Abstract
The Arabidopsis genome was searched to identify predicted proteins containing armadillo (ARM) repeats, a motif known to mediate protein-protein interactions in a number of different animal proteins. Using domain database predictions and models generated in this study, 108 Arabidopsis proteins were identified that contained a minimum of two ARM repeats with the majority of proteins containing four to eight ARM repeats. Clustering analysis showed that the 108 predicted Arabidopsis ARM repeat proteins could be divided into multiple groups with wide differences in their domain compositions and organizations. Interestingly, 41 of the 108 Arabidopsis ARM repeat proteins contained a U-box, a motif present in a family of E3 ligases, and these proteins represented the largest class of Arabidopsis ARM repeat proteins. In 14 of these U-box/ARM repeat proteins, there was also a novel conserved domain identified in the N-terminal region. Based on the phylogenetic tree, representative U-box/ARM repeat proteins were selected for further study. RNA-blot analyses revealed that these U-box/ARM proteins are expressed in a variety of tissues in Arabidopsis. In addition, the selected U-box/ARM proteins were found to be functional E3 ubiquitin ligases. Thus, these U-box/ARM proteins represent a new family of E3 ligases in Arabidopsis.
ARM repeats are short 42-amino acid motifs that were first identified in the fruitfly (Drosophila melanogaster) segment polarity protein, armadillo (Riggleman et al., 1989). ARM repeats have been subsequently identified in a wide range of eukaryotic proteins, and these proteins interact with numerous other proteins via their ARM repeats, resulting in the regulation of a variety of cellular processes (for review, see Hatzfeld, 1999). Based on the crystal structure of the mammalian armadillo homolog, β-catenin, each ARM repeat forms a trihelical structure that folds into a superhelix, and six ARM repeats are proposed to constitute a protein interaction domain (Huber et al., 1997). In animals, well-characterized ARM repeat proteins include β-catenin/armadillo involved in the Wnt/wingless signaling pathway and cadherin-mediated cell adhesion, the APC tumor suppressor protein in Wnt signaling, and several other cadherin-associated ARM repeat proteins (Hatzfeld, 1999). In addition, there is the conserved nuclear import pathway in yeast (Saccharomyces cerevisiae), plants, and animals that involves the ARM repeat protein, Importin-α, and the Importin-β protein with related HEAT repeats (Andrade et al., 2001).
More recently, a new class of ARM repeat proteins was identified in plants where the ARM repeat region is preceded by a E3 ubiquitin ligase motif called the U-box (Amador et al., 2001; Azevedo et al., 2001; Stone et al., 2003). The ubiquitination of proteins involves three enzymes: the E1 ubiquitin-activating enzyme, which forms a thioester intermediate with ubiquitin; the E2 ubiquitin-conjugating enzyme, which receives the ubiquitin molecule from the E1 enzyme; and the E3 ubiquitin ligase, which facilitates the attachment of the ubiquitin molecule from the E2 enzyme to the target protein. The E3 ligases are by far the most diverse components and are responsible for the substrate specificity in this pathway (Pickart, 2001). The attachment of ubiquitin molecules to a protein can result in proteasomal degradation or, alternately, regulate other processes such as transcription and DNA repair (Glickman and Ciechanover, 2002).
The U-box has a similar structure to the RING finger and was originally identified in the yeast UFD2 protein (Koegl et al., 1999; Aravind and Koonin, 2000; Ohi et al., 2003). UFD2 homologs are also present in Caenorhabditis elegans, Dictyostelium discoideum, fruitfly, mammals, and plants (Azevedo et al., 2001). A number of other U-box proteins are also present in these organisms, with some of these proteins now shown to function as E3 ubiquitin ligases (Hatakeyama et al., 2001; Murata et al., 2001). In plants, the Brassica ARC1 ARM repeat protein is one of the few U-box-containing proteins for which a role has been identified. ARC1 is an E3 ubiquitin ligase, which acts downstream of the S receptor kinase to promote ubiquitination and protein degradation during the rejection of self-incompatible Brassica sp. pollen (Stone et al., 2003). Other plant U-box proteins include the potato (Solanum tuberosum) PHOR1 protein, also with ARM repeats, which appears to have a role in GA signaling (Amador et al., 2001), and the parsley (Petroselinum crispum) CMPG1 protein, for which mRNA levels are rapidly up regulated in plant pathogen responses (Kirsch et al., 2001).
In this study, 108 predicted ARM repeat proteins were identified in the Arabidopsis genome (including a small number of related HEAT repeat proteins), and the U-box/ARM repeat proteins were found to make up the largest group. Previously, several Arabidopsis U-box proteins were named AtPUB (plant U-box) proteins and assigned to five different classes (Azevedo et al., 2001). In this study, both the class II (ARM repeat domain) and class III (Leu-rich domain) AtPUB proteins were found to contain ARM repeats, and an additional 11 Arabidopsis U-box/ARM repeat members were identified. Because there are other predicted Arabidopsis U-box proteins that do not contain the ARM repeat domain (Azevedo et al., 2001), we refer to this group as the AtPUB-ARM family. A subgroup of the AtPUB-ARM genes was further analyzed, and they were found to have varying expression patterns in Arabidopsis and possess E3 ubiquitin ligase activity.
RESULTS
Whole-Genome Survey of Arabidopsis ARM-Containing Sequences
To assess the diversity and abundance of the ARM repeat family in plants, we conducted a genome-wide survey on the Arabidopsis genome using two complementary approaches. The first approach identified Arabidopsis proteins that shared overall sequence similarity to all ARM repeat proteins found in the Inter Pro database. The second approach used hidden Markov models (HMMs) from the Pfam database and from alignments generated in this study. To reduce the impact of incorrectly predicted gene models, we also identified expressed sequence tags (ESTs) from GenBank and full-length cDNA sequences from SIGnAL database for the predicted ARM repeat genes. Through a comparison of the predicted coding sequences and the EST/cDNA sequences, we found that seven genes contain erroneous start/stop or missing exons. These sequences were corrected for further analysis.
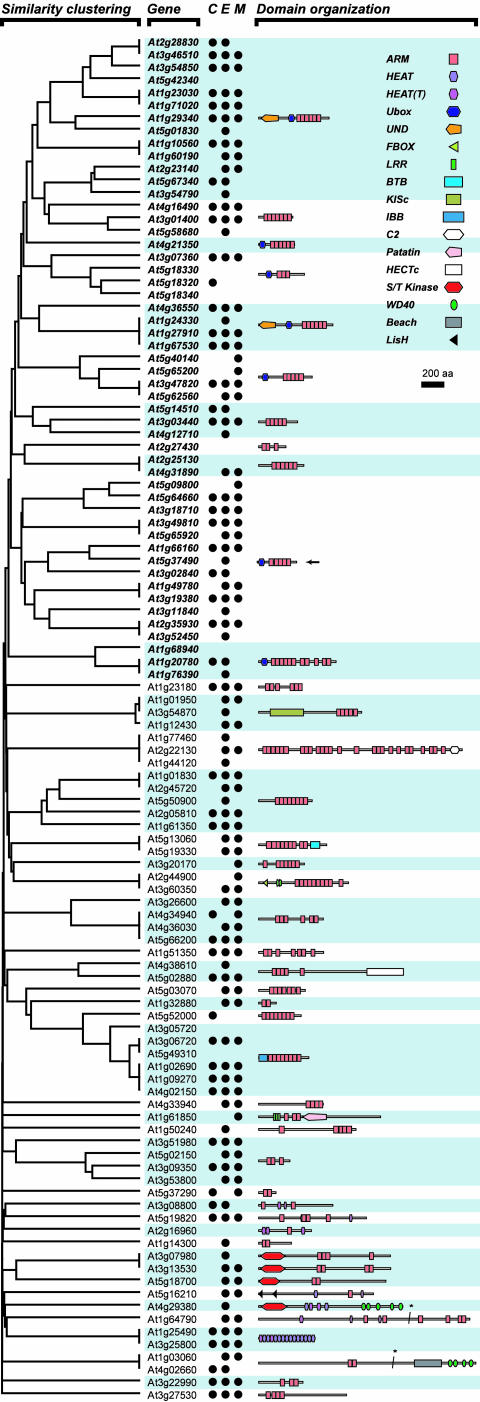
Using the combined approaches, 108 predicted Arabidopsis ARM proteins were identified, including those that would have been missed without the combination of all models (Fig. 1 and Supplemental Fig. S-1 and Table S-I. Supplemental data can be found in the online version of this article at http://www.plantphysiol.org). Despite this, the domain organizations suggest that there are likely other ARM repeats that have not been detected (Fig. 1 and Supplemental S-1). This is evident in the arrangement of ARM repeats for closely related proteins where a featureless region is present between ARM repeats in one protein but not in the other relative and the fact that ARM repeats tend to be tandemly repeated. In some cases, HEAT repeats were found to overlap with ARM repeats but were included in this analysis because ARM and HEAT belong to the same super-family of repeats (Andrade et al., 2001). Taken together, these findings indicate that this repeat family is highly divergent, not only between proteins but also between repeats within the same protein.
Figure 1.
Similarity clustering, evidence of gene expression, and overall domain organizations of Arabidopsis ARM repeat proteins. The cluster diagram on the left shows the extent of similarity between ARM repeat proteins. Higher similarity is demonstrated by shorter branch length between any two sequences. The three columns of circles indicate the presence of expression tags: C, cDNA from SIGnAL database; E, ESTs from GenBank; M, massive parallel signature sequencing (MPSS) tags from the Arabidopsis MPSS database (see “Materials and Methods”). The sequence names and their correspondence to GenBank accessions can be found in supplemental Table S-I, where the accessions for ESTs and cDNAs are also tabulated. ARM proteins with similar domain organizations are grouped together with alternate shaded boxes. The representative domain organization of each group is shown on the right. The domain names follow those in Pfam and/or SMART except the U-box N-terminal domain (UND) defined in this study. Italics, Gene names for AtPUBARM family members; arrow, divergent AtPUB-ARM members; asterisk, region where the sequence was truncated to fit the width of the graphics (no known protein domains were present in this region).
Relationships, Domain Contents, and Expression of ARM Repeat Proteins
To determine the relationships between ARM repeat proteins, we generated similarity clusters and analyzed domain contents and organizations. The Arabidopsis ARM repeat proteins can be divided into multiple groups, indicating that these proteins differ widely (Fig. 1). There are multiple other protein domains present, and as expected, proteins containing similar domain contents tend to cluster together due to overall sequence similarities. The largest class of proteins in this gene family was the U-box-containing AtPUB-ARM proteins representing 41 of the 108 ARM repeat proteins. These AtPUB-ARM proteins included 30 members previously assigned as Class II AtPUB proteins with ARM repeats and Class III AtPUB proteins with a Leu-rich region (Azevedo et al., 2001). In this study, the Leu-rich regions were found to contain ARM repeats (Fig. 1, marked with arrow) and, interestingly, had a more divergent ARM repeat detected by the ARM_HMM2 model (Alignment S-2; Supplemental Fig. S-1). Several AtPUBARM proteins were also found to have a UND region, described in more detail below. Other known proteins identified in this study were the Importin-α proteins containing the Importin-β-binding domain (IBB) followed by eight ARM repeats and the Importin-β proteins with 15 HEAT repeats (Fig. 1; Andrade et al., 2001; Merkle, 2001).
The remaining predicted ARM repeat proteins were novel and contained a wide assortment of motifs associated with the ARM repeats. These included a number of protein-protein interaction domains such as the Leu-rich repeat, BTB domain, and WD40 domain. The U-box, F-box, and HECTc domains are all implicated in ubiquitination as single or multisubunit E3 ligases (Huibregtse et al., 1995; Gagne et al., 2002; Hatakeyama and Nakayama, 2003). There were also domains with potential microtubule-associated functions such as the kinesin motor domain and the LisH domain (Block, 1998; Reiner, 2000). Finally, there were ARM repeat proteins with a predicted C2 domain that may mediate calcium-dependent phospholipid binding, a lipid acyl hydrolase (patatin) domain, and a Ser/Thr protein kinase domain, respectively (Fig. 1 and Supplemental Fig. S-1).
Expression of these predicted ARM repeat genes was investigated by searching the cDNA, EST, and MPSS databases (Fig. 1; Supplemental Table S-II). Evidence for expression was found for the majority of these ARM repeat genes. Only 10 of the predicted members did not have corresponding cDNA, EST or MPSS tags in the databases; however, seven of these predicted genes had a closely related member being expressed (Fig. 1; Supplemental Table S-II). The ARM repeat genes were generally expressed in a variety of tissues, although for several members, EST/MPSS tags were only detected in the flower samples (Table S-II). Thus, given the wide range of motifs associated with ARM repeats and their expression in various tissues, these predicted Arabidopsis proteins are likely involved in the regulation of diverse developmental processes with the ARM repeats serving as protein-protein interaction domains.
Relationships between the AtPUB-ARM Proteins
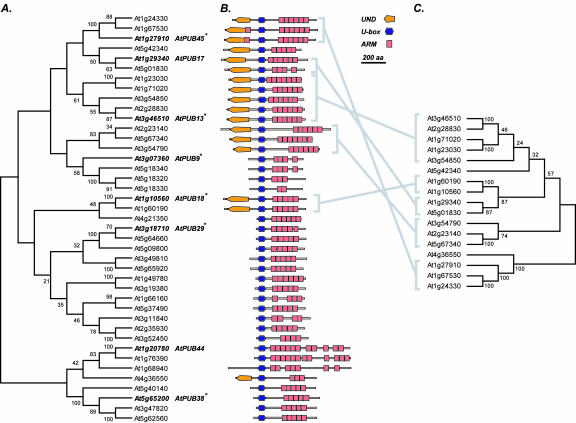
With the large number of predicted Arabidopsis AtPUB-ARM proteins, we were interested in further studying their relationships in this family. Because the AtPUB-ARM proteins contain varying numbers of ARM repeats, and these repeats are highly divergent, the phylogeny was generated using the U-box sequences (Fig. 2A). The detailed domain organizations are shown in Figure 2B, and the phylogeny of the UNDs is shown in Figure 2C. Surprisingly, instead of forming a monophyletic group, the AtPUBARM proteins that have UNDs fall into three clusters, suggesting the independent gain of the UND multiple times in the evolution of this gene family. On the other hand, clusters of AtPUB-ARM proteins with high bootstrap supports in the U-box phylogeny correspond well to those in the UND phylogeny (Fig. 2, gray lines). This finding indicates, at least in these corresponding clusters, that the domains have common origins and are most likely derived from gene duplications. Although the UND was identified based on its conservation in a subset of the AtPUBARM proteins, this region may turn out to contain subdomains with specific functions. For example, the Brassica sp. ARC1 protein has the same configuration as the Arabidopsis UND/U-box/ARM repeat proteins, and the ARC1 UND appears to have putative Leu zipper and coiled-coil motifs and a functional nuclear localization signal (Stone et al., 2003).
Figure 2.
Phylogenies of U-box and UND protein sequences. The phylogenies were generated with neighboring joining with 400 bootstrap replicates and were rooted at midpoint. The bootstrap values are shown as percentages. A, Phylogeny was generated using the U-box protein sequences from the AtPUB-ARM proteins. The database gene names for AtPUB-ARM genes that were tested for RNA expression are italicized, and their synonyms are shown on the right. Asterisks, AtPUB-ARM proteins that were further tested for E3 ligase activity. B, Domain organizations of the AtPUB-ARM proteins, ordered according to A. The color-coding schemes are the same as those in Figure 1. C, Phylogeny of UND sequences. The major groups within the UND phylogeny are linked by gray lines to those in the U-box phylogeny.
RNA Expression Analysis of the AtPUB-ARM Genes
Because all of the predicted genes in the Arabidopsis AtPUB-ARM family are novel, it was of interest to further study their expression patterns and to determine if the mRNAs for these genes corresponded to the predicted sizes, given the range of domain organizations detected. Of the 41 AtPUB-ARM genes, cDNA, EST, and/or MPSS tags were detected for 36 members (Fig. 1; Supplemental Table S-II). Several of the AtPUB-ARM genes had corresponding ESTs and MPSS tags from several different tissues, suggesting that they were broadly expressed. There were also some AtPUB-ARM genes that may have a more tissue-specific pattern of expression based on the distribution of ESTs and MPSS tags (Table S-II).
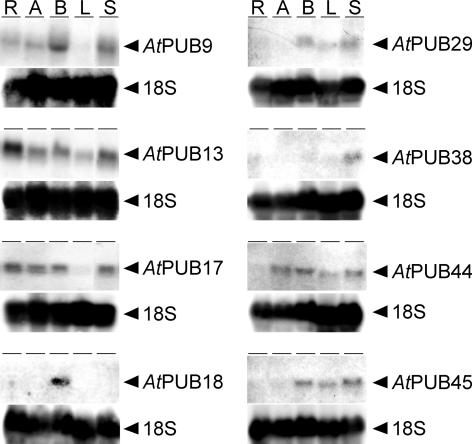
A subset of AtPUB-ARM genes from the three different clades in the phylogeny (Fig. 2A) were subjected to RNA-blot analyses (Fig. 3). AtPUB9, 29, 38, and 44 contain only the U-box/ARM domains, and AtPUB13, 17, 18, and 45 contain the UND/U-box/ARM configuration (Figs. 2B and 3). Transcripts of roughly the expected size were detected for all eight AtPUB-ARM genes supporting the gene prediction models in the databases. There were also similarities in expression patterns across members with or without UND. For example, AtPUB29 and AtPUB45 were only expressed in mature tissues (flower buds, leaves, and stems), whereas AtPUB9 and AtPUB17 were expressed in all tissues tested except for leaves. AtPUB44 is an interesting AtPUB-ARM protein because it contains a much larger number of ARM repeats but no UND, and it was found to be expressed in all tissues tested except for roots. AtPUB13 was the only AtPUB-ARM gene examined that was expressed in all tissues tested. Finally, specific expression patterns were observed for AtPUB18 in flower buds and AtPUB38 in stem tissue. Thus, the eight AtPUB-ARM genes examined showed several different patterns of expression.
Figure 3.
RNA blot analysis of selected AtPUB-ARM genes. Approximately 20 μg of total RNA isolated from Arabidopsis seedling root (R) and aerial tissues (A) and from mature Arabidopsis flower buds (B), leaves (L), and stems (S) was hybridized to labeled AtPUB-ARM cDNA probes. Blots were then hybridized to a labeled 18S rRNA probe as a control for even loading.
E3 Ubiquitin Ligase Activities of the AtPUB-ARM Proteins
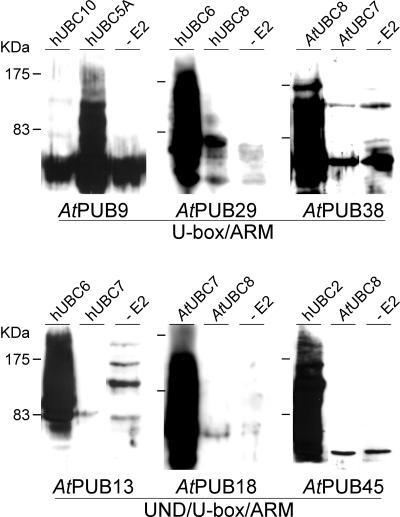
To determine if the AtPUB-ARM proteins do encode functional E3 ligases, as predicted by the U-box domain, in vitro ubiquitination assays were performed. Six AtPUB-ARM family members were tested: AtPUB9, 29, and 38 with U-box/ARM domains, and AtPUB13, 18, and 45 with UND/U-box/ARM domains (Fig. 4). These proteins were expressed with His-tags in Escherichia coli and tested for E3 ligase activity in an assay containing ubiquitin, the yeast E1 ubiquitin-activating enzyme, and different Arabidopsis (AtUBC1, 7, and 8) or human (hUBC2A, 3, 5A, 5B, 6, 7, and 10) E2 ubiquitin-conjugating enzymes. The bacterial proteins present in the E2 or E3 enzymes serve as potential substrates for ubiquitination in this assay (Lorick et al., 1999).
Figure 4.
E3 ubiquitin ligase activity of recombinant AtPUB-ARM proteins. Affinity-purified recombinant AtPUB-ARM proteins were tested for E3 ligase activity in an in vitro ubiquitination assay. The yeast E1 enzyme, E2 enzymes from Arabidopsis (AtUBC) or human (hUBC) and ubiquitin were used in the reactions. The E2 enzyme used in each reaction is indicated above each lane, and the AtPUBARM protein tested is listed below each panel. As a control, the E2 enzyme was omitted from the reaction, and as expected, no ubiquitinated proteins were observed.
All six AtPUB-ARM proteins were found to possess E3 ligase activity, and as previously seen with mammalian U-box proteins (Hatakeyama et al., 2001), the AtPUB-ARM proteins also exhibited a preference for a subset of E2 enzymes (Fig. 4). The preference for a particular E2 enzyme did not show any correlation with phylogenetic relationships or with the presence/absence of the UND. For example, AtPUB13 and At-PUB29 both showed activity with hUBC6, whereas AtPUB18 and AtPUB38 showed activity with AtUBC7 and AtUBC8, respectively. AtPUB45 showed a preference for hUBC2, whereas the ubiquitination activity of AtPUB9 was greatest with hUBC5A (Fig. 4). If the yeast E1 enzyme or the AtPUB-ARM E3 ligase was omitted, no ubiquitination was observed (data not shown). Therefore, the ubiquitination observed was due to the E2-dependent E3 ligase activity of the AtPUB-ARM proteins.
DISCUSSION
Analysis of the Arabidopsis predicted gene set has led to the identification of 108 predicted ARM repeat containing proteins with a range of two to 32 repeats detected (Fig. 1 and Supplemental Fig. S-1). Based on the three-dimensional structure of β-catenin, six ARM repeats have been predicted to form the basic superhelical structure of the protein interaction domain (Huber et al., 1997). If this is the typical requirement, then the predicted Arabidopsis proteins with fewer ARM repeats may have some undetected repeats or they may have gained new functions based on fewer ARM repeats. This motif is highly divergent, not only between ARM repeat proteins but also between ARM repeats within same protein. One explanation is that repeats such as ARMs do not have strong functional constraints. Sequence divergence is not prohibited as long as the structural requirements are not perturbed (Andrade et al., 2001). However, when comparing related ARM repeat proteins with identical number of ARMs, the repeats vary widely in their sequence similarity (data not shown), suggesting that the structural constraint may not be the only factor contributing to the observed sequence divergence. Another possibility is that the divergence may be a consequence of positive selection that drives the diversification of these repeats. Evidence for such positive selection has been found in multiple repeat families, including the Leu-rich repeats in plant R genes (Meyers et al., 1998). It remains to be determined if members of the ARM repeat families also experience such a selection regime.
The Arabidopsis ARM repeat family can be divided into several different groups based on sequence similarity and the presence of other protein domains. These associated domains provide some functional indications for these otherwise largely uncharacterized proteins and suggest that many of these ARM repeat proteins may serve as adaptors in signaling networks. Searches of the cDNA, EST, and MPSS tag databases indicated that the majority of these ARM repeat genes are expressed, and the expressed ARM repeat genes represented all the different domain organizations except for two predicted genes.
Interestingly, a large number of Arabidopsis ARM repeat proteins are implicated in protein degradation pathways as E3 ubiquitin ligases with 41 U-box proteins, two F-box proteins, and one HECTc domain protein. The ARM repeats in these proteins may involve the binding of substrates targeted for degradation by the 26S proteasome, or, alternatively, the ARM repeats may mediate protein interactions with another regulatory protein. From the characterization of animal U-box proteins, it is becoming clear that U-box proteins are functional E3 ligases. The data presented here indicate that the AtPUB-ARM proteins in Arabidopsis also represent functional E3 ligases. Interestingly, the AtPUB-ARM proteins tested for in vitro ubiquitination activity did demonstrate different preferences for E2 enzymes, and this preference may contribute to some specificity between this large group of proteins. For the mammalian U-box protein, CHIP, the U-box is proposed to recruit the E2 enzyme to promote the ubiquitination and subsequent degradation of target proteins (Wiederkehr et al., 2002). Therefore, the AtPUB-ARMs may interact with the target substrate via their ARM repeats or the UND and then recruit other components of the ubiquitination machinery through the U-box.
In Brassica sp., the ARC1 protein has been found to be a functional E3 ligase, and ubiquitination and protein degradation are part of the signaling pathway leading to the rejection of self-incompatible pollen (Stone et al., 1999, 2003). In this system, the ARC1 ARM repeats bind to the kinase domain of the S receptor kinase, and this is thought to lead to activation of ARC1 with substrate-binding sites in the UND (Gu et al., 1998; Stone et al., 2003). Therefore, it is possible that the other UND-containing AtPUB-ARM proteins may function in an analogous manner. These proteins may be activated by the related Arabidopsis S-domain receptor kinases with potential roles in growth regulation and plant-pathogen interactions (Tobias and Nasrallah, 1996; Pastuglia et al., 2002). However, because the majority of the AtPUBARM proteins lack a UND, they may have a different mode of action. Because the in vitro ubiquitination assays indicate that the UND is not required for E3 ligase activity, these proteins may simply promote the ubiquitination of proteins bound to the ARM repeat region. The diversity within the ARM repeat region and the presence/absence of UND support the notion that the AtPUB-ARM family may regulate a large and diverse number of substrates, therefore governing an array of different cellular processes. Future work focusing on the identification of substrates and elucidation of mechanisms will be needed to fully understand this divergent family.
MATERIALS AND METHODS
Retrieval of ARM-Containing Protein Sequences
Two approaches were used to retrieve ARM-containing proteins from the Arabidopsis predicted gene set. First, ARM-containing proteins were retrieved from the Inter Pro database (Apweiler et al., 2001) and used to search against Arabidopsis proteins via BLAST (Altschul et al., 1997). All entries with an E value less than or equal to 1 × 10-10 were regarded as ARM family candidates. These candidate sequences were further analyzed by querying SMART and Pfam databases (Sonnhammer et al., 1998; Schultz et al., 2000). The second approach involved constructing HMMs for identifying ARM repeats. The multiple sequence alignments of ARMs were retrieved from Pfam and further adjusted according the secondary structural requirement (Huber et al., 1997). The refined alignment was used to generate a model with HMMer (Eddy, 1998). The model generated is called ARM_HMM1 (see Alignment S-1). This model was used to screen for additional ARM family members from the Arabidopsis predicted proteins with hmmsearch in the HMMer package with a cutoff E value threshold of 1. Sequences with at least two consecutive ARMs predicted by ARM_HMM1 were regarded as ARM repeat proteins. In a subset of AtPUB-ARM members, ARM repeats were not contiguous, and the regions enclosed by the few predicted ARMs were manually aligned based on ARM signatures (Huber et al., 1997). The alignments were used to generate the second HMM model: ARM_HMM2 (see Alignment S-2). ARM_HMM2 was also used to screen for addition ARM family members. No new ARM-containing proteins were found with an E value cutoff of 1.
The predicted ARM containing proteins based on SMART, Pfam, ARM-_HMM1, and ARM_HMM2 were combined, and sequences were excluded if: (a) They contained only one ARM predicted by SMART/Pfam, or (b) they had two noncontiguous ARMs predicted by ARM_HMM1 or ARM_HMM2. Based on these rules, a finalized set of ARM family members was generated (see Supplemental Table S-I and Fig. S-1).
Classification of ARM Family Members Based on Similarity Clustering
Due to the variable number of ARMs present and the high degree of sequence divergence, it was not feasible to infer phylogenetic relationships in the finalized protein set based on ARM alignments. Therefore, different subfamilies of ARM-containing proteins were distinguished based on overall sequence similarity by conducting a BLAST search with the 108 protein sequences as the queries and the subjects. The E values were transformed logarithmically; then, absolute values were used to build a distance matrix for clustering with the UPGMA algorithm implemented in MEGA2 (Kumar et al., 2001).
Identification of Cognate ESTs, MPSS Tags, and Full-Length cDNAs and the Correction of Gene Models
The Arabidopsis ESTs available from GenBank as of August 10, 2003, were retrieved. A BLAST search was conducted using the predicted coding sequences of 108 ARM repeat genes against the retrieved EST sequences. All matches with more than 80% identity were inspected. After eliminating gaps longer than 3 from the alignments, cognate ESTs were defined as those that were top matches to the gene in question with at least 97% identity. The accessions for the matching ESTs can be found in Supplemental Table S-I. The source tissues and EST counts were tabulated in Supplemental Table S-II. The MPSS tags matching the ARM repeat genes were retrieved from the Arabidopsis MPSS database (http://dbixs001.dbi.udel.edu/MPSS4/java.html). Only tags matching exons in the crick strand with levels significantly different from 0 were regarded as evidence of expression. The MPSS tag counts were tabulated in Supplemental Table S-II. The cDNA sequences released by the SIGnAL database (http://signal.salk.edu/SSP/index.html) were retrieved from GenBank as of August 10, 2003. The predicted protein sequences of ARM repeat genes were used to search against the cDNA sequences, and the gene models were corrected based on the alignments. The cDNAs for ARM repeat genes are listed in Supplemental Table S-I. The predicted sequences were re-annotated using both ESTs and cDNAs.
Delineation of UNDs
The region that was N terminal to the U-box in several AtPUB-ARM family members was aligned using partial order alignment (Lee et al., 2002), and several stretches of sequence similarity were identified. The UND sequences were aligned with Clustal (Higgins et al., 1996) with additional adjustment of alignments to maximize similarity among sequences (see Alignment S-3). An HMM model was then generated and used to search for additional UND in other Arabidopsis proteins. No additional UND domain proteins were identified.
Cloning and RNA-Blot Analysis of Arabidopsis AtPUB-ARM Genes
The full-length cDNAs for each of the intron-containing AtPUB-ARM genes were isolated from total bud and/or leaf RNA by reverse transcriptase-PCR analysis using gene-specific primers. Intronless AtPUBARM genes were isolated from genomic DNA by PCR with gene-specific primers. The PCR fragments were cloned into pGEM-T (Promega, Madison, WI) or pTOPO2.1 (Invitrogen, Carlsbad, CA), sequenced, and the open reading frames of the correct AtPUB-ARM cDNAs were subcloned into the protein expression vector pET15b.
For RNA extractions, leaves, stems, and buds were collected from flowering Arabidopsis Colombia grown on soil in growth chambers at 22°C and 16 h of light. Root and aerial tissues were collected from 3-week-old Arabidopsis seedlings grown on one-half-strength Murashige and Skoog plates supplemented with Suc (Gibeaut et al., 1997). Total RNA was extracted as previously described (Cock et al., 1997), and 20 μg of each RNA sample was separated on a 1.2% (w/v) formaldehyde gel and transferred to nitrocellulose membranes. Hybridization and washes were carried out as previously described (Silva and Goring, 2002). Membranes were first hybridized to the 32P-labeled AtPUB-ARM cDNA and then hybridized to 32P-labeled 18S cDNA as a control for even loading.
Recombinant Protein Purification and in Vitro Ubiquitination Assays
His-tagged AtPUB-ARM proteins were expressed in Escherichia coli strain BL21 (DE3) pLysS (Novagen, Madison, WI), and purifications were carried out as previously described (Stone et al., 2003) using nickel-nitrilotriacetic acid agarose affinity purification (Qiagen USA, Valencia, CA). In vitro ubiquitination assays were performed with modifications as described by Lorick et al. (1999). The reaction mixtures (25 μL) contained the yeast (Saccharomyces cerevisiae) E1 enzyme (0.4 μg, Boston Biochem., Cambridge, MA); one of several E2 enzymes, Arabidopsis UBC1, 7, or 8 (1 μg of purified or 3 μL of lysate; Stone et al., 2003) or human UBC2A, 3, 5A, 5B, 6, 7, or 10 (0.8 μg, Boston Biochem.); the AtPUB-ARM protein as the E3 ligase (1 μg or 10 μL of lysate); 25 mm Tris-HCl (pH 7.5); 1 mm MgCl2; 120 mm NaCl; 2 mm ATP; 0.3 mm dithiothreitol; 1 μg of ubiquitin; 1 mm creatine phosphate; and 1 unit of phosphocreatine kinase (Sigma-Aldrich, St. Louis). Reactions were incubated at 30°C for 2 h and terminated by adding 4× SDS sample buffer and heating at 95°C for 5 min. The samples were separated on an 8% (w/v) SDS-PAGE gel followed by protein-blot analysis with rabbit anti-ubiquitin antibodies (Sigma-Aldrich).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Supplementary Material
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant to D.R.G. and graduate scholarship to S.L.S.), by an Ontario Premier's Research in Excellence Award (to D.R.G.), and by the National Institutes of Health (National Research Service Award grant no. 1F32GM066554–01 to S.-H.S.).
The online version of this article contains Web-only data.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador V, Monte E, Garcia-Martinez JL, Prat S (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343-354 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134: 117-131 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29: 37-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2000) The U box is a modified RING finger: a common domain in ubiquitination. Curr Biol 10: 132-134 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends Plant Sci 6: 354-358 [DOI] [PubMed] [Google Scholar]
- Block SM (1998) Kinesin: what gives? Cell 93: 5-8 [DOI] [PubMed] [Google Scholar]
- Cock JM, Swarup R, Dumas C (1997) Natural antisense transcripts of the S locus receptor kinase gene and related sequences in Brassica oleracea. Mol Gen Genet 255: 514-524 [DOI] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755-763 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski A, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519-11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373-428 [DOI] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA 95: 382-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama K (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111-33120 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI (2003) U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun 302: 635-645 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M (1999) The armadillo family of structural proteins. Int Rev Cytol 186: 179-224 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383-402 [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90: 871-882 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 2563-2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K (2001) A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J 26: 217-227 [DOI] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635-644 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244-1245 [DOI] [PubMed] [Google Scholar]
- Lee C, Grasso C, Sharlow MF (2002) Multiple sequence alignment using partial order graphs. Bioinformatics 18: 452-464 [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl. Acad Sci USA 96: 11364-11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle T (2001) Nuclear import and export of proteins in plants: a tool for the regulation of signalling. Planta 213: 499-517 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10: 1833-1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K (2001) CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2: 1133-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insights into the U-box, a domain associated with multiubiquitination. Nat Struct Biol 10: 250-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Swarup R, Rocher A, Saindrenan P, Roby D, Dumas C, Cock JM (2002) Comparison of the expression patterns of two small gene families of S gene family receptor kinase genes during the defence response in Brassica oleracea and Arabidopsis thaliana. Gene 282: 215-225 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503-533 [DOI] [PubMed] [Google Scholar]
- Reiner O (2000) LIS1: let's interact sometimes (part 1). Neuron 28: 633-636 [DOI] [PubMed] [Google Scholar]
- Riggleman B, Wieschaus E, Schedl P (1989) Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev 3: 96-113 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28: 231-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NF, Goring DR (2002) The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol Biol 50: 667-685 [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, Eddy SR, Birney E, Bateman A, Durbin R (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26: 320-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants Science 286: 1729-1731 [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CM, Nasrallah JB (1996) An S-locus-related gene in Arabidopsis encodes a functional kinase and produces two classes of transcripts. Plant J 10: 523-531 [DOI] [PubMed] [Google Scholar]
- Wiederkehr T, Bukau B, Buchberger A (2002) Protein turnover: a CHIP programmed for proteolysis. Curr Biol 12: R26-28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.