Abstract
Large-scale identification of genes expressed in roots of the model plant Arabidopsis was performed by serial analysis of gene expression (SAGE), on a total of 144,083 sequenced tags, representing at least 15,964 different mRNAs. For tag to gene assignment, we developed a computational approach based on 26,620 genes annotated from the complete sequence of the genome. The procedure selected warrants the identification of the genes corresponding to the majority of the tags found experimentally, with a high level of reliability, and provides a reference database for SAGE studies in Arabidopsis. This new resource allowed us to characterize the expression of more than 3,000 genes, for which there is no expressed sequence tag (EST) or cDNA in the databases. Moreover, 85% of the tags were specific for one gene. To illustrate this advantage of SAGE for functional genomics, we show that our data allow an unambiguous analysis of most of the individual genes belonging to 12 different ion transporter multigene families. These results indicate that, compared with EST-based tag to gene assignment, the use of the annotated genome sequence greatly improves gene identification in SAGE studies. However, more than 6,000 different tags remained with no gene match, suggesting that a significant proportion of transcripts present in the roots originate from yet unknown or wrongly annotated genes. The root transcriptome characterized in this study markedly differs from those obtained in other organs, and provides a unique resource for investigating the functional specificities of the root system. As an example of the use of SAGE for transcript profiling in Arabidopsis, we report here the identification of 270 genes differentially expressed between roots of plants grown either with NO3- or NH4NO3 as N source.
Serial analysis of gene expression (SAGE) is a sequence-based approach allowing the identification of a large number of transcripts present in tissues and the quantitative comparison of transcriptomes (Velculescu et al., 1995). The principle of SAGE is to generate a short specific tag (14 bp) from each mRNA present in a sample, resulting in the production of a SAGE tags library representative of this sample. The sequencing of these tags allows a high-throughput determination of their frequencies in the library, which are correlated with the relative amounts of the corresponding mRNAs. Thus, thousands of different transcripts can be analyzed, with a high specificity and most importantly, without any a priori on their identity. SAGE has proven to be a very powerful and robust method for investigating gene expression at the whole-genome scale (Velculescu et al., 1997; Boon et al., 2002; Liang, 2002) and to reflect the actual relative contents of mRNAs in a sample (Chrast et al., 2000, and refs. therein; Piquemal et al., 2002; Jung et al., 2003; Matsumura et al., 2003). As compared with cDNA arrays or oligochips, it has several advantages, such as the possibility to perform transcript profiling without the need of large technological investments, and the ability to obtain comprehensive transcriptomes from minute amounts of RNA (Virlon et al., 1999). The SAGE technology has been used extensively with animal systems, and more particularly in cancer research, where several hundred libraries and nearly 7 million SAGE tags have been obtained (Boon et al., 2002). Despite these developments, only very few studies have employed this methodology for transcript profiling in higher plants (Matsumura et al., 1999, 2003; Lorenz and Dean, 2002), and the first reports on SAGE in the model plant species Arabidopsis appeared only very recently (Jung et al., 2003; Lee and Lee, 2003).
At present, a major limitation of SAGE is that in most species, tag to gene assignment (e.g. the identification of the gene the transcript of which has generated the SAGE tag) is based on EST clusters or on available cDNA sequences. This results in very incomplete identification of the transcripts revealed by SAGE tags, leaving many of them without any match in the databases (Lash et al., 2000; Boheler and Stern, 2003; Pleasance et al., 2003). In mouse or man, for instance (the two species in which SAGE has been most widely employed), and despite the availability of a huge number of ESTs, the proportion of unassigned tags in SAGE libraries has been found to be as high as 46% to 76% (Zhang et al., 1997; Chrast et al., 2000; Margulies et al., 2001; Lee et al., 2002). Not surprisingly, SAGE in plants faced the same problem, and gene identification using cDNA or EST databases was not possible for up to 70% to 75% of the SAGE tags obtained from rice (Oryza sativa) plants and Arabidopsis leaves (Matsumura et al., 1999, 2003; Jung et al., 2003). Furthermore, tag to gene assignment based on ESTs only often results in erroneous gene identification, due to the fact that predicted cDNAs are not actual full-length sequences. For instance, tag to gene mismatch resulting from the use of EST databases has been estimated to be as high as 30% in fruitfly (Drososphila melanogaster; Pleasance et al., 2003).
The first aim of our work was to overcome these strong limitations and to improve gene identification in SAGE using the annotated sequence of the Arabidopsis genome. To perform tag to gene assignment, the strategy was to generate in silico a reference database containing the virtual SAGE tags corresponding to the 26,620 Arabidopsis genes annotated in Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de). This database is thus expected to associate all SAGE tags that can be theoretically found in Arabidopsis transcriptomes with their related genes. The difficulty is that correct identification of virtual SAGE tags requires accurate determination of both 5′- and 3′-UTRs, which are not easily predictable from the genome sequence in plants (Zhu et al., 2003), due in particular to the absence of consensus sequences for polyadenylation sites (Aubourg and Rouzé, 2001). Thus, to select the best procedure for large-scale gene identification in Arabidopsis transcript profiling by SAGE, we explored various options concerning both the length of the UTRs taken into account and the method for generating the reference database of virtual tags.
Our second objective was to use the SAGE technology to obtain a much more comprehensive and exhaustive view of the genes expressed in the roots of higher plants. We focused our analysis on these organs because they are strongly under-represented in EST or cDNA libraries available for Arabidopsis (Seki et al., 2002), leading to a poor knowledge of the overall root transcriptome. Therefore, six different SAGE libraries were constructed from roots of hydroponically grown Arabidopsis plants. Given the high morphological and functional specificity of roots, it was expected that root SAGE libraries will provide information on the transcription of many genes predominantly or exclusively expressed in these organs. Two examples are provided to demonstrate the ability of SAGE (a) to discriminate between homologous genes belonging to ion transporters or channels multigene families, and (b) to identify genes differentially expressed as a function of the nature of the N source supplied to the plants.
RESULTS
Summary of Sequenced SAGE Libraries
Six SAGE libraries were generated from roots of Arabidopsis plants (ecotype Col-0) subjected to various mineral nutrient supplies. After elimination of sequences from linkers and vector, duplicate ditags, and ditags shorter than 20 bp, a total of 144,083 tags was obtained from the six combined libraries, representing 52,078 different SAGE tag species. Among these, 15,964 were detected more than once and up to 830 times (the whole set of data is available at http://genoplante-info.infobiogen.fr). Tags found only once were not retained for further analysis, due to the possibility of sequencing errors generating artifactual tags. As usual with SAGE, most of the unique tag species were present at low frequency (Table I). However, 80 different tags were at copy number of 100 or higher, and more than 4,700 tags were found at least five times.
Table I.
Distribution of the experimental tags sequenced from SAGE libraries of Arabidopsis roots
| Total no. of sequenced tags: | 144,083 |
| Total no. of different tags: | 52,078 |
| Tag copy no. | No. of different tags |
| 1 | 36,114 |
| 2 | 6,786 |
| 3 | 2,842 |
| 4—5 | 2,563 |
| 6—10 | 2,059 |
| 11—20 | 964 |
| 21—99 | 670 |
| 100 and > | 80 |
Tag to Gene Assignment using EST Clusters
To identify the genes corresponding to the 15,964 different tags found at least twice, we first used EST clusters from both The Institute for Genomic Research (TIGR; http://www.tigr.org, October 2002 release) and Unigene (http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=3702, November 2002 release) databases. From each EST cluster reported either in TIGR or Unigene, we extracted the virtual tag corresponding to the 10 bp downstream from the MboI recognition site (GATC) closest to the 3′ end. This generated 31,002 and 24,456 virtual tags from TIGR and Unigene EST clusters, respectively (Table II). About 90% of these virtual tags were specific for a unique gene. However, when comparing these virtual tags to those found experimentally, a majority of the 15,964 different experimental tags did not match any EST cluster (56% and 60% for TIGR and Unigene, respectively; Table II). This finding is not unusual, and similar proportions of unmatched experimental tags were found in other species using EST clusters (see the introduction). This represents a strong limitation to the interpretation of SAGE data. Furthermore, many EST clusters do not correspond to full-length cDNAs. As a consequence, there is a risk of generating tag to gene mismatch because the virtual tag extracted from the EST cluster may not correspond to the actual SAGE tag of the gene (e.g. that corresponding to the MboI site closest to the 3′ end of the transcript). For all of these reasons, the procedure of tag to gene assignment using EST clusters was found to be unsatisfactory.
Table II.
Tag to gene assignment using TIGR or Unigene EST clusters Virtual tags were extracted from EST clusters by identifying the 10-bp sequence following the MboI site closest to the 3′ end of each cluster. The databases of virtual tags were merged with that of the 15,964 experimental tags found at least twice to determine the no. of unmatched tags.
| Database | TIGR | Unigene |
|---|---|---|
| Total no. of virtual tags | 31,002 | 24,456 |
| No. of virtual tags matching one single gene | 27,740 (89%) | 23,201 (95%) |
| No. of experimental tags with no match in the Database | 8,926 (56%) | 9,608 (60%) |
Generation of the Virtual Tags Reference Database from the Annotated Genome Sequence
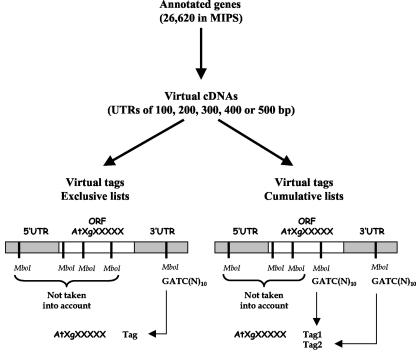
To improve, both quantitatively and qualitatively, gene identification in SAGE libraries, we chose to develop an alternative procedure using the annotated sequence of the Arabidopsis genome (ftp://ftpmips.gsf.de/cress/arabidna/arabi_genomicplus500_v111102.gz). The strategy is summarized in Figure 1. It involves gene identification by matching experimental tags with the virtual ones expected to be generated from all predictable mRNAs. In brief, 10 different virtual tags databases were generated (see “Materials and Methods”), depending on the sizes of the 5′- and 3′-UTRs that were considered for generating virtual cDNAs and on the way virtual tags were extracted from these virtual cDNAs (only the last tag for “exclusive” lists or all tags between the last one in the open reading frame [ORF] and the last one in the 3′-UTR for “cumulative” lists). The total number of virtual tags obtained ranged from 24,432 to 24,999 in the exclusive lists and from 32,052 to 59,725 in the cumulative lists when increasing the UTR length from 100 to 500 bp (Table III). The proportion of virtual tags found in the 3′-UTR strongly depends on the size of this UTR (Table IV), illustrating the fact that the various options investigated led to markedly different databases.
Figure 1.
Description of the strategy used to generate the reference database of virtual tags. For each gene annotated in MIPS (26,620 in total), five different virtual cDNAs were obtained from the genome sequence by adding 100-, 200-, 300-, 400-, or 500-bp-long 5′- and 3′-UTRs to the predicted ORF. Two different virtual tag lists were generated from each of these classes of virtual cDNAs. The first list (Exclusive list) included only the virtual SAGE tag closest to the 3′ end of the virtual cDNA. The second list (Cumulative list) included all virtual SAGE tags located between the last MboI site in the ORF and the 3′ end of the virtual cDNA (two virtual tags in the example shown).
Table III.
Total no. of different tags included in the lists of virtual tags, as a function of the length of the 5′- and 3′-UTR considered for the analysis
The exclusive lists were generated by selecting only the virtual SAGE tag closest to the 3′ end of the virtual cDNA sequence. The cumulative lists were generated by compiling all virtual SAGE tags located between the last MboI site in the ORF (or 5′-UTR in case of absence of SAGE tag in the ORF) and the 3′ end of the virtual cDNA sequence.
| Exclusive lists | |||||
| Length of 5′- and 3′-UTRs | 100 bp | 200 bp | 300 bp | 400 bp | 500 bp |
| No. of virtual tags | 24,432 | 24,678 | 24,800 | 24,975 | 24,999 |
| Cumulative lists | |||||
| Length of 5′- and 3′-UTRs | 100 bp | 200 bp | 300 bp | 400 bp | 500 bp |
| No. of virtual tags | 32,052 | 38,827 | 45,680 | 52,644 | 59,725 |
Table IV.
Location of virtual SAGE tags within virtual cDNAs as a function of the type of virtual SAGE tags lists and length of the 5′- and 3′-UTR considered for generation of virtual cDNAs
5′-UTR/ORF and ORF/3′-UTR refer to tags overlapping these regions of the virtual cDNAs. The total nos. of virtual tags differ from those in Table III because each nonspecific tag corresponding to several genes is not counted only once here, due to the fact that it may not be located in the same regions of the different virtual cDNAs it was extracted from.
| Cumulative lists | |||||
| Length of UTRs | 100 bp | 200 bp | 300 bp | 400 bp | 500 bp |
| Location | |||||
| 5′-UTR | 446 | 600 | 713 | 846 | 876 |
| 5′-UTR/ORF | 63 | 63 | 63 | 63 | 63 |
| ORF | 25,447 | 25,447 | 25,477 | 25,477 | 25,477 |
| ORF/3′-UTR | 1,823 | 1,823 | 1,823 | 1,823 | 1,823 |
| 3′-UTR | 6,964 | 14,527 | 22,353 | 30,527 | 39,066 |
| Total | 34,743 | 42,460 | 50,399 | 58,706 | 67,275 |
| % in 3′ UTR | 20.0 | 34.2 | 44.4 | 52.0 | 58.1 |
| Exclusive lists | |||||
| Length of UTRs | 100 bp | 200 bp | 300 bp | 400 bp | 500 bp |
| Location | |||||
| 5′-UTR | 121 | 209 | 226 | 206 | 176 |
| 5′-UTR/ORF | 44 | 31 | 26 | 19 | 11 |
| ORF | 18,063 | 13,704 | 10,359 | 7,805 | 5,819 |
| ORF/3′-UTR | 1,384 | 1,019 | 760 | 564 | 428 |
| 3′-UTR | 6,432 | 11,316 | 15,033 | 17,896 | 20,106 |
| Total | 26,044 | 26,279 | 26,404 | 26,490 | 26,540 |
| % in 3′-UTR | 24.7 | 43.1 | 56.9 | 67.6 | 75.8 |
Among the 10 different lists of virtual tags generated by the strategy described above, one was selected to yield the reference database for high-throughput tag to gene assignment in our SAGE libraries. Three criteria were used for this selection: lowest level of unmatched tags (experimental tags with no match in the list of virtual tags), lowest level of nonspecific tags (experimental tags matching several genes), and highest level of reliability (correct tag to gene assignment, determined on a subset of genes for which cDNA sequences were available, see below).
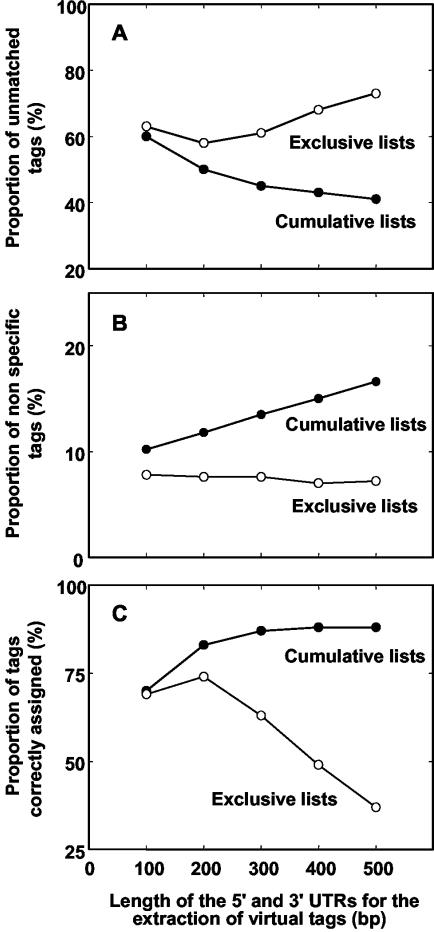
When all experimental tags (15,964 different) were matched against the virtual tag lists, marked differences were observed concerning the proportion of unmatched tags, depending on the virtual tag list (Fig. 2A). In all cases, the cumulative virtual tag lists yielded lower levels of unmatched tags, decreasing from 60% to 41% with increasing UTR length from 100 to 500 bp. With exclusive virtual tag lists, this proportion was always higher than 58%.
Figure 2.
Comparative analysis of the tag to gene assignment results obtained with the different lists of virtual tags as a function of the length of 5′- and 3′-UTRs considered for generating virtual cDNAs. A, Proportion of unmatched tags, calculated for each virtual tag list after merging the 15,964 different experimental tags with the virtual tags included in this list. B, Proportion of nonspecific tags, calculated from the total number of tags matching at least one gene. C, Proportion of tags correctly assigned to a gene, calculated from 5,340 genes with full-length cDNA sequence available, by comparing the actual SAGE tags extracted from the cDNAs sequence with the virtual tags present in the reference database.
Considering the proportion of nonspecific tags (Fig. 2B), the tendency was reversed with better results obtained with the exclusive virtual tag lists. However, this proportion remained lower than 17% with the cumulative virtual tag lists, indicating that the large majority of experimental tags matching a virtual tag in these lists can be unambiguously associated with one single gene.
Finally, the reliability of the tag to gene assignment was determined using a subset of genes for which cDNA sequences are available and experimentally confirmed. To perform this analysis, we selected 5,430 cDNA sequences (ecotype Col-0; http://signal.salk.edu), each corresponding to one gene identifier, according to the Arabidopsis Genome Initiative (AGI;http://www.arabidopsis.org/info/agi.jsp)uniformed nomenclature, and for which at least one MboI site was found. The actual SAGE tags deduced from these 5,430 cDNA sequences were compared with the virtual tags attributed to the corresponding genes in the 10 different virtual tag lists. An experimental tag was considered to be correctly assigned to a gene with our procedure when the actual tag of this gene (deduced from the cDNA sequence) is found in the virtual tag list associated with this gene. The results of this comparison led to drastically different conclusions between exclusive and cumulative virtual tag lists (Fig. 2C). For exclusive lists, the proportion of tags correctly assigned increased from 69% to 74%, with UTR length increasing from 100 to 200 bp, and markedly decreased to 37% when a 500-bp UTR was considered. In contrast, the reliability of gene identification with cumulative tag lists increased with increasing UTR length, to reach a maximum of 88% for 400- or 500-bp UTRs.
The results of this analysis clearly indicate that a compromise has to be found between the lowest level of unmatched tags and the best reliability on the one hand (obtained when gene identification is performed using cumulative virtual tag lists; Fig. 2, A and C) and the lowest level of nonspecific tags on the other hand (obtained with exclusive virtual tag lists; Fig. 2B). We favored the objective of correct tag to gene assignment, and hence selected the cumulative virtual tag list generated with a 400-bp UTR length for building the reference database. The reasons for this precise choice is that considering 3′-UTRs shorter than 400 bp did not allow us to reach the maximum level of correct tag to gene assignment (Fig. 2C), whereas UTRs of 500 bp instead of 400 bp significantly increased the proportion of nonspecific tags (Fig. 2B). Our reference database for SAGE in Arabidopsis is available at http://genoplante-info.infobiogen.fr. It includes virtual tags for 26,490 genes (130 genes among the 26,620 annotated in MIPS had no MboI site in the cDNA sequence predicted with 400-bp UTRs).
Global Analysis of the Arabidopsis Root Transcriptomes
The virtual tag reference database selected above was used to decipher the 15,964 different experimental tags identified from our SAGE libraries. From this total, 9,155 tags were assigned to one or several genes (43% of unmatched tags; Fig. 2A), with 7,776 of them matching one single gene (85% specificity; Fig. 2B). In this case, the gene identification was correct at 88% (Fig. 2C). The full list of these 7,776 genes can be viewed at http://genoplante-info.infobiogen.fr. Among them, 2,970 did not match any EST or cDNA in Unigene (November 2002 release). These genes were previously identified using gene prediction programs only, and our SAGE data thus provide strong experimental evidence for their expression in Arabidopsis. Eighty tags were found at a copy number of 100 or higher, with a majority of them (65) assigned to one or several genes (Table V). Eleven tags match several genes, but in most cases (8/11), this corresponds to two or three closely related genes, with the same function (Table V). The genes with highest levels of expression encode a late embryogenesis abundant protein (At4g02380), a thioglucosidase precursor (At3g09260), and an extensin (At1g21310). Various genes encoding ribosomal proteins and ubiquitin-related proteins were also found to be highly represented.
Table V.
List of the 80 experimental tags found 100 times or more in the sequenced libraries and tag to gene assignment
| Tag | Tag Copy No. | Gene |
|---|---|---|
| GATCGTTGGATGTA | 830 | At4g02380 late embryogenesis abundant protein-related |
| GATCCTTTGTGCCT | 636 | At3g09260 thioglucosidase three-dimensional precursor |
| GATCAATCAATGGT | 610 | At1g21310 extensin 3 (atExt3) |
| GATCAGTGTGTCAC | 565 | At1g73330 Dr4 (protease inhibitor) |
| GATCGCATGGCCTC | 551 | |
| GATCGGTCTACATA | 508 | At3g04720 hevein-like protein precursor |
| GATCGGTTAAGCCA | 454 | At4g11320 Cys proteinase-like protein |
| GATCAAAAAAAAAA | 430 | At1g14860 hypothetical protein |
| At1g51120 DNA-binding protein RAV1, putative | ||
| At2g07370 unknown protein | ||
| At2g32530 putative cellulose synthase | ||
| At3g06590 bHLH protein | ||
| At3g31470 pseudogene, putative reverse transcriptase | ||
| At4g08140 hypothetical protein | ||
| At4g09360 disease resistance protein (NBS-LRR class), putative | ||
| At5g19930 putative protein | ||
| GATCGACTCTCTTA | 406 | At3g45030 40S ribosomal protein |
| At5g60390 translation elongation factor eEF-1 α-chain (gene A4) | ||
| GATCGGAGTAATGA | 399 | |
| GATCCACTGAGATT | 395 | |
| GATCAATCAAGGAG | 386 | At2g25450 putative dioxygenase |
| GATCCTTTGTGCCA | 377 | |
| GATCGTTTAATGTT | 367 | At5g02960 unknown protein |
| GATCAGCGGATGTT | 354 | |
| GATCAAAGTTGTAC | 352 | At4g17340 membrane channel-like protein |
| GATCAACGCAGCCA | 328 | At3g11940 putative 40S ribosomal protein S5 |
| GATCGGCGCGTAAG | 319 | At2g33830 putative auxin-regulated protein |
| GATCAGGAAGCAAT | 307 | At4g21960 peroxidase prxr1 |
| GATCCGAGGAGGTG | 301 | At2g09990 40S ribosomal protein S16 |
| At5g18380 40S ribosomal protein S16 | ||
| GATCGGTGGAGACA | 293 | At4g14320 ribosomal protein |
| GATCTTTTCCCCTA | 289 | At2g30860 glutathione S-transferase |
| GATCCTCCCGGTAA | 286 | At4g25780 putative pathogenesis-related protein |
| At4g33720 pathogenesis-related protein 1 precursor, 19.3K | ||
| GATCGTCCCTTCCA | 281 | |
| GATCAAACTGCAGC | 243 | At1g07600 metallothionein-like protein |
| At1g07610 metallothionein-like protein | ||
| GATCGGGAAGAGAG | 239 | At1g56280 hypothetical protein |
| GATCTGTGCCGTCA | 236 | At1g26630 initiation factor 5A-4, putative |
| GATCAAGTTAAGAC | 233 | |
| GATCGTAGAGGAAG | 230 | At1g33120 ribosomal protein L9, putative |
| At1g33140 ribosomal protein L9, putative | ||
| GATCTGGCTAAAGG | 230 | At1g01100 acidic ribosomal protein, putative |
| GATCGCCGGAGGTA | 226 | At3g05590 putative 60S ribosomal protein L18 |
| GATCAATCAACCTT | 223 | At5g26280 unknown protein |
| GATCAAGCTGTCTT | 206 | At3g16640 translationally controlled tumor protein-like protein |
| GATCGGCATCATCT | 205 | At2g39460 60S ribosomal protein L23A |
| GATCCAGGACAAGG | 203 | At4g02890 polyubiquitin |
| At4g05050 polyubiquitin UBQ11 | ||
| At4g05320 polyubiquitin (ubq10) | ||
| GATCGGTGGTGACA | 186 | At3g23390 putative ribosomal protein |
| GATCCATTGGAGGG | 178 | |
| GATCTGTGCTTGTT | 178 | At2g27530 60S ribosomal protein L10A |
| GATCTGTTAAGACT | 171 | At1g31400 hypothetical protein |
| At5g02500 dnaK-type molecular chaperone hsc70.1 | ||
| GATCGGATAACTAT | 170 | At1g78040 phosphoglycerate mutase 1-like protein |
| GATCAGGAGGTGCC | 165 | At3g08580 adenylate translocator |
| GATCCGATTTAATA | 165 | At3g16460 putative jasmonate-inducible protein |
| GATCGCTCAAGAGG | 165 | At5g52650 unknown protein |
| GATCCGGGCGGAGG | 164 | |
| GATCGTATCATACT | 162 | At2g43150 putative extensin |
| GATCCATTTTGATA | 160 | At1g30230 elongation factor 1-β, putative |
| GATCAACGAGGTTG | 159 | At1g66410 calmodulin-4 |
| GATCCCTTTTGTTG | 158 | At5g23740 40S ribosomal protein S11 |
| GATCAATCATCACA | 149 | |
| GATCCGTAACTTCG | 149 | |
| GATCCCTTCATGCA | 147 | At1g76930 extensin 4 (atExt4) |
| GATCAGCGTCGAGG | 143 | At4g23630 unknown protein |
| GATCACATTCTCTG | 141 | At4g16260 β-1,3-glucanase class I precursor |
| GATCAGATGGGGAG | 140 | At5g64100 peroxidase ATP3a (emb CAA67340.1) |
| GATCACATTAACTA | 137 | At4g15910 drought-induced protein-like |
| GATCGGAGGTGGCG | 137 | At1g07920 elongation factor 1-α |
| At1g07930 elongation factor 1-α | ||
| At1g07940 elongation factor 1-α | ||
| GATCGTTTGAGTTT | 133 | At5g65020 annexin |
| GATCACGTGTGTAA | 130 | At5g19890 peroxidase ATP N |
| GATCATCTTCTGTT | 130 | At5g53300 ubiquitin-conjugating enzyme E2—17 kD 10 (ubiquitin-protein ligase 10; ubiquitin carrier protein 10) |
| GATCGCTGAGAAGA | 128 | At1g07890 l-ascorbate peroxidase |
| GATCGTCTTCATGT | 128 | |
| GATCAAGTAGAGAG | 125 | At3g52590 ubiquitin/ribosomal protein CEP52 |
| GATCGCGTCGTGTT | 121 | At3g11510 putative 40S ribosomal protein s14 |
| GATCGTTTAAGTAC | 120 | At2g28190 putative copper/zinc superoxide dismutase |
| GATCTTCTGAGAAG | 119 | |
| GATCCACACTCACA | 117 | At5g17920 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase |
| GATCCTTCGATGTC | 116 | |
| GATCATTTGTGTCT | 113 | At1g20450 putative cold-acclimation protein |
| GATCTACGATGTTG | 113 | At1g14320 putative 60S ribosomal protein L10 |
| GATCTTTGAAGGTG | 112 | At5g15200 40S ribosomal protein-like |
| GATCGTTGGTGTGG | 111 | At1g19570 GSH-dependent dehydroascorbate reductase 1-like protein |
| GATCGAGAGAGTCA | 110 | At3g25520 ribosomal protein, putative |
| GATCCGTAAGACGT | 107 | At3g01190 putative peroxidase |
| GATCAATTGGAGGG | 106 | |
| GATCATTCTTGTAT | 106 | At4g11650 osmotin precursor |
| GATCGACTATGTTT | 106 | At4g05050 polyubiquitin UBQ11 |
| GATCGATGTTGTTC | 105 | At4g33865 ribosomal S29 subunit |
| GATCGCCGTACCAA | 105 | At1g58983 ribosomal protein S2, putative |
| At1g59359 ribosomal protein S2, putative | ||
| GATCAGTTGGTGCT | 104 | At2g27710 60S acidic ribosomal protein P2 |
| At2g27720 60S acidic ribosomal protein P2 | ||
| GATCTGTCTCTCCT | 100 | At2g15970 similar to cold acclimation protein WCOR413 (wheat [Triticum aestivum]) |
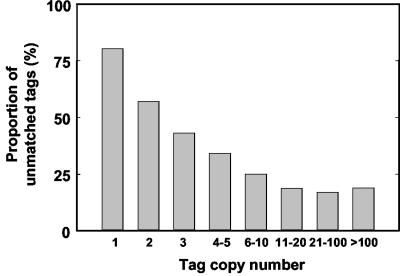
Interestingly, only 15 tags of the 80 with copy numbers of 100 or more had no match in the virtual tag database, which is much lower than expected from the proportion of unmatched tags estimated with the whole set of experimental tags (43%; Fig. 2A). The proportion of unmatched tags was clearly dependent on the tag copy number, decreasing from 57% for tags present only twice to 18% for those found more than 20 times (Fig. 3). In contrast, the proportion of nonspecific tags was not affected by the tag copy number (data not shown).
Figure 3.
Proportion of unmatched tags as a function of the tag copy number in the six combined SAGE libraries.
Expression Analysis of Ion Transporter and Channel Genes
To illustrate the power of SAGE for discriminating between various members of multigene families, the experimental tags corresponding to ion transporter or channel genes were compiled from the six SAGE libraries (Table VI). The families investigated included genes encoding nitrate, ammonium, sulfate, phosphate, potassium and iron transporters, and potassium channels. More than 50 of these genes were found to be expressed in roots. In most instances, the experimental tag was specific for one gene in the family (Table VI), even in cases where a high sequence homology was found between two or more members. The two most highly expressed genes were NRT2.1, encoding a high-affinity NO3- transporter, and At1g32450, a putative NO3- transporter, member of the large NRT1/PTR family.
Table VI.
Expression analysis of various ion transporter or channel genes in Arabidopsis roots
The tag copy nos. are the sum of those found in the six different SAGE libraries.
| Genes | Tag | Tag Copy No. |
|---|---|---|
| HAK/KUP family | ||
| At4g13420 AtHAK5 | GATCAAACAGAGTT | 3 |
| At5g14880 AtHAK8 | GATCGCTATTAACT | 8 |
| At2g40540 AtKUP2 | GATCTCTGTTTCTGa | 5 |
| At4g23640 AtKUP4 | GATCCTTTGTGAGA | 7 |
| At4g33530 AtKUP5 | GATCAAACTACTGG | 6 |
| At4g19960 AtKUP9 | GATCATGTAAAAAA | 2 |
| Shaker family | ||
| At2g26650 AtAKT1 | GATCATCTCATCTT | 12 |
| At4g32650 AtKAT3 (AtAKT4/AtKC1) | GATCGAATTCCCAA | 5 |
| At3g02850 AtSKOR | GATCCAATTGGTAGa | 8 |
| At5g37500 AtGORK | GATCAAACATAATG | 8 |
| KCO family | ||
| At5g55630 AtKCO1 | GATCGTTTCTTGAT | 14 |
| At1g02510 AtKCO4 | GATCGTAAGAGAGT | 2 |
| At4g18160 AtKCO6 | GATCGGTTTACTCTa | 14 |
| YSL family | ||
| At4g24120 AtYSL1 | GATCAGTCAAAGAG | 2 |
| At5g53550 AtYSL3 | GATCGAGTTAAAGC | 2 |
| At5g41000 AtYSL4 | GATCTGCGGAGATG | 2 |
| At1g48370 AtYSL8 | GATCTGTTGAAGAAa | 2 |
| IRT family | ||
| At4g19690 AtIRT1 | GATCTATCACATTT | 11 |
| At1g60960 AtIRT3 | GATCTCATAGCTGCa | 2 |
| ZIP family | ||
| At2g32270 AtZIP3 | GATCAGTGTCCAAG | 12 |
| NRAMP family | ||
| At1g80830 AtNRAMP1 | GATCTTCGTAGGAA | 9 |
| At2g23150 AtNRAMP3 | GATCATGAGTTCTT | 2 |
| At1g15960 AtNRAMP6 | GATCAAACATGAAG | 3 |
| Pht families | ||
| At5g43350 AtPht1;1 | GATCATGCTTGGTGa | 9 |
| At5g43370 AtPht1;2 | GATCAAATGTGGAG | 2 |
| At5g43360 AtPht1;3 | GATCATGCTTGGTGa | 9 |
| At3g26570 AtPht2;1 | GATCTTCTTCTTCTa | 7 |
| At5g14040 AtPht3;1 | GATCGGGACGTTGA | 26 |
| AMT1/AMT2 families | ||
| At1g64780 Amt1;2 | GATCAGTATGTCTT | 4 |
| At3g24300 Amt1;3 | GATCAGCTACTCCT | 6 |
| At4g13510 Amt1;1 | GATCTCCTTCTCCT | 4 |
| At2g38290 Amt2;1 | GATCAAGAAGCTGCa | 2 |
| NRT2 family | ||
| At5g60770 | GATCGAATTGCATG | 2 |
| At1g08090 NRT2.1 | GATCGCATATAAGA | 59 |
| NRT1/PTR family | ||
| At1g12110 NRT1.1 | GATCATGATGATGAa | 7 |
| At1g18880 | GATCTTTCCTTTGA | 2 |
| At1g27080 | GATCAATAGTTGAC | 3 |
| At1g32450 | GATCAGTCTTTTTC | 76 |
| At1g69850 NRT1.2 | GATCAGGTATATAT | 5 |
| At1g72130 | GATCAAACACCTTT | 3 |
| At2g02020 | GATCTCTTCTTCTGa | 4 |
| At2g02040 PTR-2B | GATCCTCACGCTCG | 3 |
| At3g45650 | GATCTTTAAGCTGG | 2 |
| At3g54450 | GATCAAAAATGTTT | 2 |
| At5g14940 | GATCAATGTGATAC | 5 |
| At5g62680 | GATCACATCTCTTT | 6 |
| Sultr families | ||
| At4g08620 Sultr1;1 | GATCATCCGTGTTG | 4 |
| At1g78000 Sultr1;2 | GATCCAGAGATGGC | 16 |
| At5g10180 Sultr2;1 | GATCGATGGGTGTG | 4 |
| At3g15990 Sultr3;4 | GATCTCTCATCAACa | 2 |
| At5g19600 Sultr3;5 | GATCAAAAAACATC | 3 |
| At5g13550 Sultr4;1 | GATCAGGAATGGTG | 7 |
| At3g12520 Sultr4;2 | GATCCTTTTGATTT | 2 |
| At1g80310 Sultr5;1 | GATCGCAGTTTGTA | 2 |
Tags with multiple gene matches
Changes in Gene Expression Associated with Two Different N Sources for Growth of the Plant
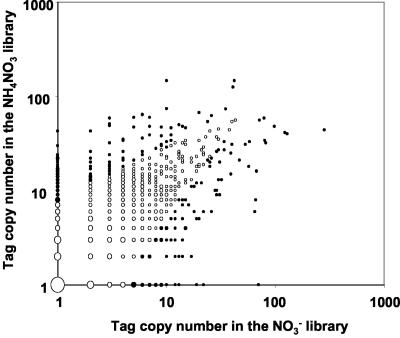
To investigate the usefulness of SAGE for identifying global modifications of gene expression triggered by environmental changes, we compared two libraries obtained from plants grown with either NO3- or NH4NO3 as N source (Fig. 4). The numbers of sequenced tags were 20,886 and 31,354 for the NO3- and NH4NO3 libraries, respectively. This yielded a total of 6,715 different tags found at least twice in the two combined libraries, among which 4,001 matched one single gene. Statistical analysis of the data indicated that 270 of these 4,001 genes were differentially expressed at P < 0.01 between NO3- and NH4NO3 libraries. A large variety of functions are associated with these genes, and a significant proportion of them (26%) encode putative, hypothetical, or unknown proteins. As a limited example, Table VII presents a subset of selected genes for which a response was found between the two conditions (the whole set of data is available at http://genoplanteinfo.infobiogen.fr). These include genes involved in nitrogen metabolism, carbon metabolism, and water transport. Gene expression of the NR1 isoform of nitrate reductase was strongly repressed on NH4NO3 as compared with NO3-, as is also the case for the high-affinity nitrate transporter NRT2.1. In contrast, transcript accumulation of two Glu dehydrogenase isoforms increased when NH4+ was present in addition to NO3- in the nutrient solution. Several genes encoding malate dehydrogenases, malic enzyme, and NAD-dependent isocitrate dehydrogenase were also induced in presence of NH4+. Finally, at least four genes encoding aquaporins were found to be strongly overexpressed with NH4NO3 as a N source.
Figure 4.
Scatter-plot of the comparison of tag frequency between SAGE libraries obtained from roots of plants grown for 6 weeks either on 1 mm NO3- or 1 mm NH4NO3. The tags with no occurrence in one library were set at a copy number of one in this library to enable their representation on a logarithmic scale. The size of the data points is correlated with the number of different tags with the same coordinates. The closed symbols correspond to tags with frequencies significantly different (P < 0.01) between the two libraries.
Table VII.
Differential expression of genes involved in nitrogen or carbon metabolism and water transport between roots of Arabidopsis plants grown either on NO3- or NH4NO3 as N source
The tag frequencies in each SAGE library are calculated by dividing the tag copy number by the total number of sequenced tags.
| Gene | Function | SAGE Tag | Tag Frequency in the NO3- Library (10-3) | Tag Frequency in the NH4NO3 Library (10-3) |
|---|---|---|---|---|
| At1g08090 | High-affinity nitrate transporter NRT2.1 | GATCGCATATAAGA | 0.28 | 0.09 |
| At1g77760 | Nitrate reductase NR1 | GATCTATGTTGTTG | 0.72 | 0.13 |
| At5g07440 | Glu dehydrogenase GDH2 | GATCCAGATGCTGA | 0.10 | 0.83 |
| At5g18170 | Glu dehydrogenase GDH1 | GATCTCCGGATGGG | 0.05 | 0.32 |
| At1g04410 | Malate dehydrogenase like | GATCTCCAAACACT | 0.10 | 0.83 |
| At1g53240 | Mitochondrial NAD-malate dehydrogenase | GATCATAAGTGTGG | 0.24 | 0.61 |
| At2g13560 | Malic enzyme | GATCCAAAAGCTGG | 0 | 0.41 |
| At4g35260 | NAD isocitrate dehydrogenase | GATCCAAGCACAAC | 0 | 0.16 |
| At2g37170 | Aquaporin PIP2;2 | GATCCTTCAGAAGT | 0.48 | 0.92 |
| At2g45960 | Aquaporin PIP1;2 | GATCTTCGCTCTCG | 0 | 1.37 |
| At3g53420 | Aquaporin PIP2;1 | GATCTCTCTGTACA | 0.43 | 0.86 |
| At5g47450 | Aquaporin TIP2;3 | GATCCGAGTGTAAT | 0.10 | 0.45 |
DISCUSSION
The Advantages of Tag to Gene Assignment Based on the Annotated Genome Sequence
Our initial attempts to perform gene identification for SAGE transcript profiling using EST/cDNA databases resulted in a large proportion of unmatched tags (Table II), as always reported in other species (Lee et al., 2002; Zhang et al., 1997; Matsumura et al., 1999, 2003; Chrast et al., 2000; Lash et al., 2000; Margulies et al., 2001; Boheler and Stern, 2003). To circumvent this limitation, we explored the possibility to perform tag to gene assignment using the annotated sequence of the Arabidopsis genome. The strategy, already used by others in yeast (Kal et al., 1999), Caenorhabditis elegans (Jones et al., 2001), and fruitfly (Pleasance et al., 2003), was to merge experimental tags with the list of virtual tags compiling all SAGE tags that can be theoretically found in libraries. The originality of our work was: (a) to investigate several options for generating the virtual tag database, and (b) to quantify the reliability of our procedures for gene identification on the whole-genome scale (more than 26,000 genes) by comparing our results with those obtained using 5,430 available full-length cDNA sequences. The main problem for identifying virtual SAGE tags relates to the length of the 3′-UTR taken into account. In yeast and C. elegans, 500- and 460-bp 3′-UTRs were considered to compile virtual tags, respectively (Kal et al., 1999; Jones et al., 2001). However, the consequences of these choices concerning the quality of tag to gene assignment were not examined. In plants, no reliable polyadenylation signal can be used to predict the 3′ end of a cDNA (Aubourg and Rouzé, 2001). Thus, assumptions have to be made concerning 3′-UTRs for generation of virtual tags at the genomic scale. We explored 10 different options (Fig. 1), thus leading to 10 different virtual tag lists. The data presented in Figure 2 show that these options lead to markedly different results concerning gene identification for the 15,964 experimental tags found at least twice. This is easily explained by the observation that many virtual tags are located in the 3′-UTRs added in silico to the available ORF sequences (Table IV), which has major consequences on the respective contents of the 10 virtual tag lists.
Tag to gene assignment based on the exclusive virtual tag lists was found not to be satisfactory because less than 40% of the experimental tags could be assigned to one or several genes (Fig. 2A) and because this assignment was in many cases not in agreement with that performed using cDNA sequences, especially when UTRs longer than 200 bp were considered (Fig. 2C). Interestingly, the best results with exclusive lists were obtained with 200-bp UTRs (Fig. 2, A and C). This is consistent with the idea that the mean length of 3′-UTRs for Arabidopsis genes is 210 ± 95 bp (Mathé, 2000). Increasing the 3′-UTR length from 200 to 500 bp results in replacing the actual tags of many genes by virtual ones outside the gene sequence. This obviously leads to increasing erroneous tag to gene assignment with increasing length of 3′-UTR (Fig. 2C).
Much more exhaustive and correct gene identification was obtained with cumulative virtual tag lists, when 3′-UTRs of 200 bp or longer were considered (Fig. 2, A and C). This can be explained by the fact that the actual tags of most genes are already present in those selected with 200-bp UTRs. On the other hand, including the virtual tags found between 200 and 500 bp after the stop codon in the list improved tag to gene assignment both qualitatively and quantitatively because it allowed us to take into account correctly genes with unusually long 3′-UTRs (Fig. 2, A and C). However, cumulative virtual tag lists gave less satisfactory results than exclusive ones concerning the specificity of tag to gene assignment (Fig. 2B). This is understandable because in cumulative lists, several virtual tags are generally associated with each gene (from 1.2 to 2.3 on average with 3′-UTR from 100 to 500 bp). Thus, this increases the possibility that an experimental tag matches more than one gene.
When considering all data, we made a compromise and chose the cumulative virtual tag list with 400-bp UTRs for merging with the list of experimental tags. In this case, the percentage of unassigned tags was limited to 43%. This allowed matching of 9,155 experimental SAGE tags to one or several genes, instead of 7,038 or 6,356 using TIGR or Unigene EST clusters, respectively. In agreement with other studies (Jones et al., 2001; Margulies et al., 2001; Pleasance et al., 2003), we found that most experimental SAGE tags matching gene sequences are specific for one single gene. Our calculation of 15% of nonspecific tags indicates that SAGE is efficient to investigate a large number of genes unambiguously. Accordingly, specific analysis of the expression of ion transporter or channel genes shows that SAGE has the potential to distinguish between most of the genes belonging to quite large families (Table VI). Furthermore, the development of the “long SAGE” (SAGE tags of 21 bp) is now expected to solve most of the remaining nonspecificity problem (Saha et al., 2002).
Clearly, the use of the annotated sequence of the Arabidopsis genome markedly improves gene identification in SAGE, as found also in C. elegans or fruitfly (Pleasance et al., 2003). However, our results demonstrate that an in-depth critical analysis of the procedure for tag to gene assignment is strictly required for a reliable transcript profiling by SAGE. Such an analysis, carried out by comparing 10 different options for building the virtual tag reference database, had never been done before. As a result, we reached the highest level of reliability in gene identification reported to date for SAGE studies (88% of correct tag to gene assignment, as compared with 85% in fruitfly; see Pleasance et al., 2003). Thus, we provide in this study an improved computational resource for SAGE in Arabidopsis that was not available before. Our reference database will be implemented and improved, together with the advances in the annotation of the genome of this model species.
Why Are So Many SAGE Tags Unassigned to Genes?
The finding of a fairly large proportion (more than 40%) of experimental tags that could not be assigned to any gene deserves detailed discussion. This was commonly observed in SAGE studies, even when using full genome sequence for gene identification (Pleasance et al., 2003). There are two hypotheses to explain this striking observation: either the unassigned tags are artifactual, or they are true SAGE tags, but originating from transcripts with an incorrect/absent virtual tag in the reference database.
Artifactual tags can arise at many steps of the SAGE procedure. A first cause for generating artifactual tags is production of internal tags upstream from the correct SAGE tag. This may be due either to incomplete digestion by the anchoring enzyme (MboI in our case) or to mispriming of the oligo(dT) to internal stretches of poly(A) during cDNA synthesis (Welle et al., 1999; Jones et al., 2001). We have evidence that this problem occurred in our libraries, because for highly expressed genes, tags upstream from the actual SAGE tag were found (data not shown). However, as in other studies (Piquemal et al., 2002), these artifactual tags are only a very small proportion of the correct ones (2% on average, data not shown). Thus, they cannot explain alone the high percentage of unassigned tags. Second, errors during PCR and sequencing steps of the SAGE procedure do generate artifactual tags. However, sequencing errors are generally not frequent enough to explain such a large proportion of unmatched tags (Wang, 2003; see also “Materials and Methods” for PHRED quality scores of our tag sequences), and excluding from our analysis the experimental tags found only once allowed us to discard the large majority of these erroneous tags (Lash et al., 2000; Piquemal et al., 2002). The fact that experimental tags found more than 20 times remain unassigned clearly demonstrates that the lack of gene identification is not solely the result of PCR or sequencing errors (Fig. 3). Taken together, these considerations support the hypothesis that most of the unassigned SAGE tags are not artifactual (Chen et al., 2002).
The second option, i.e. that unassigned tags are true tags originating from transcripts with an incorrect/absent virtual tag in the reference database, is much more likely. This may also have several causes. First, our virtual tag database is most certainly not fully correct, in particular because it relies on the assumption that 5′- or 3′-UTRs are not longer than 400 bp. In fact, this database matches that generated from the available sequences of 5,430 cDNAs for only 88% of the genes, suggesting that 12% of the virtual tags included in our list may be incorrect. However, it is not clear whether all selected cDNA sequences correspond to truly full-length cDNAs, leaving the possibility that wrong virtual tags can also be extracted from partial cDNA sequences. Second, errors in the Arabidopsis genome sequence can generate wrong virtual tags if the error is located in the GATC MboI site or in the downstream 10 bp. Polymorphism can also explain part of the lack of tag to gene assignment as well, if SNPs are located in the areas corresponding to SAGE tags. This should be limited in our case because we used Col-0 ecotype for our study, which is the one selected for sequencing the genome. Furthermore, polymorphism seems to be quite limited, even between different Arabidopsis ecotypes (Haas et al., 2002). However, sequence variation between the various Col-0 stocks available worldwide cannot be totally excluded. Finally, another explanation for the absence of gene identification for SAGE tags is that these tags correspond to unknown or incorrectly annotated genes. This hypothesis is supported by recent studies, showing that in human SAGE libraries, a very significant proportion of unmatched tags (70% or higher) originate from actual transcripts that can be experimentally identified through real-time PCR (Saha et al., 2002), Rapid Analysis of unknown SAGE Tags by PCR (van den Berg et al., 1999), or the Generation of Longer Fragments for Gene Identification procedure (Chen et al., 2002; Lee et al., 2002). These transcripts revealed alternative splicing (this is expected to be very limited in plants; see Haas et al., 2002; Zhu et al., 2003), but also genes that were incorrectly annotated (wrong exon/intron prediction, wrong or multiple polyadenylation site, gene merging, and gene disruption), genes transcribed in antisense orientation (Jones et al., 2001), and fully novel genes. Accordingly, analysis of full-length cDNA libraries of Arabidopsis (Haas et al., 2002; Seki et al., 2002) or complete EST mapping in this species (Zhu et al., 2003) also led to the identification of a large number of transcripts originating from genes that have been incorrectly annotated or are unannotated. In agreement with most other reports (Zhang et al., 1997; Matsumura et al., 1999; Chrast et al., 2000; Margulies et al., 2001; Piquemal et al., 2002; Pleasance et al., 2003; Wang, 2003), our data suggest that, if they correspond to actual transcripts, the unmatched tags mainly arise from genes expressed at low levels (Fig. 3). This may explain why these genes have not been correctly identified to date, because the structure of genes with low expression level is often not supported by any cDNA or EST, but mostly results from computer predictions, which are far from being fully reliable (Aubourg and Rouzé, 2001; Haas et al., 2002; Zhu et al., 2003). These considerations strongly support the view that SAGE, due to the absence of any a priori concerning the genes investigated, is an important tool to characterize all transcripts actually present in a sample and hence to improve genome annotation when these transcripts do not match any predicted gene (Chen et al., 2002; Saha et al., 2002; Boheler and Stern, 2003). It is thus hypothesized that further investigation of our unmatched SAGE tags may help to unravel a significant number of novel transcripts and genes in Arabidopsis.
The Root Transcriptome, Its Specificity, and Its Changes in Response to Two Different N Sources for the Plant
Our set of 144,083 sequenced tags represents to date the largest set of SAGE data available in plants. It allowed us to identify 7,776 genes expressed in roots, with nearly 3,000 of them with no EST or cDNA match in Unigene. This constitutes an important advance in the characterization of the Arabidopsis root transcriptome. This transcriptome appears to be quite specific. At the exception of a few genes encoding late embryogenesis abundant protein, glutathione S-transferase, and methallothionein, none of the genes highly expressed in the roots (Table V) were found to be significantly represented in SAGE libraries of Arabidopsis leaves (Jung et al., 2003) or pollen (Lee and Lee, 2003).
As a first example to illustrate the usefulness of SAGE for transcript profiling in roots, with respect to the key function of these organs in nutrient acquisition from the soil, we show that SAGE allows specific investigation of the expression levels of the various members of ion transporter multigene families (Table VI). This point remains one of the shortcoming of investigations based on cDNA arrays. The two transporter genes with the highest expression levels, At1g08090 and At1g32450, belong to NRT families of putative nitrate transporters. At1g08090 is AtNrt2.1, which has been found to encode a major component of the high-affinity nitrate uptake system (Cerezo et al., 2001; Filleur et al., 2001). At1g32450 belongs to the large NRT1/PTR family of putative nitrate/peptide transporters. This gene has not been functionally characterized to date, and the strong accumulation of its transcript in the roots suggests that it may play an important role in these organs. The SAGE tags for several other transporter genes were also found at significant copy numbers (around 10 or higher). These data are in agreement with previous studies, showing that these genes are significantly or predominantly expressed in the roots. This is in particular true for AtAKT1 (Lagarde et al., 1996), AtKCO1 (Very and Sentenac, 2003), AtIRT1 (Vert et al., 2002), AtNRAMP1 (Curie et al., 2000), and AtSultr1.2 (Shibagaki et al., 2002).
The second example relates to the characterization of differential gene expression in roots in response to environmental changes such as a modification of the N source supplied to the plants. Among the various genes found to be differentially expressed between NO3- and NH4NO3 libraries, several were already known to be affected by the nature of the N source and allow at least partial validation of our data. For instance, NO3- uptake and assimilation is known to be markedly repressed in the presence of NH4+, and our observation that both nitrate reductase and the high-affinity nitrate transporter NRT2.1 were down-regulated at the transcript level in NH4NO3-grown plants (Table VII) is in complete agreement with previous studies with these genes (Vincentz et al., 1993; Cerezo et al., 2001). Also, genes encoding enzymes involved in metabolism of carboxylic acids such as malate dehydrogenase, malic enzyme, or isocitrate dehydrogenase were found to be strongly regulated by nitrogen. Our finding that the tags to these genes were more highly represented in the NH4NO3 library fits with the hypothesis of a strong stimulation of carboxylic acid synthesis in response to increased N assimilation (Stitt, 1999). Because carboxylic acids are the carbon skeletons used for synthesis of amino acids, this stimulation is strictly required to meet the demand associated with the exclusive assimilation of NH4+ in the root system, as opposed to NO3-, which is mostly assimilated in the shoot in herbaceous species (Beevers and Hageman, 1980). Clearly, these coordinated changes in gene expression are fully consistent with some of the well-documented physiological modifications associated with the supply of a reduced N source such as NH4+ to the plant. The fact that several aquaporin genes were up-regulated in NH4NO3-grown plants is more original and has never been reported before. It is known at the functional level that supply of nitrogen in the external medium markedly modifies the hydraulic conductivity of the root system (Hoarau et al., 1996). The effect of N source on aquaporin gene expression observed from our study may then provide interesting molecular hypotheses to explain the interactions between N and H2O transport in root cells and will deserve further attention.
MATERIALS AND METHODS
Plant Culture and RNA Isolation
The Arabidopsis plants (ecotype Col-0) were grown hydroponically as described previously (Lejay et al., 1999) in a controlled growth chamber with 8-h/16-h day/night cycle, at 24°C/20°C. Light intensity during the light period was at 250 μmol m-2s-1. In brief, seeds were sown directly on the surface of wet sand in modified 1.5-mL microcentrifuge tubes, with the bottom replaced by a metal screen. The tubes supporting the seeds were placed on polystyrene floating rafts, on the surface of a 10-L tank filled with tap water. After 1 week, the tap water was replaced by basal nutrient solution (Gansel et al., 2001), containing either 1 mm NO3- or 1 mm NH4NO3 as N source. The plants were harvested after 4 to 5 additional weeks of growth, and the roots were separated from the shoot. Six SAGE libraries were obtained from six different root samples, originating from plants subjected to various treatments concerning mineral nutrition. These treatments involved N, P, or K limitation, or modification of the N source provided to the plant (NO3- or NH4NO3). This was done to obtain a wide collection of transcripts from genes regulated by the nutrient status of the plant. Total RNA extraction was performed as described previously (Lobreaux et al., 1992), using a guanidine containing extraction buffer and lithium chloride for RNA precipitation.
SAGE Library Synthesis and Tag Sequencing
To obtain the six SAGE libraries, we followed the SADE protocol (SAGE Adaptation for Downsized Extracts; a SAGE variant) described by Virlon et al. (1999), with the difference that the anchoring enzyme was MboI (New England Biolabs, Beverly, MA) instead of Sau3AI. In our hands, the use of MboI improved digestion efficiency during preliminary experiments performed to validate the linkers. Poly(A) RNAs were isolated from 100 μg of total RNA using Dynabeads mRNA direct kit (Dynal Biotech France, Compiègne, France) based on oligo(dT)25 bound covalently to magnetic beads. cDNA was synthesized directly on the beads, and all enzymatic steps needed before digestion by BsmFI were performed on cDNA linked to the beads. All oligonucleotides, with sequences and modifications identical to Virlon et al. (1999), were obtained from Eurobio (Les Ulis, France).
Final concatemers were cloned in pBluescript II KS(-) from Stratagene (La Jolla, CA), digested by EcoRV, dephosphorylated, and purified on agarose gel. Ligation was performed overnight at 16°C and ElectroMAX DH10B Escherichia coli cells (Invitrogen, Rockville, MD) were then used for transformation by electroporation. Plasmids were prepared using the R.E.A.L. Prep 96 Plasmid kit (Qiagen, Courtaboeuf, France). The six SAGE libraries were sequenced separately. Cycle sequencing reactions were carried out using the ABI PRISM Dye Terminator kit (Applied Biosystems, Foster City, CA), and the products were run on an Applied Biosystems Prism 3100 DNA sequencer.
SAGE Data Analysis
The raw sequences obtained from concatemer clones were analyzed using PHRED (Ewing et al., 1997) and trimmed for quality to eliminate as much as possible erroneous tags. The mean PHRED score of the 144,083 tags retained for analysis was 43.8, with only 1.1% of them below 20. Contaminating vector sequences or SAGE tags derived from linkers were then discarded using CROSS-MATCH software (http://www.phrap.org). Extraction of experimental tag sequences was performed using DIGITAG (Piquemal et al., 2002). This software was written in PERL and implemented on a UNIX operating workstation for automatic tag detection and counting. DIGITAG analyzes all concatemer sequences to discard ditags that are duplicated or shorter than 20 bp between the two GATC. Then, for each concatemer sequence, DIGITAG generates the reverse complement, adds it to the initial sequence, and extracts all GATC plus the 10 following bases to obtain the tag sequences, and determine their copy number in each library. The reference database of virtual SAGE tags was obtained from the 26,620 ORFs available from MIPS (ftp://ftpmips.gsf.de/cress/arabidna/arabi_genomicplus500_v111102), including those corresponding to mitochondrial and plastid genes. To generate virtual cDNAs, the ORF sequence was spliced in silico, and genomic sequences upstream from the initiation codon and downstream from the stop codon were added at the 5′ and 3′ ends of the ORF, respectively. Five options were explored to generate virtual cDNAs, with 5′- and 3′-UTR lengths of 100, 200, 300, 400 and 500 bp, respectively (equal length for 5′- and 3′-UTRs). For each of these five options, two different procedures were employed to extract the virtual tag(s) of any given gene. The first was named “exclusive,” in which one single virtual tag was extracted, corresponding to that closest to the 3′ end of the sequence. In this case, each gene corresponds to one virtual tag (Fig. 1). This generated five “exclusive” lists of virtual tags, one for each option of the length of 5′- and 3′-UTRs. The second procedure was named “cumulative,” in which all tags located between the last MboI site of the ORF (or of the 5′-UTR when no MboI site was found in the ORF) and the end of the 3′-UTR were collected and included in the list. In this case, each gene can be associated with several virtual tags (Fig. 1). This also generated five “cumulative” lists of virtual tags. Thus, the whole procedure generated 10 different virtual tags lists.
The statistical analysis of SAGE data for identification of genes differentially expressed was performed as described by Piquemal et al. (2002), using a modified procedure of Audic and Claverie (1997).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Note Added in Proof
In a very recent paper, Yamada et al. (2003) reported on the identification in Arabidopsis thaliana of more than 2,000 genes that were not previously annotated. Among them, 181 match unassigned SAGE tags found in our libraries. This supports our hypothesis that part of the SAGE tags lacking gene attribution reveal actual but yet uncharacterized transcripts.
Acknowledgments
We gratefully acknowledge the technical assistance of Michèle Laudié and Christel Llauro for sequencing of the SAGE libraries.
The work was supported by Génoplante (project nos. Af 1999 064 and Bi 1999 065) and by the Montpellier LR Génopole.
References
- Aubourg S, Rouzé P (2001) Genome annotation. Plant Physiol Biochem 39: 181-193 [Google Scholar]
- Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986-995 [DOI] [PubMed] [Google Scholar]
- Beevers L, Hageman RH (1980) Nitrate and nitrite reduction. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 115-168 [Google Scholar]
- Boheler KR, Stern MD (2003) The new role of SAGE in gene discovery. Trends Biotechnol 21: 55-57 [DOI] [PubMed] [Google Scholar]
- Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ et al. (2002) An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA 99: 11287-11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO3- uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127: 262-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Lee S, Zhou G, Rowley JD, Wang SM (2002) Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc Natl Acad Sci USA 99: 12257-12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, Davisson MT, Schmidt C, Estivill X, Dierssen M et al. (2000) The mouse brain transcriptome by SAGE: differences in gene expression between P30 brains of the partial trisomy 16 mouse model of Down syndrome (Ts65Dn) and normals. Genome Res 10: 2006-2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749-755 [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier LD, Wendl M, Green P (1997) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8: 175-185 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220-224 [DOI] [PubMed] [Google Scholar]
- Gansel X, Muños S, Tillard P, Gojon A (2001) Differential regulation of the NO3- and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26: 143-155 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3: research 0029.1-0029.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau J, Barthes L, Bousser A, Deléens E, Prioul JL (1996) Effect of nitrate on water transfer across roots of nitrogen pre-starved maize seedlings. Planta 200: 405-415 [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA (2001) Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res 11: 1346-1352 [DOI] [PubMed] [Google Scholar]
- Jung SH, Lee JY, Lee DH (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol 52: 553-567 [DOI] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B et al. (1999) Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell 10: 1859-1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C (1996) Tissue specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J 9: 195-203 [DOI] [PubMed] [Google Scholar]
- Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF (2000) SAGEmap: a public gene expression resource. Genome Res 10: 1051-1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee DH (2003) Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol 132: 517-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Clark T, Chen J, Zhou G, Scott LR, Rowley JD, Wang SM (2002) Correct identification of genes from serial analysis of gene expression tag sequences. Genomics 79: 598-602 [DOI] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509-519 [DOI] [PubMed] [Google Scholar]
- Liang P (2002) SAGE Genie: a suite with panoramic view of gene expression. Proc Natl Acad Sci USA 99: 11547-11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Massenet O, Briat JF (1992) Iron induces ferritin synthesis in maize plantlets. Plant Mol Biol 19: 563-575 [DOI] [PubMed] [Google Scholar]
- Lorenz WW, Dean JF (2002) SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol 22: 301-310 [DOI] [PubMed] [Google Scholar]
- Margulies EH, Kardia SL, Innis JW (2001) A comparative molecular analysis of developing mouse forelimbs and hindlimbs using serial analysis of gene expression (SAGE). Genome Res 11: 1686-1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathé C (2000) Analyse in silico des gènes d'Arabidopsis thaliana: description, classification, et modélisation pour aider à la prédiction des gènes. PhD thesis. Paris VII University, Paris
- Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J 33: 425-434 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R (1999) Transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J 20: 719-726 [DOI] [PubMed] [Google Scholar]
- Piquemal D, Commes T, Manchon L, Lejeune M, Ferraz C, Pugnere D, Demaille J, Elalouf J, Marti J (2002) Transcriptome analysis of monocytic leukemia cell differentiation. Genomics 80: 361-371 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Marra MA, Jones SJM (2003) Assessment of SAGE in transcript identification. Genome Res 13: 1203-1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE (2002) Using the transcriptome to annotate the genome. Nat Biotechnol 20: 508-512 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141-145 [DOI] [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies J (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475-486 [DOI] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178-186 [DOI] [PubMed] [Google Scholar]
- van den Berg A, van der Leij J, Poppema S (1999) Serial analysis of gene expression: rapid RT-PCR analysis of unknown SAGE tags. Nucleic Acids Res 27: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484-487 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE Jr, Hieter P, Vogelstein B, Kinzler KW (1997) Characterization of the yeast transcriptome. Cell 88: 243-251 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Very AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575-603 [DOI] [PubMed] [Google Scholar]
- Vincentz M, Moureaux T, Leydecker MT, Vaucheret H, Caboche M (1993) Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J 3: 315-324 [DOI] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet AJ, Elalouf JM (1999) Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci USA 96: 15286-15291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM (2003) Response: the new role of SAGE in gene discovery. Trends Biotechnol 21: 57-58 [DOI] [PubMed] [Google Scholar]
- Welle S, Bhatt K, Thornton CA (1999) Inventory of high-abundance mR NAs in skeletal muscle of normal men. Genome Res 9: 506-513 [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SC, Hruban RH, Hamilton SR, Volgelstein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276: 1268-1272 [DOI] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132: 469-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]