Abstract
Background:
Extended-spectrum beta-lactamase (ESBL)-producing Gram-negative bacteria are emerging and impacting significantly on the management of patients and hospital costs. Besides, they are not being routinely sought after in diagnostic laboratories thus contributing to treatment failure.
Materials and Methods:
Bacterial isolates from wounds of 45 patients were identified using commercial identification kits and antibiotic susceptibility was evaluated by the Bauer-Kirby method. Screening and phenotypic confirmation of ESBL production were done as prescribed by the Clinical and Laboratory Standards Institute. The conjugation experiment was performed by the mating assay in broth between the ESBL producers and E. coli ATCC 25922 as the recipient.
Results:
Out of 102 Gram-negative bacteria isolated, 36 were positive for ESBL mainly of the Enterobacteriaceae family (33) and the rest were oxidase-positive bacilli (3). The predominant bacteria were Klebsiella spp. and E. coli. Others were Serratia rubidae, Citrobacter freundii, Morganella morgannii, Proteus spp., Providencia stuartii, and Enterobacter spp. There was a significant association between treatment with third-generation cephalosporins (3GCs) and isolation of ESBLs (P=0.0020). The ESBL producers were multiply resistant and moderately sensitive to colistin. The conjugation experiment showed that the ESBL gene was transferred horizontally and tetracycline, cotrimoxazole, nitrofurantoin, gentamicin, and aztreonam resistance genes were co-transferred. No mortality was recorded but the mean length of stay in the hospital was 82 days.
Conclusion:
The development and spread of ESBL among Gram-negative bacteria and possible horizontal transfer calls for concern, especially in view of treatment failure, high treatment cost, and consequent discomfort to patients.
Keywords: Extended-spectrum beta-lactamase, Gram-negative bacteria, Horizontal gene transfer
INTRODUCTION
Wound infection could result in prolonged hospital stay, increased trauma care, and high treatment costs if not properly attended to. Gram-negative bacteria have been reported to be the major etiologies of wound infections.[1–3] Extended-spectrum beta-lactamases (ESBL) are β-lactamases capable of conferring bacterial resistance to the penicillins, first, second, and 3GCs, and aztreonam (but not the cephamycins or carbapenems)[4] and are usually encoded on plasmids which frequently carry genes encoding resistance to other classes of antibiotics. Members of the Enterobactericeae family are fast emerging as important agents for the spread of ESBL genes and strains of K. pneumoniae, K. oxytoca, and E. coli have been reported.[5] A considerable geographical spread of these β–lactamase-producing bacteria is being reported and is becoming a global problem in treating infections with 3GC antibiotics. That ESBLs are encoded on plasmids and are, therefore, easily transmissible from one organism to another is a therapeutic challenge for physicians as resistance genes for other antimicrobials such as aminoglycosides, tetracyclines, and trimethoprim/sulfamethoxazole are often present on the same plasmid,[6–8] thereby contributing further to the narrowing of the already limited treatment alternatives of choice of antibiotics.
There is currently scanty data on ESBL-producing organisms from wound infections in Nigeria and we have not come across documented report on the association of ESBL-producing bacilli with orthopedic wound infections in the Orthopedic Ward of the OAUTHC, Ile-Ife, Nigeria. We carried out a study to investigate the incidence of ESBL-producing Gram-negative bacteria in orthopedic wound infections here in Ile-Ife, Nigeria, and to determine the possible genetic medium of transfer of the ESBL-gene in vitro.
MATERIALS AND METHODS
Study design and sample collection
The study population comprised of patients presenting with orthopedic wounds in the Adult Orthopedic Ward of the Obafemi Awolowo University Teaching Hospitals Complex, Ile-Ife, Nigeria, located at latitude N 07° 30.246’ and longitude E 004° 34.247’. Seventy-three specimens were collected from 45 patients whose wounds had purulent discharges and complaints of pain around the site of the wounds. Pre-wound dressing swabs were collected with all aseptic precautions and were immediately transported to the research laboratory in sterile normal saline. Biopsies of soft tissue were obtained with the use of a sterile scalpel and a forceps and suspended in tryptic soy broth (Biolab, Hungary). All specimens were immediately transported to the laboratory for microbiological analysis.
Ethical approval
Clearance of research involving human subjects or patient records was granted by the Ethics and Research Committee of the Obafemi Awolowo University Teaching Hospitals’ Complex (OAUTHC), Ile-Ife, Nigeria, for this study.
Bacterial isolation and identification
The tissue biopsies and wound swab specimens were inoculated on blood and MacConkey agar plates (Oxoid, England) and were incubated at 37°C for 24 h. Identification of bacterial isolates was done using colony and microscopic morphology as described by Cheesbrough[9] and conventional biochemical tests using the GNB 24E Microbact Kit (Oxoid, England) for Gram-negative bacilli. Pure cultures in broth were stored in sterile glycerol at –20°C as stock and on tryptic soy agar (Biolab, Hungary) slants for further studies.
Antibiotic susceptibility testing of isolates
The antibiotic susceptibility testing was performed by the Bauer-Kirby Disc Diffusion method.[9] The inoculum was prepared from an overnight culture in tryptic soy broth. From this culture, a 0.5 (625 nm) McFarland standard suspension was prepared in sterile normal saline, 0.85% NaCl (w/v) using a colorimeter (WPA, UK). This standard contains approximately 107 CFU/ml. A sterile cotton wool swab was inserted into each test-tube containing the standardized inoculum suspension, rotated with firm pressure on the inside wall of the test-tube to remove excess fluid and then used to inoculate the entire surface of a dry sterile Mueller Hinton agar plate (Oxoid, England). The antibiotics used in the testing included aztreonam (30 μg), cefotaxime (30 μg), cefoxitin (30 μg), gentamicin (10 μg), nalidixic acid (30 μg), nitrofuratoin (300 μg), tetracycline (25 μg), cotrimoxazole (1.25/23.75 μg), streptomycin (25 μg), colistin (25 μg) and ampicillin (25 μg) on discs. All plates were incubated at 37°C for 24-36 h. The diameters of zones of inhibition were measured to the nearest millimeter using a ruler. Approved CLSI[10] susceptibility zone diameter interpretative standards were used.
Extended-spectrum β-lactamase detection
ESBL resistance in Gram-negative bacilli was screened for using cefotaxime (30 μg) and aztreonam (30 μg) discs alone and confirmed by the CLSI method using cefotaxime and clavulanic acid combination disc on Mueller Hinton agar. A greater than or equal to a 5 mm increase in the zone diameter for cefotaxime tested in combination with clavulanate versus its zone diameter when tested alone confirmed an ESBL-producing organism.[10]
In vitro conjugation experiment
Minimum inhibitory concentrations of the antibiotics (cefotaxime, tetracycline, nitrofurantoin, cotrimoxazole, and gentamicin) used in the conjugation experiments were determined by agar dilution method as prescribed by CLSI.[10] Conjugation experiment was performed by the mating assay using the method of Sambrook et al.,[11] in tryptic soy broth for six of the ESBL isolates as donors (all confirmed sensitive to nalidixic acid) and E. coli ATCC 25922 as the recipient which was confirmed as resistant to nalidixic acid and sensitive to all other antibiotics except ampicillin. A suspension of each organism was made in the sterile tryptic soy broth and adjusted to 0.5 McFarland Standard. The donor and the recipient were then mixed in a ratio 1:9 (50 μl of the donor to 450 μl of the recipient) and incubated on a MacConkey agar plate containing 128 μg/ml of nalidixic acid (selective for the recipient) at 37°C for 2 h. Transfer of resistance was read by observation of recovery of the recipient colonies on MacConkey agar containing the corresponding antibiotics at different concentrations (cefotaxime 8 μg/ ml, tetracycline 12.5 μg/ ml, cotrimoxazole 1.25/23.75 μg/ml, nitrofurantoin 100 μg/ml and gentamicin 5 μg/ml).
DNA extraction
DNA extraction was performed using the Wizard Genomic Extraction Kit (Promega, Madison, USA) according to the manufacturer's specifications. The extracted plasmid DNA preparations were separated on 0.8% agarose gel (Oxoid, Basingstoke, England) in a 1X TBE buffer and a 100 bp ladder (Promega, Madison, USA) was used as standard.
Quality control
All prepared media were checked for sterility for 24 h. E. coli ATCC 25922 was used as a quality control strain for antibacterial susceptibility testing. The E. coli strain was also used as a negative control in the screening and phenotypic confirmatory tests of ESBL-producing Gram-negative rods.
Data analysis
Data are presented as frequencies and/or percentages. Statistical analysis was performed using the chi-square (χ2) Fischer's exact test in PAST software package by Hammer et al.(2001).[12]
RESULTS
The 45 orthopedic patients consisted of 31 (68.9%) males and 14 (31.1%) females approximately in the ratio 2:1 and which reflects the risk of wounds resulting from automobile accidents. The most commonly administered antibiotics to the patients were the second and 3GC (cefuroxime and ceftriaxone), followed by gentamicin which was used in combination with the 3GC antibiotics.
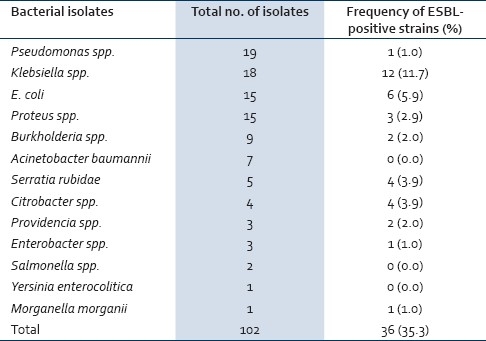
Out of the 73 samples that were collected, 22 yielded multiple isolates (n=52; swab 22, biopsy 30) while 50 yielded single isolates (n=50; Swab 28, Biopsy 22); 1 sample yielded no isolate. Out of the 102 Gram-negative bacteria isolates recovered (n=36, 35.3%) were positive for ESBL made up mainly of the members of the Enterobacteriaceae family (n=33, 32.4%), while the rest constituted the oxidase positive bacilli (n=3, 2.9%) [Table 1]. The predominant bacteria among the ESBL producers were the Klebsiella spp. (12) and E. coli (6), Serratia rubidae, and Citrobacter freundii (4 each). Others were Proteus spp. (3), Providencia stuartii (2), Enterobacter spp., Morganella morganii, and Pseudomonas spp. (1 each). Isolates of Acinetobacter baumannii, Salmonella spp., and Yersinia enterocolitica were negative for ESBL production.
Table 1.
Frequencies of ESBL-producing strains among Gram-negative bacteria isolated from orthopedic wounds
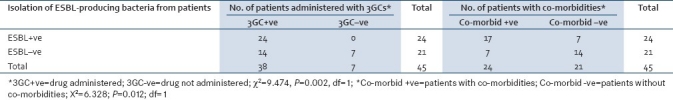
A significant association was seen to exist between treatment with 3GC and the isolation of ESBL from patients (P= 0.0020); the presence of co-morbidities (malaria, diabetes mellitus, hypertension, hospital acquired pneumonia, hemorrhage, septicemia, renal failure and epilepsy) was also significantly related to the acquisition of ESBL-producing bacterial isolates (P= 0.0120) [Table 2]. The minimum and maximum lengths of stay in the hospital were 7 days and 365 days respectively, while the mean was 82 days. Thirty-nine (86.7%) of the 45 patients were eventually discharged without full healing to be followed up at the outpatient clinic. The final outcome of their clinical status was not investigated.
Table 2.
Isolation of ESBL producers in relation to administration of 3GC to the patients and the presence of co-morbidities in the patients
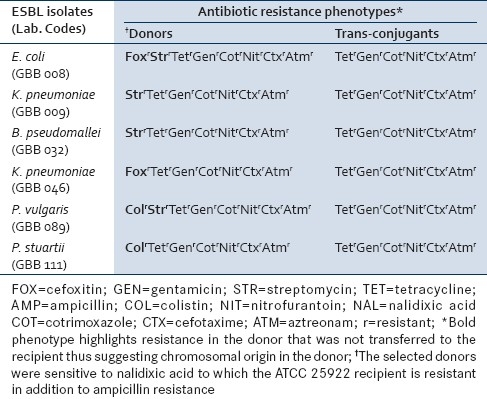
More than 50.0% of the ESBL isolates were resistant to all the tested antibiotics except colistin (36.0%). All were resistant to ampicillin [Table 3]. The mating assay showed that six (6) of the multiple resistance phenotypes observed for the ESBL isolates were transferred within 2 h at 37°C and confirmed in the trans-conjugants except for streptomycin, colistin, and cefoxitin in different combinations and which are suspected to be of chromosomal origin [Table 4]. The frequency of transfer of the cefotaxime-resistance phenotype ranged from 2.70 to 3.60×10–6 of transconjugants per recipient cells. The resistance phenotype was picked up by all the colonies of the recipient within 2 h.
Table 3.
Frequencies of antibiotic resistance phenotypes among the ESBL-producing bacterial isolates
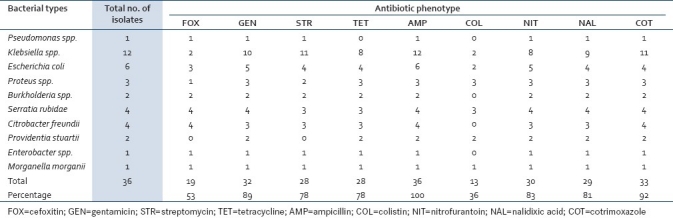
Table 4.
Comparison of the antibiotic resistance phenotypes of the ESBL donors and the corresponding trans-conjugants
The gel electrophoretic mobilities of plasmid DNA of the ESBL isolates and the corresponding trans-conjugants confirmed three plasmids of approximately 8.872 kb, 9.772 kb, and 10.765 kb sizes among the isolates [Figure 1].
Figure 1.
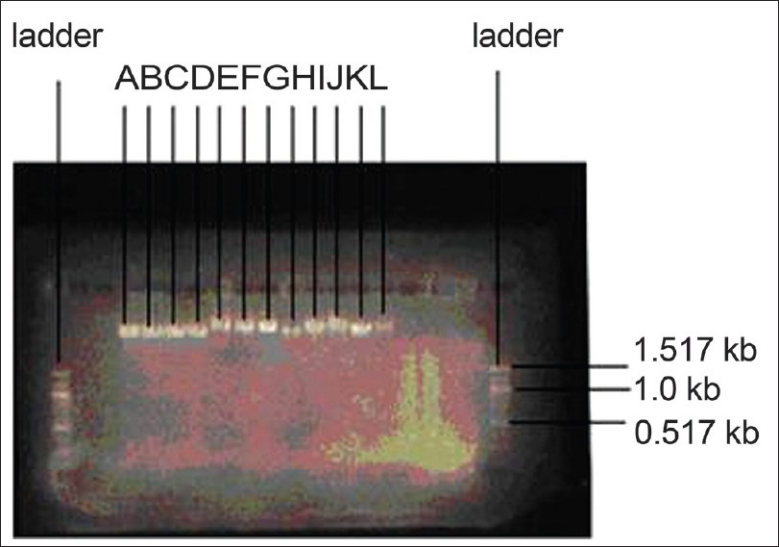
Plasmid DNA sizes of the donors and corresponding trans-conjugants. A=P. stuartii (8.872 kb), B=trans-conjugant of A (8.872 kb), C=P. vulgaris (8.872 kb), D=trans-conjugant of C (8.872 kb), E=K. pneumoniae (10.765 kb), F=trans-conjugant of E (10.765 kb), G=B. pseudomallei (10.765 kb), H=trans-conjugant of G (10.765 kb), I=K. pneumoniae (9.772 kb), J=trans-conjugant of I (9.772 kb), K=E. coli (9.772 kb), L=trans-conjugant of K (9.772 kb)
DISCUSSION
This study has demonstrated the existence of ESBL-producing Gram-negative bacteria here in Ile-Ife, Nigeria. The incidence rate in wound samples was about 35%, higher than 18% reported in India[13] but much lower than 77% in Saudi Arabia.[14] Klebsiella spp. and E. coli were consistently the most encountered in all the studies.
Infection of wounds by microorganisms is most often associated with prolonged hospital stay with the attendant risk of acquisition of multiple resistant organisms from medical devices[4] and hospital environment. Our study has demonstrated a significant relationship between co-morbidities and acquisition of ESBL-producing Gram-negative bacteria. The long stay in the hospital calls for concern as the patients from whom most of the ESBL isolates were recovered spent longer periods in the hospital in spite of persistent treatment with antibiotics in different combinations. The treatment failure was at high costs to the patients most of whom were eventually discharged for the lack of ability to meet up with hospital bills.
The observed association between therapeutic administration of 3GCs and isolation of ESBL producers from patients has also been reported in other studies[15,16] as well as fluoroquinolone and aminoglycosides.[17] However, about 65% of the ESBL isolates were sensitive to colistin. Bishara et al.,[18] and Urbanek et al.,[19] have reported that 100% of their ESBL isolates were sensitive to colistin. Colistin was not administered to any of the patients in this study which again raises the question of the need for continual routine screening for antibiotic sensitivity of bacteria during therapy and which could have reduced the level of treatment failure observed.
Studies have shown that resistance to 3GCs was accompanied by resistance to other groups of antibiotics[20,21] and were borne on plasmids. ESBL resistance genes in our isolates were consistently found co-migrating with other five antibiotics, constituting an additional challenge for effective treatment of other infections in which these bacteria might be involved.
CONCLUSION
The detection of multiple antibiotic resistances among the ESBL-producing Gram-negative bacteria in this study is a challenge that has serious consequences on infection control and hospital management of patients, and calls for proactive mitigation strategies. There has been treatment failure with the attendant high cost and discomfort to the patients. These problems have to be addressed. For enhanced therapeutic management of bacterial infections, there is need to encourage routine continual monitoring and evaluation of antibiotic treatment regimen and choice of drugs. It would appear that an epidemiological clone of an ESBL gene co-existing with resistant genes to other five antibiotics on the same plasmid DNA exists and is circulating here in Ile-Ife, Nigeria. We intend to investigate this further.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Sule AM, Thanni LO, Sule OO, Olusanya O. Bacterial pathogens associated with infected wounds in Ogun State University Teaching Hospital, Sagamu, Nigeria. Afr J Clin Exp Microbiol. 2002;3:13–6. [Google Scholar]
- 2.Agrawal AC, Jain S, Jain RK, Raza HK. Pathogenic bacteria in an orthopaedic hospital in India. J Infect Dev Ctries. 2008;2:120–3. [PubMed] [Google Scholar]
- 3.Yishak A, Biruk LW. Microbial susceptibility of bacteria isolated from open fracture wounds presenting to the Err of Black-Lion Hospital, Addis Ababa University, Ethiopia. Afr J Microbiol Res. 2009;3:939–51. [Google Scholar]
- 4.Paterson DL, Bonomo RA. Extended-Spectrum β-Lactamases: A clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathisuwan S, Burgess DS, Lewis JS., 2nd Extended-spectrum beta-lactamases: Epidemiology, detection and treatment. Pharmacotherapy. 2001;21:920–8. doi: 10.1592/phco.21.11.920.34529. [DOI] [PubMed] [Google Scholar]
- 6.Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, Bush K, et al. Multiple antibiotic resistant Klebsiella pneumoniae and Escherichia in nursing homes. JAMA. 1999;281:517–23. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 7.Jones RN. Resistance patterns among nosocomial pathogens: Trends over the past few years. Chest. 2001;119(2 Suppl):397S–404S. doi: 10.1378/chest.119.2_suppl.397s. [DOI] [PubMed] [Google Scholar]
- 8.Graffunder EM, Preston KE, Evans AM, Venezia RA. Risk factors associated with extended-spectrum beta-lactamase-producing organisms at a tertiary care hospital. J Antimicrob Chemother. 2005;56:139–45. doi: 10.1093/jac/dki180. [DOI] [PubMed] [Google Scholar]
- 9.Cheesbrough M. District laboratory practice in tropical countries, part 2. United Kingdom: Cambridge University Press; 2002. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. Approved standard M2-A9 and M7-A7.Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. Clin Lab Stand Inst. 2008;28:M100–S18. [Google Scholar]
- 11.Sambrook JE, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 12.Hammer Ø, Harper DA, Ryan PD. PAST: Palaeontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica. 2001;4:9. [Google Scholar]
- 13.Tsering DC, Das S, Adhiakar L, Pal R, Singh TS. Extended spectrum beta-lactamase detection in Gram-negative Bacilli of Nosocomial origin. J Glob Infect Dis. 2009;1:87–92. doi: 10.4103/0974-777X.56247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kader AA, Kumar AK. Prevalence of extended-spectrum β-lactamase among multidrug resistant Gram-negative isolates from a general hospital in Saudi Arabia. Saudi Med J. 2004;25:570–4. [PubMed] [Google Scholar]
- 15.Asensio A, Oliver A, Gonzœlez-Diego P, Baquero F, Pirez-Dvaz JC, Ros P, et al. Outbreak of a multi resistant Klebsiella pneumoniae strain in an intensive care unit: Antibiotic use as a risk factor for colonization and infection. Clin Infect Dis. 2000;30:55–60. doi: 10.1086/313590. [DOI] [PubMed] [Google Scholar]
- 16.Sarma JB, Ahmed GU. Prevalence and risk factors for colonisation with extended spectrum β-lactamase producing enterobacteriacae vis-à-vis usage of antimicrobials. Indian J Med Microbiol. 2010;28:217–20. doi: 10.4103/0255-0857.66476. [DOI] [PubMed] [Google Scholar]
- 17.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: Risk factors for infection and impact of resistance on outcomes. Clin Infect Dis. 2001;32:1162–71. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- 18.Bishara J, Livne G, Ashkenazi S, Levy I, Pitlik S, Ofir O, et al. Antibacterial susceptibility of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli. Isr Med Assoc J. 2005;7:298–301. [PubMed] [Google Scholar]
- 19.Urbanek K, Kolar M, Loveckova Y, Strojil J, Santava L. Influence of third-generation cephalosporin utilization on the occurrence of ESBL-positive Klebsiella pneumoniae strains. J Clin Pharm Ther. 2007;32:403–8. doi: 10.1111/j.1365-2710.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 20.Bella JM, Turnidge JD, Gales AC, Pfaller MA, Jones RN. Sentry APAC Study Group. Prevalence of extended-spectrum beta-lactamase (ESBL)-producing clinical isolates in the Asia-Pacific region and South Africa. Regional results from SENTRY antimicrobial surveillance program (1998-99) Diagn Microbiol Infect Dis. 2002;42:193–8. doi: 10.1016/s0732-8893(01)00353-4. [DOI] [PubMed] [Google Scholar]
- 21.Shukla I, Tiwari R, Agrawal M. Prevalence of extended spectrum β-lactamase-producing Klebsiella pneumoniae in a tertiary care hospital. Indian J Med Microbiol. 2004;22:87–91. [PubMed] [Google Scholar]


