) caused by Verticillium lecanii. Mycoses. 1988;31:208–212. doi: 10.1111/j.1439-0507.1988.tb03868.x. [DOI] [PubMed] [Google Scholar]
- 33.Prince VE, Price AL, Ho RK. Hox gene expression reveals regionalization along the anteroposterior axis of the zebrafish notochord. Dev Genes Evol. 1998;208:517–522. doi: 10.1007/s004270050210. [DOI] [PubMed] [Google Scholar]
- 34.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perlberg ST, Diamant A, Ofir R, Zilberg D. Characterization of swim bladder non-inflation (SBN) in angelfish, Pterophyllum scalare (Schultz), and the effect of exposure to methylene blue. J Fish Dis. 2008;31:215–228. doi: 10.1111/j.1365-2761.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, Daugherty B, Keise LL, Wei Z, Foley JP, et al. Heterogeneity of claudin expression by alveolar epithelial cells. Am J Respir Cell Mol Biol. 2003;29:62–70. doi: 10.1165/rcmb.2002-0180OC. [DOI] [PubMed] [Google Scholar]
- 38.Van Itallie C, Rahner C, Anderson JM. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest. 2001;107:1319–1327. doi: 10.1172/JCI12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1166–1178. doi: 10.1152/ajplung.00182.2003. [DOI] [PubMed] [Google Scholar]
- 40.Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, et al. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci. 2006;119:4819–4827. doi: 10.1242/jcs.03233. [DOI] [PubMed] [Google Scholar]
- 41.Goolish EM, Okutake K. Lack of gas bladder inflation by the larvae of zebrafish in the absence of an air-water interface. Journal of Fish Biology. 1999;55:1054–1063. [Google Scholar]
- 42.Ostrander G, Bullock G, Bunton T. Academic press; 2000. The laboratory fish. [Google Scholar]
- 43.Finney JL, Robertson GN, McGee CA, Smith FM, Croll RP. Structure and autonomic innervation of the swim bladder in the zebrafish (Danio rerio). J Comp Neurol. 2006;495:587–606. doi: 10.1002/cne.20948. [DOI] [PubMed] [Google Scholar]
- 44.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panettieri RA, Tan EM, Ciocca V, Luttmann MA, Leonard TB, et al. Effects of LTD4 on human airway smooth muscle cell proliferation, matrix expression, and contraction In vitro: differential sensitivity to cysteinyl leukotriene receptor antagonists. Am J Respir Cell Mol Biol. 1998;19:453–461. doi: 10.1165/ajrcmb.19.3.2999. [DOI] [PubMed] [Google Scholar]
- 46.Johnson PR, Black JL, Carlin S, Ge Q, Underwood PA. The production of extracellular matrix proteins by human passively sensitized airway smooth-muscle cells in culture: the effect of beclomethasone. Am J Respir Crit Care Med. 2000;162:2145–2151. doi: 10.1164/ajrccm.162.6.9909111. [DOI] [PubMed] [Google Scholar]
- 47.Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc Natl Acad Sci U S A. 2002;99:7414–7419. doi: 10.1073/pnas.102185399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268–275. doi: 10.1016/j.tcb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 52.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 53.Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, et al. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 54.Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, et al. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 55.Lei H, Juan AH, Kim MS, Ruddle FH. Identification of a Hoxc8-regulated transcriptional network in mouse embryo fibroblast cells. Proc Natl Acad Sci U S A. 2006;103:10305–10309. doi: 10.1073/pnas.0603552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei H, Wang H, Juan AH, Ruddle FH. The identification of Hoxc8 target genes. Proc Natl Acad Sci U S A. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCabe CD, Spyropoulos DD, Martin D, Moreno CS. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes AL. Evolution of the lung surfactant proteins in birds and mammals. Immunogenetics. 2007;59:565–572. doi: 10.1007/s00251-007-0218-6. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan LC, Daniels CB, Phillips ID, Orgeig S, Whitsett JA. Conservation of surfactant protein A: evidence for a single origin for vertebrate pulmonary surfactant. J Mol Evol. 1998;46:131–138. doi: 10.1007/pl00006287. [DOI] [PubMed] [Google Scholar]
- 60.Ballard PL, Merrill JD, Godinez RI, Godinez MH, Truog WE, et al. Surfactant protein profile of pulmonary surfactant in premature infants. Am J Respir Crit Care Med. 2003;168:1123–1128. doi: 10.1164/rccm.200304-479OC. [DOI] [PubMed] [Google Scholar]
- 61.Orgeig S, Daniels CB, Johnston SD, Sullivan LC. The pattern of surfactant cholesterol during vertebrate evolution and development: does ontogeny recapitulate phylogeny? Reprod Fertil Dev. 2003;15:55–73. doi: 10.1071/rd02087. [DOI] [PubMed] [Google Scholar]
- 62.Daniels CB, Orgeig S, Smits AW. The composition and function of reptilian pulmonary surfactant. Respiration Physiology. 1995;102:121–135. doi: 10.1016/0034-5687(95)00039-9. [DOI] [PubMed] [Google Scholar]
- 63.Prem C, Salvenmoser W, Wurtz J, Pelster B. Swim bladder gas gland cells produce surfactant: in vivo and in culture. Am J Physiol Regul Integr Comp Physiol. 2000;279:R2336–2343. doi: 10.1152/ajpregu.2000.279.6.R2336. [DOI] [PubMed] [Google Scholar]
- 64.Fange R. Hughes GM, editor. Gas exchange in the swim bladder. Respiration of amphibious vertebrates London: Academic Press. 1976. pp. 189–211.
- 65.Morris SM, Albright JT. Ultrastructure of the swim bladder of the goldfish, Carassius auratus. Cell Tissue Res. 1979;198:105–117. doi: 10.1007/BF00234838. [DOI] [PubMed] [Google Scholar]
- 66.Torday JS, Rehan VK. Deconvoluting lung evolution using functional/comparative genomics. Am J Respir Cell Mol Biol. 2004;31:8–12. doi: 10.1165/rcmb.2004-0019TR. [DOI] [PubMed] [Google Scholar]
- 67.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam SH, Wu YL, Vega VB, Miller LD, Spitsbergen J, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24:73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- 71.van Ruissen F, Ruijter JM, Schaaf GJ, Asgharnegad L, Zwijnenburg DA, et al. Evaluation of the similarity of gene expression data estimated with SAGE and Affymetrix GeneChips. BMC Genomics. 2005;6:91. doi: 10.1186/1471-2164-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Diehn M, Sherlock G, Binkley G, Jin H, Matese JC, et al. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan D, Prodduturi N, Zhang B. WebGestalt2: an updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11(Suppl 4):10. [Google Scholar]
- 75.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 76.Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of sequencing results for the zebrafish swimbladder, brain, heart and head kidney.
(DOC)
Detailed results of Gene Ontology slim classification of the entire swimbladder transcriptome.
(DOC)
Complete list of zebrafish genes expressed in the swimbladder. The genes are annotated based on Unigene cluster ID and ranked by RPKM. Swimbladder enriched genes are indicated in bold.
(XLS)
Enrichment of Gene Ontology terms in the swimbladder enriched gene list. The counts are presented in Unigene cluster counts. The percentage for each GO term represents the percentage of Unigene clusters in the GO term in the total transcript entries identified in the DAVID database. P-values represent a modified Fisher’s exact t-test. Only GO terms with p-value<0.01 were shown in the table.
(DOC)
KEGG pathway analysis of the swimbladder enriched gene list. The counts are presented in Unigene cluster counts. The percentage for each GO term represents the percentage of Unigene clusters in the GO term in the total transcript entries identified in the DAVID database. P-values represent a modified Fisher’s exact t-test. Only GO terms with p-value<0.05 were shown in the table.
(DOC)
GSEA analysis of the swimbladder enriched gene list. Gene sets that are statistically enriched with nominal p-value (NP) are shown. The sizes of the gene sets mean the number of the genes from the pre-defined canonical pathway database which are identified from the swimbladder enriched gene list. Values of normalized enrichment score (NES) indicate the activities of the enriched gene sets.
(DOC)
GSEA leading edge genes between zebrafish swimbladder and human lung.
(DOCX)