Abstract
The dwarf ucu (ultracurvata) mutants of Arabidopsis display vegetative leaves that are spirally rolled downwards and show reduced expansion along the longitudinal axis. We have previously determined that the UCU1 gene encodes a SHAGGY/GSK3-like kinase that participates in the signaling pathways of auxins and brassinosteroids. Here, we describe four recessive alleles of the UCU2 gene, whose homozygotes display helical rotation of several organs in addition to other phenotypic traits shared with ucu1 mutants. Following a map-based strategy, we identified the UCU2 gene, which was found to encode a peptidyl-prolyl cis/trans-isomerase of the FK506-binding protein family, whose homologs in metazoans are involved in cell signaling and protein trafficking. Physiological and double mutant analyses suggest that UCU2 is required for growth and development and participates in auxin and brassinosteroid signaling.
Plant growth and development are ruled by environmental and endogenous signals, which are integrated through genetic networks that finally act on the division, expansion, and differentiation of cells to generate specific developmental patterns (Tsiantis and Langdale, 1998). Brassinosteroids (BRs) are steroid hormones belonging to the group of endogenous signals required for plant growth and organogenesis, controlling processes such as cell expansion, vascular differentiation, etiolation, reproductive development, and stress responses (Bishop and Yokota, 2001; Bishop and Koncz, 2002).
The biosynthetic pathway of brassinolide, the most active BR, has been dissected thanks to the genetic and molecular analysis of Arabidopsis dwarf mutants (Altmann, 1999; Li and Chory, 1999; Bishop and Yokota, 2001). Little is known, however, about the genes involved in BR signal transduction (Altmann, 2001; Friedrichsen and Chory, 2001; Bishop and Koncz, 2002; Clouse, 2002). Among the BR-insensitive dwarf mutants isolated until now, all those carrying recessive mutations were shown to be affected in the BRI1 (BRASSINOSTEROID INSENSITIVE1) gene, which codes for a cell-surface Leu-rich repeat receptor Ser/Thr kinase (Li and Chory, 1997; He et al., 2000; Wang et al., 2001). BRI1 transduces the BR signal into the nucleus through a mostly unknown phosphorylation cascade (Friedrichsen et al., 2000), leading to the differential expression of target genes, some of which are assumed to control cell expansion (Müssig et al., 2002). On the contrary, BR-insensitive semidominant dwarf mutants carry mutations in the UCU1 (ULTRACURVATA1; Berná et al., 1999) gene, also named BIN2 (BRASSINOSTEROID INSENSITIVE2; Li et al., 2001a), which encodes a member of the SHAGGY/GSK3 family of intracellular protein kinases (Li and Nam, 2002; Pérez-Pérez et al., 2002), whose metazoan homologs are involved in the negative regulation of several developmental pathways, such as those of insulin or Wingless/Wnt signaling (Kim and Kimmel, 2000; Cohen and Frame, 2001). More recently, screenings based on the use of a specific BR-biosynthesis inhibitor have led to the identification of dominant mutations in the BZR1 (BRASSINAZOLE RESISTANT1; Wang et al., 2002) gene and its homolog BES1 (BRI1-EMS-SUPPRESSOR1), also named BZR2 (Yin et al., 2002), the protein products of which are positive regulators of the BR signaling pathway and cooperatively regulate the expression of BR target genes. BIN2 (UCU1) has been shown to be a negative regulator of the BR signaling pathway that destabilizes BZR1 and BES1 by phosphorylation, in an analogous way to that of SHAGGY/GSK3 over β-catenin in the Wingless/Wnt pathway (He et al., 2002; Yin et al., 2002).
In this work, we present the genetic and molecular analyses of four loss-of-function, recessive alleles of the UCU2 gene, the phenotype of which is extremely similar to that of plants homozygous for the semidominant alleles of the UCU1 gene, sharing most phenotypic traits such as dwarfism and circinate leaves. In addition, ucu2/ucu2 plants display helical rotation of some of their organs. We identified the UCU2 gene by a map-based approach and found that it encodes an FK506 binding-like protein (FKBP), named AtFKBP42. FKBPs are a class of peptidyl-prolyl cis/trans-isomerases (PPIases) that play critical roles in regulating cellular processes through protein activation (Harrar et al., 2001; Schiene-Fischer and Yu, 2001). The twd1 (twisted dwarf1) mutant, which is phenotypically indistinguishable of the ucu2 mutants, is also defective in AtFKBP42 (B. Schulz, unpublished data; quoted in Kamphausen et al., 2002), which indicates that TWD and UCU2 are the same gene. The responses of ucu2 mutants to exogenous plant hormones and the genetic analysis of ucu2 alleles suggest that UCU2 participates in auxin and BR signaling.
RESULTS
Isolation of ultracurvata2 Mutants
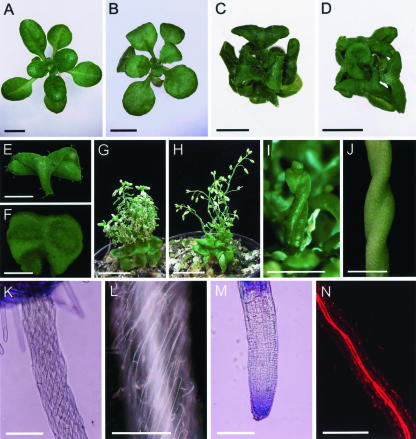
In a large-scale screening for Arabidopsis mutants displaying abnormally shaped or sized leaves (Berná et al., 1999; Serrano-Cartagena et al., 1999; Robles and Micol, 2001), one of the strongest leaf phenotypes found was that of ucu (ultracurvata) mutants, whose vegetative leaves are rolled spirally downwards, in a circinate manner (Fig. 1, B-D). Five ucu lines were identified among our mutants, which were found to fall into two complementation groups. On the one hand, the UCU1 gene was represented by one recessive (Fig. 1B) and two semidominant ethyl methanesulfonate-induced alleles (Fig. 1C; Pérez-Pérez et al., 2002). On the other hand, the UCU2 complementation group included two recessive alleles, ucu2-1 (Fig. 1D) and ucu2-3, which were respectively induced by fast neutron bombardment (P. Robles and J.L. Micol, unpublished data) and T-DNA insertional mutagenesis (this work). Although it was isolated from a collection of lines carrying T-DNA insertions, we found the ucu2-3 mutation to be untagged (data not shown). Another line displaying the Ultracurvata phenotype, CS3397, initially named inl (invalida) by Rédei, was obtained from the ABRC and found to be a double mutant carrying the ucu2-2 and gi-2 (gigantea-2; Rédei, 1962) recessive mutations, the latter conferring an extremely late flowering phenotype, both of them putatively induced by x-ray mutagenesis. After backcrossing the CS3397 line to Columbia-0 (Col-0), F2 plants that displayed the Ultracurvata phenotype and normal flowering time were isolated, and the absence of the gi-2 mutation was confirmed in their F3 progeny.
Figure 1.
The Ultracurvata phenotype. A to D, Rosettes from a Landsberg erecta (Ler) wild-type individual (A) and several ucu mutants: B, ucu1-3/ucu1-3; C, ucu1-2/ucu1-2; and D, ucu2-1/ucu2-1. Vegetative (E, third node) and cauline (F, eighth node) leaves from an ucu2-1/ucu2-1 individual. G and H, Inflorescence of ucu2-1/ucu2-1 (G) and ucu2-3/ucu2-3 (H) plants. I and J, Helical rotation in some organs of ucu2-1/ucu2-1 plants: elongated pistil (I) and mature silique (J). K and L, Root epidermis and root tip (M), visualized by light microscopy. N, Root vascular tissues, visualized by confocal microscopy. Pictures were taken 23 (A-D and K-N), 30 (E and F), and 45 (G-J) d after sowing. Scale bars = 4 mm (A-D and G-I), 2 mm (E, F, and J), 200 μm (K, M, and N), and 100 μm (L).
Pleiotropy of the Phenotype of ucu2 Mutants
All the ucu2 mutant alleles studied in this work displayed complete penetrance and only minor variations in expressivity. The rosette phenotypes of ucu2-1, ucu2-2, or ucu2-3 homozygotes are indistinguishable (Fig. 1D) and very similar to the homozygotes for the ucu1-1 and ucu1-2 semidominant alleles of the UCU1 gene (Fig. 1C).
Although the ucu mutants were isolated on the basis of the abnormal shape and size of their vegetative leaves, they display a pleiotropic phenotype. The ucu2/ucu2 plants are dwarf, with shorter roots, hypocotyls, stems, and fruits than the wild type (Table I), compact dark-green rosettes (Fig. 1D), and reduced inflorescence length with partial loss of apical dominance (Fig. 1, G and H), traits also displayed by the homozygotes for the semidominant ucu1 alleles (Fig. 1C; Table I; Pérez-Pérez et al., 2002). In addition, the three ucu2 mutant alleles cause helical rotation of several radial organs, such as roots, hypocotyls, stems, and pistils, a trait that is clearly visible at whole-organ (Fig. 1, I and J) and epidermal cell level (Figs. 1, K and L). Confocal microscopy of hypocotyls and roots allowed us to visualize in addition the rotation of their internal tissues, such as the vasculature (Fig. 1N). Such spiral habits of growth in ucu2/ucu2 plants caused the distortion of roots and stems, which are wavy and disorganized (Fig. 1H). Their flowers are small, and their pedicels, stamens, pistils, and siliques are twisted (Fig. 1, I and J). Some homeotic transformations of sepals and stamens into pistil tissues were also found. Fertility is reduced in these mutants, which is likely to be due to all the above-mentioned flower abnormalities.
Table I.
Some body parameters of representative ucu mutants
Values are means of at least 20 measurements ± SD. Lengths and widths are indicated in millimeters, and weights are indicated in milligrams.
| Body parameters | Ler | ucu1-2/ucu1-2 | ucu2-1/ucu2-1 |
|---|---|---|---|
| Root lengtha | 64.3 ± 9.0 | 27.5 ± 9.3 | 30.5 ± 9.4 |
| Hypocotyl length (grown in the dark)a | 25.5 ± 5.0 | 5.1 ± 1.4 | 15.3 ± 3.0 |
| Number of vegetative leavesb | 9.4 ± 0.6 | 9.5 ± 0.8 | 9.9 ± 0.9 |
| Rosette diameterb | 21.9 ± 4.3 | 7.6 ± 1.6 | 7.4 ± 1.6 |
| Fresh weightb | 24.4 ± 4.0 | 12.0 ± 3.7 | 9.4 ± 3.4 |
| Dry weightb | 1.9 ± 0.4 | 1.1 ± 0.5 | 0.8 ± 0.3 |
| Petiole lengthb | 4.4 ± 0.9 | 1.9 ± 0.5 | 1.3 ± 0.4 |
| Lamina lengthb | 7.3 ± 1.0 | 3.5 ± 0.5 | 2.8 ± 0.4 |
| Lamina widthb | 5.7 ± 0.9 | 5.3 ± 0.5 | 4.0 ± 0.8 |
| Pedicel lengthc | 3.5 ± 1.1 | 1.6 ± 0.9 | 1.4 ± 1.2 |
| Silique lengthc | 8.5 ± 1.0 | 4.7 ± 0.9 | NDd |
| Seeds per siliquec | 34.1 ± 5.2 | 13.4 ± 5.0 | NDd |
| Primary stem lengthc | 197.8 ± 30.9 | 59.9 ± 8.9 | 36.4 ± 12.6 |
| No. of secondary stemsc | 2.1 ± 0.7 | 3.9 ± 0.9 | 6.2 ± 1.8 |
Measurements were taken from plant material collected 11 d after sowing
Measurements were taken from plant material collected 21 d after sowing
Measurements were taken from plant material collected 49 d after sowing (see “Materials and Methods”)
ND, Not determined
The gravitropic response of the ucu2 roots is similar to that displayed by the wild-type ones, as already shown in the ucu1 mutants (Pérez-Pérez et al., 2002). Root length is severely reduced, whereas root width is only slightly increased in the ucu2 mutants (Table I), probably due to the helical rotation of this organ. The number of lateral roots of ucu2/ucu2 plants was not found to be different from that of the wild type (data not shown). The helical rotation of primary and secondary ucu2 roots is similar to that described above for the hypocotyls, as clearly shown in epidermal cells (Fig. 1, K and L) and vascular tissues (Fig. 1N). The apical region of ucu2 roots does not display helical rotation of epidermal cells (Fig. 1M).
Leaf Architecture in the ucu2 Mutants
The vegetative leaves of ucu2/ucu2 plants show reduced expansion along the proximodistal axis (Fig. 1, D and E; Table I), which affects both the petiole and the lamina, and are indistinguishable from those of ucu1-1 or ucu1-2 homozygous plants (Fig. 1C; Table I). Their cauline leaves are reduced in length, wide, and very wrinkled (Fig. 1F) but do not display the circinate morphology of those of ucu1/ucu1 plants.
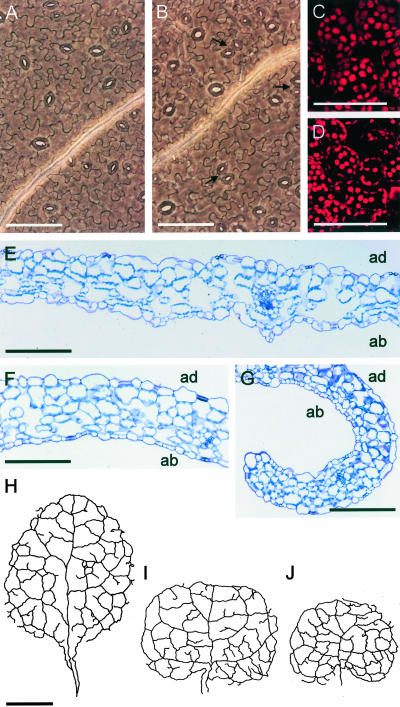
When ucu2/ucu2 leaves are stretched on a slide for observation under a microscope, their adaxial surface wrinkles, as occurred in the semidominant ucu1/ucu1 mutants (Pérez-Pérez et al., 2002). The abaxial pavement cells of the ucu2/ucu2 mutants (Fig. 2, B, F, and G), however, are only slightly smaller than those of the wild type (Fig. 2, A and E), unlike that observed in the ucu1/ucu1 plants (Pérez-Pérez et al., 2002). No obvious differences with the wild type were found in the inner tissues of ucu2/ucu2 leaves (Fig. 2, C-G), in contrast to ucu1/ucu1 mutants, which showed a reduced number of air spaces (Pérez-Pérez et al., 2002). In addition, ucu2/ucu2 leaves displayed many more clustered stomata than the wild-type leaves on both the adaxial and abaxial epidermis (Fig. 2, A and B).
Figure 2.
Ultrastructure of ucu mutant leaves. A and B, Micrographs of the abaxial epidermis. C and D, Confocal microscopy sections of third node vegetative leaves from the wild-type Ler (A and C) and ucu2-1/ucu2-1 (B and D) plants. Arrows indicate clustering of stomata in the abaxial epidermis of ucu2-1/ucu2-1 leaves. E to G, Longitudinal sections of third node vegetative leaves of Ler (E) and ucu2-1/ucu2-1 (F and G) plants (F, basal region of the leaves; and G, apical regions of the leaves). Camera lucida drawings of the venation pattern of third node vegetative leaves of Ler (H), ucu1-2/ucu1-2 (I), and ucu2-1/ucu2-1 (J) plants. Samples were collected 23 d after sowing. Scale bars = 100 μm (A-G) and 1 mm (H-J). ad, Adaxial, ab, abaxial.
Bending of the ucu2/ucu2 vegetative leaves is more pronounced along the primary vein, whose length is clearly reduced compared with the wild type (Fig. 2, H and J), whereas the higher order veins and the complexity of the venation pattern seem to be much less affected, similar to that already observed in plants homozygous for the ucu1 semidominant alleles (Fig. 2I).
Physiological Analyses
The ucu1 mutants display a de-etiolated phenotype similar to that of BR synthesis or response mutants (Pérez-Pérez et al., 2002). On the contrary, ucu2/ucu2 plants grown in the dark displayed a wild-type photomorphogenic response because no leaves were developed (Table I; Fig. 3B). Although ucu2/ucu2 hypocotyls were shorter than those of the wild type, their epidermal cells are not noticeably reduced in length along the proximodistal axis of the hypocotyls or in their width (Fig. 3A; data not shown).
Figure 3.
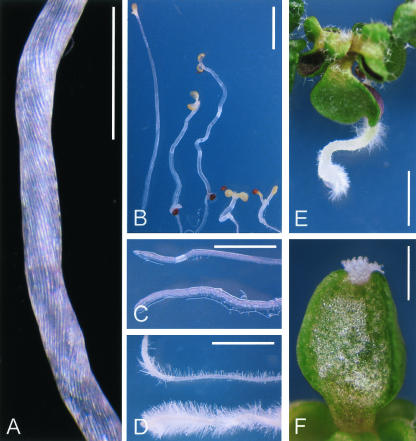
Physiological analyses of the ucu2 mutants. A, ucu2-3/ucu2-3 de-etiolated hypocotyl showing helical rotation of the epidermis. B, Photomorphogenic response of ucu2 mutants when grown for 21 d in the dark. Left to right, Wild-type Col-0, ucu2-1/ucu2-1, ucu2-3/ucu2-3, ucu1-2/ucu1-2, and det2-1/det2-1. Although ucu2-1/ucu2-1 and ucu2-3/ucu2-3 display distorted hypocotyl growth, ucu1-2/ucu1-2 and det2-1/det2-1 show a characteristic deetiolated phenotype. C, Roots of Col-0 (top) and ucu2-3/ucu2-3 (bottom) plants. D, Col-0 (top) and ucu2-3/ucu2-3 (bottom) roots grown on medium supplemented with 10 μm 1-N-naphthyl-phthalamic acid (NPA). E, ucu2-3/ucu2-3 seedling grown on medium supplemented with 10 μm NPA. F, ucu2-3/ucu2-3 heart-shaped cotyledon grown on medium supplemented with 10 μm 2,3,5-triiodobenzoic acid (TIBA), displaying apical tissue overgrowths. Pictures were taken 13 d after sowing. Scale bars = 1 mm (A and B) and 2 mm (C-F).
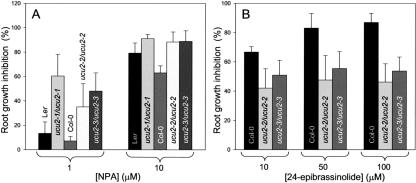
We have shown previously that ucu1 mutants are hypersensitive to auxin (Pérez-Pérez et al., 2002). The reduction in root length displayed by ucu2/ucu2 plants grown in auxin-supplemented medium (20 or 50 nm 2,4-dichlorophenoxyacetic acid [2,4-D]) was similar to that found for the wild type (data not shown). Some phenotypic traits of the ucu2/ucu2 plants, as the helical rotation displayed by some organs, are shared by mutants with disrupted auxin transport, such as lop1 (lopped1; Carland and McHale, 1996). Hence, we analyzed the responses of ucu2/ucu2 mutants to auxin transport inhibition by specifically blocking auxin influx with 1-naphthoxyacetic acid (1-NOA; Parry et al., 2001) or auxin efflux with NPA and TIBA (Fujita and Syono, 1996), in this way disrupting net polar auxin transport. Root growth was more inhibited by 1 μm NPA in the ucu2/ucu2 mutants than in their wild-type backgrounds (Fig. 4A). Similar results were obtained when using 1 μm TIBA (data not shown). When grown in the presence of these auxin efflux inhibitors, ucu2/ucu2 mutants also displayed an increase in root width and in the number and length of root hairs compared with the wild type. In addition, root hairs developed closer to the root apex in the ucu2/ucu2 mutants than in the wild type (Fig. 3D). We found that these traits can be phenocopied in wild-type seedlings exposed to 50 nm 2,4-D. The aerial parts of ucu2/ucu2 seedlings also displayed hypersensitivity to auxin efflux inhibitors (Fig. 3, E and F).
Figure 4.
Response of the ucu2 mutants to hormones. A, Root growth response to the auxin transport inhibitor NPA. B, Root growth response to 24-epibrassinolide. Each point in G and H represents mean data (n > 15) of the reduction in root length displayed by plants grown on hormone-supplemented media, compared with those grown on non-supplemented media.
The aux1/aux1 mutant plants, which are affected in the AUX1 permease-like protein, display agravitropic roots and disrupted auxin uptake (Bennett et al., 1996; Marchant et al., 1999). Phenocopies of aux1/aux1 mutants can be obtained by treating wild-type plants with the auxin influx inhibitor 1-NOA, which does not affect root elongation (Parry et al., 2001). Although small concentrations of 1-NOA up to 10 μm abolish gravitropism in wild-type seedlings, thus phenocopying aux1/aux1 mutants, this trait is unaffected in ucu2/ucu2 seedlings. In addition, we found that the ucu2/ucu2 mutants display a moderately reduced sensitivity to 24-epibrassinolide in root elongation assays (Fig. 4B), as already shown for the weak ucu1-3 recessive allele of the UCU1 gene (Pérez-Pérez et al., 2002).
Double Mutant Analysis
We obtained ucu1/ucu1;ucu2/ucu2 double mutants (Fig. 5, A and B; Tables II and III) whose overall phenotype was in all cases the same irrespectively of the strength of the ucu1 allele involved. Their rosette phenotype was strongly different from that of each ucu/ucu single mutant and quite similar to that of bri1/bri1 plants (Fig. 5G). The BRI1 BR-receptor (Li and Chory, 1997) is upstream of the UCU1 (BIN2) protein (Li and Nam, 2002; Pérez-Pérez et al., 2002) in the BR signal transduction pathway. The extreme rosette phenotype of the ucu1-1/ucu1-1;ucu2/ucu2 double mutant (Fig. 5A) makes it difficult to determine whether or not it indicates additivity or synergy. However, the extreme rosette phenotype of the ucu1-3/ucu1-3;ucu2/ucu2 double mutants (Fig. 5B) is clearly synergistic, given that it cannot be expected from the weak leaf phenotype of ucu1-3/ucu1-3 single mutant plants (Fig. 1B).
Figure 5.
Genetic interactions between UCU2 and the UCU1, DET2, and DIM1 genes. Rosettes from ucu2-1/ucu2-1;ucu1-1/ucu1-1 (A) and ucu2-1/ucu2-1;ucu1-3/ucu1-3 (B) double mutants. Inflorescence (C) and detail (D) of a helically rotated pistil of a ucu2-1/ucu2-1;ucu1-1/ucu1-1 plant. ucu2-1/ucu2-1;dim1-1/dim1-1 (E) and ucu2-1/ucu2-1;det2-1/det2-1 (F) double mutant plants, which are extremely similar to the bri1-1/bri1-1 mutant (G). Pictures were taken 23 (A, B, and E-G) and 45 (C and D) d after sowing. Scale bars = 4 mm (A, B, and E-G), 1 mm (D), and 1 cm (C).
Table II.
Phenotypic segregations in the F2 progeny of crosses involving ucu mutants
Figures indicate the no. of plants belonging to a given phenotypic class. WT, Wild-type phenotype. UcuX, Ucul, and UcuW indicate extreme, intermediate, and weak Ultracurvata phenotype, respectively. UcuXH, Extreme Ultracurvata phenotype and helical rotation of some organs.
| Cross (♀ × ♂)
|
F1
|
F2
|
||||||
|---|---|---|---|---|---|---|---|---|
| WT | Ucul | UcuX | UcuXH | UcuW | Putative double mutants | χ2 | ||
| ucu2-1/ucu2-1 × ucu1-1/ucu1-1 | 38 Ucul | 47 | 111 | 54 | 51 | — | 13 | 2.135a |
| ucu2-1/ucu2-1 × ucu1-2/ucu1-2 | 9 Ucul | 54 | 104 | 60 | 56 | — | 14 | 1.778a |
| ucu2-1/ucu2-1 × ucu1-3/ucu1-3 | 30 WT | 147 | — | — | 57 | 58 | 11 | 4.001b |
The χ2 values represent the fit of the data (only families segregating for the putative double mutant are shown) to an expected 3:6:3:3:1 phenotypic segregation
The χ2 values represent the fit of the data (only families segregating for the putative double mutant are shown) to an expected 9:3:3:1 phenotypic segregation. Corresponds to a modified 3:6:3:1:2:1 segregation, given that the ucu1-1/UCU1; ucu2-1/ucu2-1 and the ucu1-1/ucu1-1; ucu2-1/ucu2-1 individuals were phenotypically indistinguishable
Table III.
Phenotypic segregations in the F3 progeny of crosses involving ucu mutants
See Table 2 legend.
| Cross (♀ × ♂)
|
F2
|
F3
|
|||
|---|---|---|---|---|---|
| UcuX | UcuW | Putative double mutants | χ2 (3:1) | ||
| ucu2-1/ucu2-1 × ucu1-1/ucu1-1 | UcuX | 42 | — | 12 | 0.099 |
| UcuX | 45 | — | 19 | 0.521 | |
| ucu2-1/ucu2-1 × ucu1-2/ucu1-2 | UcuX | 50 | — | 12 | 0.774 |
| UcuX | 26 | — | 8 | 0.000 | |
| ucu2-1/ucu2-1 × ucu1-3/ucu1-3 | UcuW | — | 98 | 36 | 0.159 |
| UcuW | — | 100 | 40 | 0.771 | |
Several dwarf mutants have been described as affected in BR biosynthesis (Altmann, 1999). These include dim1 (Takahashi et al., 1995) and det2 (Li et al., 1996), which are specifically blocked in the early steps of the BR biosynthetic pathway. The ucu2/ucu2; dim1/dim1 (Fig. 5E) and ucu2/ucu2;det2/det2 (Fig. 5F) double mutants that we obtained displayed additive phenotypes similar to those of bri1/bri1 mutants (Fig. 5G). The helical rotation characteristic of ucu2/ucu2 organs was also displayed by all the double mutants obtained (Fig. 5D).
BR and Auxin-Induced Gene Expression
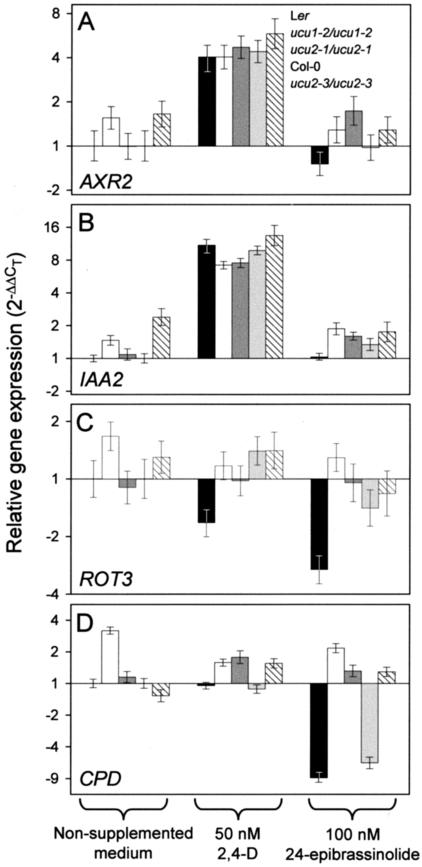
We tested whether exogenous 2,4-D or 24-epibrassinolide modulate the expression of several genes in the ucu2 mutants (Fig. 6). To this end, several genes were chosen, all of them already described as regulated by plant hormones. This group of genes included the following (Table IV): IAA7 (also named AUXIN RESISTANT2; AXR2) and IAA2, which are overexpressed in BR-deficient mutants and encode proteins of the Aux/indole-3-acetic acid (IAA) family (Nagpal et al., 2000; Liscum and Reed, 2002; Müssig et al., 2002) induced by IAA (Abel et al., 1995). CPD (CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM; Szekeres et al., 1996) and ROT3 (ROTUNDIFOLIA3; Kim et al., 1998), which encode two cytochrome P450 enzymes involved in BR biosynthesis, both of them negatively regulated by BR (Mathur et al., 1998; Bancos et al., 2002); CycD3 (CyclinD3), the product of which is involved in the progression of the cell cycle; and BEE1 (BRASSINOSTEROID ENHANCED EXPRESSION1), which encodes a transcription factor. CycD3 and BEE1 are known to be induced by BR (Friedrichsen et al., 2002; Müssig et al., 2002). We studied also the expression of a gene that has no known relationship with BR or auxin signaling, NCED3, which codes for the 9-cisepoxycarotenoid dioxygenase, one of the enzymes involved in abscisic acid (ABA) biosynthesis, whose expression is positively regulated by ABA (Xiong et al., 2002). Transcription of the UCU1 gene was also studied.
Figure 6.
Real-time reverse transcriptase (RT)-PCR quantification of the expression of several genes in the ucu1 and ucu2 mutants. The expression of AXR2 (A), IAA2 (B), ROT3 (C), and CPD (D) genes was studied in the presence of exogenous auxin (50 nm 2,4-D) or BR (100 nm 24-epibrassinolide) and compared with their expression under noninductive conditions in the wild type. Relative expression was normalized as regards to that of the OTC gene (see “Materials and Methods”).
Table IV.
Quantitative RT-PCR primer pairs used in this work
| Gene
|
Bacterial Artificial Chromosome (BAC)
|
Oligonucleotide Sequences (5′ → 3′)
|
PCR Product Size (bp)
|
|
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| CPD (At5g05690) | M3J3 | CTCAACTCAAGGAAGAGCATGA | GTGTGAATGGCATTGACTTGTAAT | 93 |
| CycD3 (At1g43680) | F28A23 | TCTGTAATCTCCGATTCAAGATT | GCTCTATAATTCGCATCATGGTA | 79 |
| ROT3 (At1g43680) | F24P17 | AAAGTCTTTATACGGGAAAGTGTT | ACCACTTTATTCACCTCTGCAT | 87 |
| BEE1 (At1g18400) | F15H18 | CACTGAAACAGGTTCTTTGAGAA | GGCCTCTTCTGGCTCTCACA | 104 |
| IAA2 (At3g23030) | MXC7 | TACACCTCCTACCAAAACTCAA | CTTTCACGTAGCTCACACTGTT | 86 |
| UCU1 (At4g18710) | F28A21 | GAGATCTAAAGCCTCAAAATCTT | TGGCTTCACCTTTAACGAGCT | 99 |
| AXR2 (At3g23050) | MXC7 | GCTCCTTTACTATGGGAAACTAT | TACTCAGAGCTATTCAGCAGATT | 91 |
| NCED3 (At3g14440) | MOA2 | CGGTGGTTTACGACAAGAACAA | CAGAAGCAATCTGGAGCATCAA | 103 |
| OTC (At1g75330) | F1B16 | TGAAGGGACAAAGGTTGTGTATGTT | CGCAGACAAAGTGGAATGGA | 95 |
Expression of the above-mentioned genes was analyzed by means of quantitative, real-time RT-PCR in ucu2-1/ucu2-1 and ucu2-3/ucu2-3 plants, together with their respective wild-type backgrounds Ler and Col-0. In addition, we also studied ucu1-2/ucu1-2 plants, the genetic background of which is Ler. RNA was extracted from plants grown in liquid media containing 50 nm 2,4-D or 100 nm 24-epibrassinolide and quantified by means of the comparative threshold cycle (CT) method (Livak and Schmittgen, 2001), using the expression of the housekeeping OTC (ORNITINE TRANSCARBAMILASE) gene as an internal reference. We found no significant differences in the expression of the NCED3, CycD3, BEE1, and UCU1 genes in the wild-type plants after auxin and BR treatment (data not shown). Hence, activity of these genes was not analyzed in the ucu mutants.
The ucu1 and ucu2 mutations do not affect induction by auxin of the AXR2 and IAA2 genes, as indicated by the similar strong effects of exogenous 2,4-D on the wild-type and mutant plants studied (Fig. 6, A and B). No remarkable effect of 24-epibrassinolide on AXR2 and IAA2 gene expression was observed. As expected, ROT3 and CPD were found down-regulated by brassinolide in the wild types studied, an effect that is abolished or extremely reduced by the ucu2 mutations (Fig. 6, C and D). On the other hand, the expression of ROT3 and CPD in the ucu1 mutant studied was higher than in the wild type in noninductive conditions and did not change significantly by auxin or brassinolide treatment.
Isolation and Molecular Characterization of the UCU2 Gene
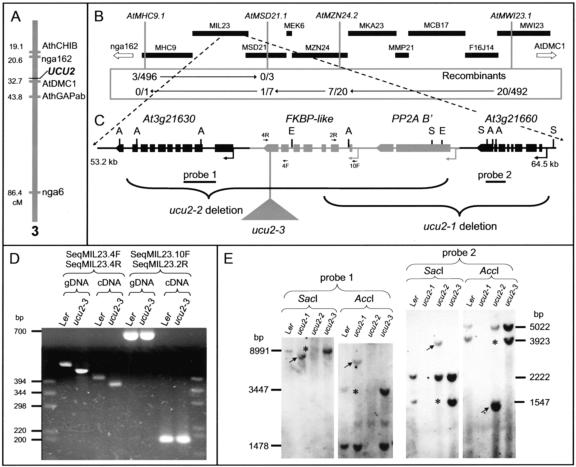
Using an F2 mapping population obtained from three ucu2-1/ucu2-1 × Col-0 crosses, we determined the map position of the UCU2 gene, which was found to be closely linked to the AtDMC1 (Klimyuk and Jones, 1997) cleaved-amplified polymorphic sequence marker (1.92 ± 0.57 cM, n = 574) in a region of 12.11 cM (1.9 Mb), flanked by the nga162 (8.35 ± 1.76 cM, n = 278) and AthGAPab (11.72 ± 2.12 cM, n = 252) markers (Fig. 7A). Allelism between UCU2 and the DIM (Takahashi et al., 1995) and AXR2 (Wilson et al., 1990) genes, the latter two mapping within the candidate interval, was ruled out after the corresponding complementation tests.
Figure 7.
Positional cloning of the UCU2 gene. A, Map position of the UCU2 gene as determined by low-resolution mapping. B, Physical map of the BAC contig assumed to contain the UCU2 gene. A total of 23 recombination events were identified relative to four molecular markers (those shown in italics) within this region. C, Candidate region with indication of the mutations found in the ucu2 mutants. Boxes, Exons; lines between boxes, introns. A, S, and E, Positions of restriction sites for AccI, SacI, and EcoRI, respectively, for the enzymes used for the Southern blot shown in E. D, Amplification products corresponding to the At3g21640 gene, obtained using the primers indicated at the top and genomic DNA (gDNA) or cDNA as a template, from wild-type (Ler) and ucu2-3 homozygous (ucu2-3) plants. The positions of the primers used within the region containing the At3g21640 gene are indicated as 4R, 4F, 2R, and 10F in C. E, Gel blots of genomic DNA from the ucu2 mutants hybridized with probes 1 and 2. The results shown were obtained by reprobing a single membrane. The absence of some bands is indicated by asterisks, and the differential bands present in the DNA of the mutants are indicated by arrows.
Based on the genome sequence available at Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de/proj/thal/db/index.html), we developed new simple sequence length polymorphism (SSLP) markers within the candidate region (Fig. 7B; Table V), which were used to screen for informative recombinants. The linkage analysis of 1,292 chromosomes performed using these new markers delimited the UCU2 gene to an interval of 160 kb, encompassing three BAC clones (Fig. 7B). One of the candidate genes within this region, At3g21650, included in the MIL23 BAC clone, encodes a putative B′-regulatory subunit of a protein phosphatase 2A (PP2A B′-ζ; Terol et al., 2002), which displays 54% identity and 71% similarity to the human B56 PP2A protein delta isoform (NP_006236). The latter is known to inhibit Wnt signaling in a similar way to that in which it reduces the signaling of SHAGGY/GSK3 metazoan kinases, by reducing the transcription of Wnt-responsive genes by means of β-catenin degradation through ubiquitinylation (Seeling et al., 1999; Li et al., 2001b). Given the similarities between the phenotypes of ucu1 and ucu2 mutants and that UCU1 encodes a SHAGGY/GSK3-like kinase (Pérez-Pérez et al., 2002), we considered At3g21650 a strong candidate. Hence, several primer pairs were designed to PCR amplify the genomic region containing the At3g21650 gene, but no amplification products were obtained from ucu2-1/ucu2-1 or ucu2-2/ucu2-2 genomic DNA. Because these results suggested a deletion, several primer pairs were designed for the adjacent gene, At3g21640, which codes for a putative FK506-binding protein (FKBP); once again, no PCR products were obtained. On the contrary, PCR products from both the At3g21640 and At3g21650 genes were obtained from the DNA of ucu2-3/ucu2-3 plants, and no differences were found with the wild type in the coding region of the At3g21650 transcription unit or in its 5′- and 3′-flanking regions. However, a rearrangement of 40 bp, including two small deletions in the 3′ region of the At3g21640 gene, was shown to cause a frameshift and to induce a premature stop codon in the ucu2-3/ucu2-3 mutant. In addition, we found by RT-PCR that the At3g21640 gene is expressed in ucu2-3/ucu2-3 plants, although its transcript is smaller than that of the wild type (Fig. 7D).
Table V.
Novel SSLP primer sets used in this work
| Marker
|
BAC
|
Position in BAC
|
Oligonucleotide sequences (5′ → 3′)
|
PCR Product Size (bp)
|
||
|---|---|---|---|---|---|---|
| Forward labeled primer | Reverse primer | Ler | Col-0 | |||
| AtMHC9.1 | MHC9 (AP001305) | 3,316—3,716 | ACAGAGGAACAATTGATCTCATCa | TCACAGGAAACTCTGAGAAACG | 448 | 400 |
| AtMSD21.1 | MSD21 (AB025634) | 29,271—29,544 | GTAGATATTATACATGTAACATTCGb | TTAAAAACGGTAAGGTAGTTATTAC | 247 | 274 |
| AtMZN24.2 | MZN24 (AB028622) | 69,687—69,707 | ATCAACTGGAAGCACTTGCGCc | TGAGGTGTTTGCCGATCTACG | 230 | 240 |
| AtMW123.1 | MW123 (AB022223) | 52,271—52,588 | GAAGCCTTAGTAATGCACCGGb | GATTGTGCATCCTTTGGCTGC | 318d | 354d |
Primer pair included an oligonucleotide labeled with 4,7,2′,4′,5′,7′-hexachlo-0-6-carboxyfluorescein phosphoramidites
Primer pair included an oligonucleotide labeled with 6-carboxyfluorescein phosphoramidites
Primer pair included an oligonucleotide labeled with 4,7,2′,7′-tetrachloro-6-carboxyfluorescein phosphoramiotes. Primers were designed based on the Col-0 genomic sequence data available at MIPS (http://mips.gsf.de/proj/db/index.html)
Sizes were determined by sequencing the corresponding amplification products, which were found to contain 15 and nine TGGAGG repeats in the Col-0 and Ler accessions, respectively. In contrast to the Col-0 genomic sequence annotated at MIPS, which has been reported not to be different from that of Ler
To determine the nature of the alterations carried by the ucu2-1/ucu2-1 and ucu2-2/ucu2-2 mutants, we performed a Southern blot using three probes spaced along the region of the MIL23 BAC clone including At3g21640 and At3g21650 (probe 3) and their two flanking genes, At3g21630 (probe 1) and At3g21660 (probe 2; Fig. 7E). No differences with the wild type were found for the genomic DNA of ucu2-3/ucu2-3 plants (Fig. 7E). In regards to ucu2-1, no hybridization bands were detected with probes 2 and 3, whereas the bands obtained with probe 1 were different from those of the wild type, suggesting the deletion of about 6 kb, which removes the At3g21640, At3g21650, and At3g21660 genes. In regards to ucu2-2, no hybridization bands were detected when using probes 1 and 3, but probe 2 revealed bands different from those of the wild type, suggesting that this mutant suffers a deletion encompassing the At3g21630, At3g21640, and At3g21650 genes. In addition, RT-PCR analyses confirmed the absence of the At3g21640 transcription product (data not shown) in ucu2-1/ucu2-1 and ucu2-2/ucu2-2 plants (data not shown).
Complementation of the Mutant Phenotype of ucu2-3/ucu2-3 Plants
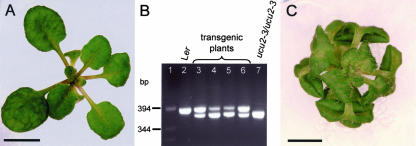
With the aim of confirming that the lack of function of the At3g21640 gene is responsible for the phenotype of ucu2 mutants, a segment of 2,441 bp of the genomic region harboring this gene, including its whole coding region, which contains 1,235 bp of the 1,360-bp full-length cDNA, was cloned into the binary vector pART27 (see “Materials and Methods”) under the control of a 35S promoter. Homozygous ucu2-3 plants were transformed with the 35S: At3g21640:ocs construct, and the T2 progeny obtained was sown on kanamycin-supplemented medium. Six transformants were isolated, which unambiguously displayed wild-type vegetative and cauline leaf phenotypes, normal length of the stem and siliques, and no helical rotation of organs (Fig. 8A). The phenotypic segregations observed in their T3 progeny (Table VI) suggested that the T2 plants were as expected, hemizygous for the transgene. Furthermore, the presence of the 35S:At3g21640:ocs transgene in the T2 plants was confirmed by amplification of the segment of the At3g21640 gene that is deleted in the ucu2-3 allele (Fig. 8B).
Figure 8.
Functional complementation of an ucu2 mutation. Wild-type phenotype of ucu2-3/ucu2-3;35S:At3g21640:ocs transgenic plants (A). B, PCR amplification products obtained using primers flanking the At3g21640 transcription unit and DNA extracted from: lane 2, Ler; lane 7, ucu2-3/ucu2-3; and lanes 3 to 6, ucu2-3/ucu2-3;35S:At3g21640:ocs transgenic plants. C, ucu2-4/ucu2-4 mutant plant. Pictures were taken 23 d after sowing. Scale bars = 4 (A) and 2 (C) mm.
Table VI.
Segregation of kanamycin resistance in the progeny of ucu2-3/ucu2-3 plants subjected to transformation
See Table 2 legend. KanR and KanS, Kanamycin-resistant and -sensitive individuals, respectively.
| T3
|
|||||||
|---|---|---|---|---|---|---|---|
| Transgene Used | T2 | Non-supplemented medium
|
Supplemented medium
|
||||
| WT | UcuX | χ2 (3:1) | KanR | KanS | χ2 (3:1) | ||
| 35S:At3g21640:ocs | WT | 77 | 34 | 1.589 | 142 | 63 | 0.063 |
| WT | 76 | 38 | 3.789 | 100 | 58 | 2.364 | |
| WT | 70 | 32 | 1.882 | 98 | 48 | 4.898 | |
| WT | 140 | 50 | 0.112 | 84 | 47 | 1.528 | |
| WT | 59 | 24 | 0.486 | 63 | 23 | 3.172 | |
| WT | 21 | 8 | 0.011 | 48 | 19 | 0.065 | |
In addition, we screened T-DNA collections for additional alleles of the UCU2 gene. The Salk_012836 insertion line from the Salk Institute Genomic Analysis Laboratory (SIGnAL) collection carries a T-DNA insertion in the fifth exon of At3g21640. Its mutant phenotype is indistinguishable of that of ucu2 mutants and was inherited as a recessive trait (Fig. 8C). We named this allele ucu2-4.
The UCU2 Gene Encodes the FKBP42 of Arabidopsis
The At3g21640 gene codes for a protein with homology to the FKBP type of PPIases (or rotamases; E.C. 5.2.1.8). Its mRNA is 1,360 nucleotides long (AJ224640), with an open reading frame of 1,095 nucleotides that gives rise to a protein of 365 amino acids and 42 kD, nine amino acids different from the At3g21640 gene, as annotated in the MIPS genome project Web page (http://mips.gsf.de/proj/thal/db/index.html), probably as the result of the incorrect deduction of the splicing site. The UCU2 gene consists of eight exons, and the UCU2 protein shows significant similarity with the FKBP-type immunophillins (Galat, 2000), which are characterized by a conserved FKBP-like domain carrying the PPIase activity (Schiene-Fischer and Yu, 2001). When we compared the UCU2 amino acid sequence with those of already known FKBPs, we found a single PPIase domain that includes amino acids 58 to 156. The C-terminal region of UCU2 contains three putative domains of the tetratricopeptide repeat (TPR) type, a 34-amino acid domain involved in protein-protein interactions, which mediates association into multiprotein complexes (Blatch and Lassle, 1999). When the amino acid sequence of UCU2 was compared with those of proteins found in databases, the highest degree of similarity was observed with other FKBPs: 50% similarity and 34% identity with the FKBP1 Rotamase (ROF1; Vucich and Gasser, 1996; S72485) and 46% similarity and 28% identity with PASTICCINO1 (AL132960), both of them of Arabidopsis; 50% similarity and 32% identity with a 77-kD FKBP of wheat (Triticum aestivum; wFKPB77; T06489), and 52% similarity and 29% identity with a 59-kD FKBP of mouse (Mus musculus) (AAH03447). All of these FKBPs contain three PPIase domains, in contrast to the single PPIase domain of UCU2, which includes five putative phosphorylation sites and several N-myristoylation sites that may correspond to posttranslational modifications. An unusual characteristic of the UCU2 protein when compared with already known immunophilins of the FKBP type is a putative transmembrane region in its C terminus that is absent from other FKBP-like proteins.
The small DNA rearrangement of the UCU2 gene found in the ucu2-3 allele causes a frameshift that abolishes the third TPR domain and the putative transmembrane domain at the C terminus, as a consequence of the appearance of a premature stop codon. Because the ucu2-3 mutation does not affect transcription of the UCU2 gene, the similarity between the phenotypes of ucu2-1 and ucu2-2, which are null alleles, and ucu2-3 clearly indicates that the C-terminal region is required for the activity of UCU2.
DISCUSSION
Morphology of the ucu Mutants
Here, we present a genetic and molecular analysis of four recessive alleles of the UCU2 gene, which cause the mutant plants to be dwarf and to display circinate vegetative leaves and helical rotation of some organs. The circinate leaf phenotype of the ucu1 and ucu2 mutants is presumably the consequence of a perturbation in the coordination of the growth of the adaxial and abaxial tissues along the proximodistal leaf axis. Histological analyses of ucu2/ucu2 leaves indicated that they might be defective in the differentiation of the primary vein. In fact, leaf morphology is severely altered in venation pattern mutants (Koizumi et al., 2000; Semiarti et al., 2001) and in wild-type plants treated with auxin transport inhibitors (Mattsson et al., 1999; Sieburth, 1999). A reduction in primary vein length was also observed in the ucu1 mutants (this work), which is consistent with the abaxial origin of these tissues. Although we have shown previously that the ucu1 mutants display differences in cell size between the abaxial and adaxial epidermis (Pérez-Pérez et al., 2002), no such differences were found in the mutant leaves of ucu2/ucu2 plants.
The small size of most organs is a phenotypic trait shared by the ucu mutants and BR-deficient or -insensitive mutants. The leaf morphology of ucu mutants, however, is rather different from that of all other dwarf mutants affected in BR synthesis or perception, whose small rounded leaves suffer a significant reduction in both cell size and cell number in all the tissues studied (Nakaya et al., 2002). Both the vegetative and cauline leaves of ucu mutants are reduced in length but retain a normal width, which indicates that the expansion of the organ in these mutants is reduced along the proximodistal axis but not along the mediolateral axis. In addition, the ucu2 mutants displayed an etiolated phenotype when grown in the dark, contrary to that shown by BR-deficient and -insensitive dwarf mutants, which indicates that ucu2 mutants are not affected in the repression by darkness through the BR signaling pathway of the light-induced developmental program.
Only recently has the genetic analysis of Arabidopsis mutants shed light on the molecular components of handedness in plants, by the identification of genes involved in establishing left-right asymmetry patterns (Hashimoto, 2002). A number of mutants with a helical habit of growth have been identified, including lefty1 and lefty2, which are affected in the genes that encode the TUA6 and TUA4 α-tubulins, both of them involved in cortical microtubule orientation (Thitamadee et al., 2002). These results, together with the observation that proper orientation of microtubule arrays is disrupted in the spiral mutants (Furutani et al., 2000), indicate the importance of microtubule organization in establishing helical handedness in Arabidopsis. Plant hormones such as auxins and BR, among others, can perturb plant morphology through microtubule reorientation (Catterou et al., 2001). It must be noted, however, that the helical rotation found in the ucu2 mutants only affects organs that present radial symmetry in the wild type, whereas all organs are rotated in other helical mutants. No helical rotation has been shown in any BR-deficient mutants described to date (Catterou et al., 2001).
The helical rotation displayed by ucu2 mutants might be related to that caused by mutant alleles of the LOP1 (LOPPED1) gene (Carland and Hale, 1996), also named TRN1 (TORNADO1; Cnops et al., 2000). Such lop1 and trn1 mutants display a pleiotropic phenotype, which is characterized by a perturbation in leaf vasculature and abnormal patterns of cell expansion that cause helical rotation of the root and deformed leaves, including twisting of the petiole (Carland and McHale, 1996). The phenotype of the lop1 mutants seems to be caused by deficiencies in the basipetal transport of IAA (Carland and McHale, 1996). The maize rs2 (rough sheath2) mutant, which displays aberrant leaf vasculature development and dwarfism, was also found to be defective in the basipetal transport of auxin in the shoot (Tsiantis et al., 1999).
The UCU2 Gene Encodes an FKBP
To gain insight into the molecular nature of the UCU2 function, we positionally cloned the UCU2 gene, which was found to encode an Arabidopsis FKBP. FKBPs belong to the PPIase family of folding proteins, also named rotamases, whose homologs are found in prokaryotes, such as Escherichia coli, and eukaryotes such as yeast, invertebrates, mammals, and plants (Galat, 2000). Low-Mr FKBPs participate in signal transduction pathways through association with transmembrane receptors and usually include a single PPIase domain, similar to that found in the UCU2 protein. An example of this is mammalian FKBP12, which has been found to be associated with the TGF-β type II receptor and calcium channels (Schiene-Fischer and Yu, 2001). Multidomain PPIases are characterized by their C-terminal domain, which binds calmodulin and interacts with multiple partners (Schiene-Fischer and Yu, 2001). The multidomain PPIase FKBP52 (52-kD mammalian FKBP) is associated with Hsp90 through its TPR domain and participates in the activation of the cytoplasmic glucocorticoid receptor complex and its migration into the nucleus (Galigniana et al., 2001; for review, see Pratt et al., 2001). It has been demonstrated recently that plants hold the entire heterocomplex assembly machinery that links the glucocorticoid receptor complex to dynein, the motor protein that guides the complex to the nucleus (Harrell et al., 2002). Furthermore, wheat plants overexpressing wFKBP77 showed dwarfism, sterility, and altered leaf shape (Harrar et al., 2001). The UCU2 protein is a high-Mr FKBP that includes three TPR domains but only one PPIase domain, hence sharing structural properties with both previously known classes of FKBPs.
We found the UCU2 protein to be the same as AtFKBP42, some of whose functional characteristics have been reported recently (Kamphausen et al., 2002), including its localization in the plasma membrane and in the tonoplast and the interaction of its TPR domain with the COOH-terminal region of the heat shock protein, Hsp90. Such an interaction with Hsp90 is similar to that already shown for other FKBPs (Owens-Grillo et al., 1996; Young et al., 1998) as the mammalian PPIases, FKBP51 and FKBP52, which is necessary for the maturation of nonactivated steroid hormone receptor complexes (Richter and Buchner, 2001). AtFKBP42 also may be part of an analogous steroid hormone receptor complex in Arabidopsis, probably through their interaction with the BRI1 BR receptor. No PPIase activity has been detected for AtFKBP42 (Kamphausen et al., 2002), in contrast to that found for mammalian FKBP51 and FKBP52 (Schiene-Fischer and Yu, 2001). At least another FKBP, FKBP71, is known to be involved in the development of Arabidopsis. Mutations in the PAS1 (PASTICCINO1) gene, which encodes the FKBP71 (Vittorioso et al., 1998), display altered cell division and elongation, probably as a consequence of the perturbation of cytoquinin signaling (Faure et al., 1998).
All the ucu2 alleles studied here are likely to be null, given that the ucu2-1 and ucu2-2 mutants carry a deletion of the whole At3g21640 gene and that their mutant phenotypes are indistinguishable from that of ucu2-3 and ucu2-4. The deletion of the At3g21630 and At3g21650 genes in the ucu2-1 mutant and that of At3g21650 and At3g21660 in the ucu2-2 mutant do not cause a visible phenotype, possibly due to gene redundancy. The At3g21650 gene, for instance, codes for the PP2A B′-ζ, which has at least seven paralogs in the genome of Arabidopsis (Terol et al., 2002).
Auxin and BR Responses Are Perturbed in the ucu Mutants
A large body of evidence supports the hypothesis of a connection between the auxin and BR signaling pathways. Some auxin-regulated genes, such as Aux/IAA and ARF (AUXIN RESPONSE FACTOR) genes, have been found overexpressed in BR-deficient mutants (those of ARF7, AXR3 [IAA17], SHY2 [IAA3], IAA2, AXR2 [IAA7], and IAA22 genes), or expressed in response to BR (such as IAA3 and IAA19; Müssig et al., 2002). In addition, altered responses to auxins have been described in some BR-deficient or -insensitive dwarf mutants, such as in the curl-3 tomato (Lycopersicon pimpinellifolium) mutant, which is hypersensitive to 2,4-D and is affected in a homolog of the BRI1 receptor (Koka et al., 2000; Bishop and Koncz, 2002), in the sax1 (hypersensitive to ABA and auxin1) mutant, whose hypersensitivity to auxin is restored after BR treatment (Ephritikhine et al., 1999a; 1999b) or in the ucu mutants (Pérez-Pérez et al., 2002; this work). A reduction of IAA levels was also observed in the BR-deficient lkb mutants of pea (Pisum sativum; Nomura et al., 1997).
Our results on the morphological, physiological, and genetic analysis of the ucu mutants of Arabidopsis indicate the implication of the UCU genes in the signaling pathways of auxin and BRs. Further support to this hypothesis is given by the quantitative differences found between the ucu1 and ucu2 mutants and the wild types studied here with regard to the effects of 24-epibrassinolide on the expression of the CPD and ROT3 genes, which are involved in BR biosynthesis. Down-regulation by exogenous BR of CPD and ROT3 is abolished by the ucu1 and ucu2 mutations, which indicates that UCU1 and UCU2 are required for BR-regulated gene expression.
Our auxin transport inhibition analyses suggest that UCU2 might be involved in the regulation of the transport of auxin. Contrary to that expected from the hypersensitivity displayed by the ucu1 mutants to auxin (Pérez-Pérez et al., 2002) and by the ucu2 mutants to auxin transport inhibitors (this work), auxin-induced expression of AXR2 and IAA2 genes was found to be similar in the wild-type and in the ucu mutants. Further analyses will be required to ascertain whether or not the growth responses of the ucu mutants to exogenous auxin or auxin transport inhibitors are mediated by the AXR2 and IAA2 genes.
It has been proposed that ATP-binding cassette (ABC) plant proteins, which are related to those of the multidrug resistance (MDR) family of animal proteins (often referred to as P-glycoproteins), could be involved in auxin transport in plants (Gaedeke et al., 2001; Noh et al., 2001; Luschnig, 2002). Support for this hypothesis is provided by mutant alleles of the AtMDR1 gene, which are defective in basipetal auxin transport and cause epinastic cotyledons and wrinkled leaves (Noh et al., 2001). Furthermore, the phenotype of a double mutant lacking both AtMDR1 and its closest homolog AtPGP1 (Noh et al., 2001) is similar to that of the ucu2 mutants described in this work, with a dwarf phenotype, leaves curled down, and a distorted habit of growth.
AtMDR1 and AtPGP1 are likely to be required for auxin efflux, preventing excessive auxin accumulation (Luschnig, 2002). In a similar way, the ucu2 epinastic leaf phenotype can be explained by the accumulation of intracellular auxin as a result of a defect in its polar transport, either directly if UCU2 participates in the auxin transport complex or indirectly through the posttranslational modification of the AtMDRP1 and AtPGP1 transporters. The analysis of ucu2/ucu2;atpgp1/atpgp1;atmdr1/atmdr1 triple mutants would help to test such a hypothesis. It is known that FK506 immunosuppressant drugs are able to inhibit P-glycoprotein-mediated MDR in mammals, which suggests that FKBPs might positively regulate ABC transporter function (Hemenway and Heitman, 1996). Increased auxin levels have been found in the atmdr5 mutants of Arabidopsis. The wild-type ABC transporter AtMRP5 is also able to transport organic anions when expressed in yeast, such as estradiol-17-(β-d-glucuronide), a product of the catabolism of steroids in animals (Gaedeke et al., 2001). Furthermore, a role as a steroid transporter has been reported for the human P-glycoprotein (Ueda et al., 1992).
UCU1 and UCU2 Function and Interaction
The similarity between the leaf phenotypes of ucu1 and ucu2 mutants, together with their insensitivity to BR, prompted us to consider the possibility that the UCU1 and UCU2 genes participate in some developmental process related with BR signal transduction. We found that the phenotype of ucu1 ucu2 double mutants is indistinguishable from that of bri1 single mutants, which suggests that UCU1 and UCU2 act in convergent pathways, both probably downstream of the BRI1 receptor, to cooperatively trigger BR responses. The UCU2 protein can be a positive regulator of BR signaling, acting by controlling hormone transport through ABC transporters. Another possibility is that UCU1 and UCU2 participate in the same pathway and that the semidominant, gain-of-function ucu1 alleles interfere with the function of UCU2.
Recessive mutations in the AXR1 gene confer resistance to auxin and cause morphological alterations, such as wrinkled leaves. AXR1 encodes a protein related to the ubiquitin-activating enzyme E1, which is a key component in a complex that controls ubiquitin-mediated protein stability (Leyser et al., 1993). In addition, it is known that alterations in the ubiquitin pathway, caused by overexpressing a modified ubiquitin that inhibits protein degradation, could originate curled down leaves in tobacco (Nicotiana tabacum; Bachmair et al., 1990). Furthermore, the auxin efflux carrier EIR1 (also named AGR or AtPIN2), which is involved in the gravitropic response of roots, is posttranscriptionally modified and requires the function of AXR1, suggesting that auxin transport is regulated through protein stability (Sieberer et al., 2000). UCU1 (BIN2) is likely to be a negative regulator of the BR transduction pathway, acting through the destabilization of the BES1 and BZR1 transcription factors (He et al., 2002; Yin et al., 2002). Such negative regulation may be dependent on protein degradation mediated by ubiquitin, which is analogous to protein degradation mediated by of SGG/GSK3 in the Wingless/Wnt pathway of metazoans (Barker et al., 2000; Clouse, 2002). On the other hand, metazoan SGG/GSK3 kinases regulate signaling pathways other than that of Wingless/Wnt, such as those of insulin or cell cycle control (for review, see Cohen and Frame, 2001). It has been described recently that BRI1 interacts with BAK1, another Leu-rich repeat receptor-like protein kinase, which suggests a conserved downstream activation pathway between BR signal transduction in plants and the TGF-β signaling cascade in metazoans (Li et al., 2002). In fact, the interplay between the TGF-β and Wingless/Wnt signaling pathways is important for the specification of cell fates during animal development (Capdevila and Izpisúa-Belmonte, 2001). If a similar cross talk between pathways occurs in plants, the phenotype of ucu1 ucu2 double mutants might suggest the implication of both UCU1 and UCU2 in an interaction between the BRI1 pathway and the activation of the ABC transporters. This would explain the similarities of ucu1 and ucu2 leaf phenotypes and their phenotypic differences with BR-deficient or -insensitive mutants.
A role for the UCU2 gene can be proposed in the cross talk between the auxin and BR signal transduction pathways, which would be achieved primarily by regulation of auxin homeostasis through activation of one or both of the AtMDR1 and AtPGP1 ABC transporters or by the regulation of the BRI1 and BAK1 heterocomplex. Further molecular analyses will be required to gain insight into such a mechanism of cross talk between plant hormones.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds from Arabidopsis Ler and Col-0 accessions were supplied by the Nottingham Arabidopsis Stock Center (Loughborough, UK). The CS3397 line and the DIM1/dim1-1 (CS8100), det2-1/det2-1 (CS6159), BRI1/bri1-1 (CS3723), aux1-7/aux1-7 (CS3074), and axr2-1/axr2-1 (CS3077) mutants were supplied by the Arabidopsis Biological Resource Center (Ohio State University, Columbus). The ucu2-1 and ucu2-3 alleles, respectively, were isolated after fast neutron mutagenesis in a Ler background (P. Robles and J.L. Micol, unpublished data) and T-DNA mutagenesis in a Col-0 background (this work). The N512836 insertion line (ucu2-4/ucu2-4) was supplied by the Nottingham Arabidopsis Stock Center and is described at the SIGnAL Web site (http://signal.salk.edu). The nomenclature rules of Meinke and Koornneef (1997) for phenotypes, genes, alleles, mutations, and proteins are followed in this paper.
Cultures were performed as described by Ponce et al. (1998), at 20°C ± 1°C and 60% to 70% relative humidity under continuous illumination of 7,000 luxes. Crosses were performed as described by Berná et al. (1999).
Photomicrography and Morphometric Analysis
Morphometric analyses of ucu2 mutants were carried out as described previously (Candela et al., 1999; Pérez-Pérez et al., 2002). Photographs were taken with a Cybershot FV-505 digital camera (Sony, Tokyo) using a resolution of 1,024 × 768 pixels. Photomicrography and confocal microscopy of whole leaves and leaf sections were performed as described by Serrano-Cartagena et al. (2000) and Pérez-Pérez et al. (2002). Images were digitally processed with the Adobe Photoshop 6.0 program (Adobe Systems Incorporated, San Jose, CA).
Physiological Analyses and Hormone Treatment
Photomorphogenic and gravitropic responses were studied as described by Pérez-Pérez et al. (2002). Photomorphogenic development was monitored 13 and 21 d after sowing.
For the root elongation assays of hormone-treated plants, at least 30 seedlings of each genotype were grown in vertically oriented agar plates supplemented with 0, 10, 50, or 100 nm epibrassinolide (no. E1641, Sigma, St. Louis), a mixture of synthetic BR; 0, 10, 20, or 50 nm 2,4-D (no. 11215-019, Life Technologies/Gibco-BRL, Cleveland), a synthetic auxin; 0, 1, 5, or 10 μm TIBA (no. T5910, Sigma) or NPA (no. 34361, Riedel-de-Häen, Seelze, Germany), two auxin efflux transport inhibitors; and 0, 1, 5, or 10 μm 1-NOA (no. 25,541-6, Aldrich, Milwaukee, WI), an auxin influx transport inhibitor. To determine root lengths, primary roots grown in the above-mentioned supplemented media were stretched by forceps and photographed 13 d after sowing. Statistical analysis of the data was performed with the SPSS version 7.5 software package (Statistical Products & Service Solutions, Chicago), and plots were obtained with the SigmaPlot 2000 version 6.0 program (Statistical Products & Service Solutions).
Hormone-Induced Gene Expression Analyses
Plants grown on agar plates as described above were collected 20 d after sowing and transferred to flasks containing 50 mL of one-half-strength Murashige and Skoog (no. M 0221, Duchefa Biochemie B.V., Haarlem, The Netherlands) liquid medium, which were incubated for 24 h at 225 rpm, 20°C ± 1°C, and 60% to 70% relative humidity under continuous illumination of 7,000 luxes. Ethanol solutions of 2,4-D or 24-epibrassinolide were then added to the liquid cultures to reach a final concentration of 50 or 100 nm, respectively. After incubation for 12 h in the same conditions, plants were immediately frozen in liquid nitrogen and stored at -80°C. RNA was extracted from frozen samples of 50 to 100 mg, using the Qiagen RNeasy Plant Mini Kit (no. 74904, Qiagen USA, Valencia, CA), following the instructions of the manufacturer. The eluate was incubated for 30 min at 37°C with 40 units of DNaseI. After inactivation of DNaseI at 70°C for 15 min, RNA was ethanol precipitated and resuspended in 40 μL of RNase-free water. About 4 μg of the RNA extracted from each sample was reverse transcribed using the SuperScript II RNase H RT following the instructions of the manufacturer (no. 18064-022, Invitrogen, Carlsbad, CA). The cDNA solution obtained in this way was diluted by adding 60 μL of distilled water.
Real-Time Quantitative PCR
Reaction mixes of 25 μL were prepared including 12.5 μL of the SYBR Green PCR Master Kit (no. 4309155, Applied Biosystems, Foster City, CA) containing the AmpliTaq Gold polymerase, 0.4 pmol of a primer pair, and 1 μL of cDNA. PCR amplifications were carried out in 96-well optical reaction plates on the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Three independent amplifications were performed from each cDNA sample. The thermal cycling program started with a step of 2 min at 50°C and 10 min at 95°C to activate the polymerase, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Dissociation kinetics analyses of the amplification products were performed to check their specificity. Only the expected products were found to be amplified. The primer pairs used and the sizes of the expected amplification products are shown in Table IV. One of the primers of each pair contained the sequences of the ends of two contiguous exons to avoid amplification of genomic DNA.
Relative quantitation of gene expression was carried out using the 2-ΔΔCT method (Livak and Schmittgen, 2001). Expression levels of the target genes were normalized using the CT values obtained for the OTC (ORNITINE TRANSCARBAMILASE) housekeeping gene (Quesada et al., 1999), which was used as an endogenous reference. The relative amount of target gene transcripts was calculated as the difference in CTs for the target gene products and the reference gene products, given by 2-ΔΔCT, where ΔΔCT was calculated as: (CT, target gene- CT, reference gene)mutant - (CT, target gene - CT, reference gene)wild type, in noninductive conditions; or (CT, target gene - CT, reference gene)induced - (CT, target gene - CT, reference gene)noninduced, in supplemented media. Confidence intervals were obtained by evaluating the expressions 2-(ΔΔCy + SD) and 2(ΔΔCT + SD), where sd is the sd of the ΔΔCT value. Data were represented in Figure 6 as the relative gene expression normalized to an internal reference (OTC) and relative to gene expression in noninductive conditions in the wild type.
Positional Cloning
Low-resolution gene mapping was performed by using SSLP (Bell and Ecker, 1994) and cleaved-amplified polymorphic sequence (Konieczny and Ausubel, 1993) markers. Homozygous ucu2-1 plants were crossed to Col-0, and ucu2-2 and ucu2-3 homozygotes were crossed to Ler. Phenotypically mutant F2 individuals were collected and frozen to extract their DNA as described by Ponce et al. (1999). Multiplex PCR amplification conditions and automated fragment sizing of amplified microsatellites were as described by Ponce et al. (1999). Map distances were determined using Kosambi's map function (Kosambi, 1944). Novel SSLP markers developed in this work are described in Table V.
Sequencing reactions were carried out on PCR amplification products, previously purified by ethanol precipitation, using ABI PRISM BigDye Terminator Cycle Sequencing version 2.0 Ready Reaction kits (Applied Biosystems) according to the instructions provided by the manufacturer, in a reaction volume of 5 μL, and GeneAmp PCR System 2400 (Applied Biosystems) thermal cycler. Sequencing electrophoresis and detection were performed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
The sequences obtained were assembled with the Multiple Alignment Construction & Analysis Workbench program (version 2.0.5, Greg Schuler, National Center for Biotechnology Information, 1995). Sequences with homology to that of the UCU2 gene were identified in the GenBank database (Benson et al., 2002; http://www.ncbi.nlm.nih.gov/) using the BLAST program (Altschul et al., 1997). Analyses of amino acid sequences were under-taken by using the tools available at the PROSITE database (Falquet et al., 2002; http://www.expasy.org/prosite/).
Southern-Blot Analysis
Genomic DNA was isolated from whole rosette leaves as previously reported (Dellaporta et al., 1983). Five micrograms of DNA was digested with SacI or AccI and electrophoresed (15 h at 30 V in a 0.8% [w/v] agarose gel in 0.5× Tris-borate/EDTA buffer) and then treated with depurination (no. N1907, Sigma), denaturation (no. N1531, Sigma), and neutralization (no. N1532, Sigma) solutions following the protocol provided by the manufacturer. Transference to a Hybond N+ membrane (Amersham, Buckinghamshire, UK) was performed overnight in 2× SSC buffer (no. S6639, Sigma). Before prehybridization, the membrane was oven dried (10 min at 80°C) and cross linked (15 s under 70,000 μJ cm-2). Prehybridization (1 h at 46°C) and hybridization (18 h at 46°C) were carried out in 20 and 30 mL, respectively, of a water solution of DIG Easy Hyb Granules (no. 1,796,895, Boehringer Mannheim/Roche, Basel), prepared as indicated by the manufacturer, in a hybridization oven. The membrane was hybridized with 30 ng mL-1 denatured (10 min at 95°C) digoxigenin 11-dUTP (no. 1,570,552, Boehringer Mannheim/Roche) probe, which had been synthesized as previously described (Ponce et al., 1998). The primers used to PCR synthesize the probes were the following: SeqMIL23.13F (5′-TCCTGAAGACAGTCTCAGTTC-3′) and SeqMIL23.10R (5′-CAGGTTGACATGATGTACACGC-3′) for probe 1, and SeqMIL23.11F (5′-GTCAGCTATAGAAAGATCTGAGC-3′) and SeqMIL23.11R (5′-AGCCTTCTCGATGGTTTGGTC-3′) for probe 2. Washes and detection were performed using the DIG Wash and Block buffer set (no. 1,585,762, Boehringer Mannheim/Roche) as indicated by the manufacturer. Detection was performed with anti-DIG-AP Fab fragments (no. 1,093,274, Boehringer Mannheim/Roche) and the chemiluminiscent phosphatase CDP-Star substrate (no. 1,685,627, Boehringer Mannheim/Roche). Autoradiograms were obtained on Hyperfilm ECL (Amersham-Pharmacia Biotech, Uppsala) exposed for about 2 h. Membranes were reprobed after being washed with water to remove the substrate and incubated twice in 0.3 m NaOH and 0.1% (w/v) SDS for 20 min at 37°C and rinsed in 2× SSC buffer to remove the probe.
Functional Complementation
To clone candidate genes in the pART7 vector (Gleave, 1992), the following oligonucleotides were designed. All had a 5′ tail (shown in lowercase), including restriction sites for KpnI or XbaI (in bold): ClonMIL23.10F, 5′-gccggtaccTCTTCGTGAGTGACTTCGACG-3′; and ClonMIL23.4R, The ClonMIL23.10F and ClonMIL23.4R primers flanked a 2,441-bp fragment of the At3g21640 gene that contains its whole open reading frame. The PCR product obtained was digested with KpnI and XbaI and cloned into the pART7 vector (Gleave, 1992). Competent Escherichia coli DH5α cells were transformed, and the transformants were isolated on Luria-Bertani broth plates supplemented with 100 μg mL-1 ampicillin and later tested by PCR amplification for the presence of the transgene. Plasmid DNA was isolated and digested with NotI, and the restriction fragments were cloned into the pART27 vector in DH5α cells. Positive clones were selected on Luria-Bertani broth agar plates supplemented with 25 μg mL-1 streptomycin, 5% (w/v) X-Gal, and 50 mg mL-1 isopropylthio-β-galactoside, and tested by PCR for the presence of the construct. Plasmid DNA was isolated from these clones and used to transform chemical competent Agrobacterium tumefaciens C58C1 cells. Transformant A. tumefaciens cells were selected as described above and used to obtain liquid cultures that were used for in planta transformations of Arabidopsis wild-type and mutant plants (Bechtold and Pelletier, 1998). The T2 seeds obtained were sown in agar plates supplemented with 50 μg mL-1 kanamycin. PCR amplifications using primers flanking the At3g21640 transcription unit were performed to confirm the presence of the transgene in the kanamycin-resistant T2 plants.
Gene Expression Analysis
To determine whether or not the expression of candidate genes was affected in the ucu2 mutants, RT-PCR amplifications of RNA extracted from leaves and flower buds were performed as described by Ponce et al. (2000). The primer pairs used corresponded to the 5′ region (SeqMIL23.10F, 5′-TCTT-CGTGAGTGACTTCGACG-3′; and SeqMIL23.2R, 5′-CTCTTGAGATGGCT-CACTATG-3′) or the 3′ region (SeqMIL23.4F, 5′-GAAGCAATTGGTCACT-GCAAC-3′; and SeqMIL23.4R, 5′-GGTAGATCTTTCACGTTGGTC-3′) of the At3g21640 gene. Only the SeqMIL23.4F and SeqMIL23.4R primer pair was found useful for distinguishing between wild-type and ucu2-3 sequences.
Acknowledgments
The authors thank María Alonso-Peral, José María Barrero, María Asun-ción Brotons-Gil, Héctor Candela, Rebeca González-Bayón, Andrea Hricová, Sara Jover, Francisca María Lozano, Victor Quesada, and Pedro Robles for comments on the manuscript; José Miguel Martínez-Zapater for providing the T-DNA seed collection from which ucu2-3 was isolated; and José Manuel Serrano for his expert technical assistance. The pART7 and pART27 vectors were kindly supplied by Chris Winefield, and the NCED3 and OTC real-time RT-PCR primers by were supplied by José María Barrero and Héctor Candela, respectively. We thank SIGnAL for providing the sequence-indexed Arabidopsis T-DNA insertion mutant.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.032524.
This work was supported by the Ministerio de Educación y Cultura and the Ministerio de Ciencia y Tecnología of Spain (grant nos. PB98-1389 and BMC2002-02840) and by the Conselleria de Cultura, Educació i Ciència of the Generalitat Valenciana (fellowship to J.M.P.-P.).
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533-549 [DOI] [PubMed] [Google Scholar]
- Altmann T (1999) Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208: 1-11 [DOI] [PubMed] [Google Scholar]
- Altmann T (2001) Brassinosteroid signaling in plants. Trends Endocrinol Metab 12: 398-402 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JZ, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Becker F, Masterson RV, Schell J (1990) Perturbation of the ubiquitin system causes leaf curling, vascular tissue alterations and necrotic lesions in a higher plant. EMBO J 9: 4543-4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnar G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Morin PJ, Clevers H (2000) The Yin-Yang of TCF/beta-catenin signaling. Adv Cancer Res 77: 1-24 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259-266 [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137-144 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948-950 [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL (2002) GenBank. Nucleic Acids Res 30: 17-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152: 729-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C (2002) Brassinosteroids and plant steroid hormone signaling. Plant Cell 14: S97-S110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T (2001) Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol 42: 114-120 [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21: 932-939 [DOI] [PubMed] [Google Scholar]
- Candela H, Martínez-Laborda A, Micol JL (1999) Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev Biol 205: 205-216 [DOI] [PubMed] [Google Scholar]
- Capdevila J, Izpisüa-Belmonte JC (2001) Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol 17: 87-132 [DOI] [PubMed] [Google Scholar]
- Carland FM, McHale NA (1996) LOP1: a gene involved in auxin transport and vascular patterning in Arabidopsis. Development 122: 1811-1819 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS (2001) Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana: II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 212: 673-683 [DOI] [PubMed] [Google Scholar]
- Clouse S (2002) Brassinosteroid signaling: novel downstream components emerge. Curr Biol 12: R485-R487 [DOI] [PubMed] [Google Scholar]
- Cnops G, Wang X, Linstead P, Van Montagu M, Van Lijsebettens M, Dolan L (2000) Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development 127: 3385-3394 [DOI] [PubMed] [Google Scholar]
- Cohen P, Frame S (2001) The renaissance of GSK3. Nat Rev Mol Cell Biol 2: 769-776 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 4: 19-21 [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H (1999a) The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J 18: 303-314 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H (1999b) The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 18: 315-320 [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30: 235-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JD, Vittorioso P, Santoni V, Fraisier V, Prinsen E, Barlier I, Van Onckelen H, Caboche M, Bellini C (1998) The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125: 909-918 [DOI] [PubMed] [Google Scholar]
- Friedrichsen D, Chory J (2001) Steroid signaling in plants: from the cell surface to the nucleus. Bioessays 23: 1028-1036 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Syono K (1996) Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol 37: 1094-1101 [DOI] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T (2000) The SPIRAL genes are required for directional control of cell elongation in Arabidopsis thaliana. Development 127: 4443-4453 [DOI] [PubMed] [Google Scholar]
- Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Muller A, Ansorge M, Becker D, Mamnun Y, Kuchler K, Schulz B et al. (2001) The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J 20: 1875-1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A (2000) Sequence diversification of the FK506-binding proteins in several different genomes. Eur J Biochem 267: 4945-4959 [DOI] [PubMed] [Google Scholar]
- Galigniana MD, Radanyi C, Renoir JM, Housley PR, Pratt WB (2001) Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J Biol Chem 276: 14884-14889 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203-1207 [DOI] [PubMed] [Google Scholar]
- Harrar Y, Bellini C, Faure JD (2001) FKBPs: at the crossroads of folding and transduction. Trends Plant Sci 6: 426-431 [DOI] [PubMed] [Google Scholar]
- Harrell JM, Kurek I, Breiman A, Radanyi C, Renoir JM, Pratt WB, Galigniana MD (2002) All of the protein interactions that link steroid receptorhsp90-immunophilin heterocomplexes to cytoplasmic dynein are common to plant and animal cells. Biochemistry 41: 5581-5587 [DOI] [PubMed] [Google Scholar]
- Hashimoto T (2002) Molecular genetic analysis of left-right handedness in plants. Philos Trans R Soc Lond B Biol Sci 357: 799-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185-10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J (2000) Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360-2363 [DOI] [PubMed] [Google Scholar]
- Hemenway CS, Heitman J (1996) Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J Biol Chem 271: 18527-18534 [DOI] [PubMed] [Google Scholar]
- Kamphausen T, Fanghanel J, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32: 263-276 [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12: 2381-2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Kimmel AR (2000) GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Genet Dev 10: 508-514 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Jones JD (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J 11: 1-14 [DOI] [PubMed] [Google Scholar]
- Koizumi K, Sugiyama M, Fukuda H (2000) A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: calling the auxin signal flow canalization hypothesis into question. Development 127: 3197-3204 [DOI] [PubMed] [Google Scholar]
- Koka CV, Cerny RE, Gardner RG, Noguchi T, Fujioka S, Takatsuto S, Yoshida S, Clouse SD (2000) A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol 122: 85-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403-410 [DOI] [PubMed] [Google Scholar]
- Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12: 172-175 [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161-164 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929-938 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1999) Brassinosteroid actions in plants. J Exp Bot 50: 275-282 [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398-401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299-1301 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001a) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213-222 [DOI] [PubMed] [Google Scholar]
- Li X, Yost HJ, Virshup DM, Seeling JM (2001b) Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J 20: 4122-4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387-400 [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402-408 [DOI] [PubMed] [Google Scholar]
- Luschnig C (2002) Auxin transport: ABC proteins join the club. Trends Plant Sci 7: 329. [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066-2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593-602 [DOI] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T (1999) Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979-2991 [DOI] [PubMed] [Google Scholar]
- Meinke D, Koornneef M (1997) Community standards for Arabidopsis genetics. Plant J 12: 247-253 [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241-1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Tsukaya H, Murakami N, Kato M (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol 43: 239-244 [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441-2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Nakayama M, Reid JB, Takeuchi Y, Yokota T (1997) Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Phisiol 113: 31-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P (1996) Binding of immunophilins to the 90 kDa heat shock protein (hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochemistry 35: 15249-15255 [DOI] [PubMed] [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ (2001) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25: 399-406 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL (2002) The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol 242: 161-173 [DOI] [PubMed] [Google Scholar]
- Ponce MR, Pérez-Pérez JM, Piqueras P, Micol JL (2000) A multiplex reverse transcriptase-polymerase chain reaction method for fluorescence-based semiautomated detection of gene expression in Arabidopsis thaliana. Planta 211: 606-608 [DOI] [PubMed] [Google Scholar]
- Ponce MR, Quesada V, Micol JL (1998) Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J 14: 497-501 [DOI] [PubMed] [Google Scholar]
- Ponce MR, Robles P, Micol JL (1999) High-throughput genetic mapping in Arabidopsis thaliana. Mol Gen Genet 261: 408-415 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Krishna P, Olsen LJ (2001) Hsp90-binding immunophilins in plants: the protein movers. Trends Plant Sci 6: 54-58 [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (1999) OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett 461: 101-106 [DOI] [PubMed] [Google Scholar]
- Rédei GP (1962) Supervital mutants of Arabidopsis. Genetics 47: 443-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Buchner J (2001) Hsp90: chaperoning signal transduction. J Cell Physiol 188: 281-290 [DOI] [PubMed] [Google Scholar]
- Robles P, Micol JL (2001) Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol Genet Genomics 266: 12-19 [DOI] [PubMed] [Google Scholar]
- Schiene-Fischer C, Yu C (2001) Receptor accessory folding helper enzymes: the functional role of peptidyl prolyl cis/trans isomerases. FEBS Lett 495: 1-6 [DOI] [PubMed] [Google Scholar]
- Seeling JM, Miller JR, Gil R, Moon RT, White R, Virshup DM (1999) Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283: 2089-2091 [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771-1783 [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena J, Candela H, Robles P, Ponce MR, Pérez-Pérez JM, Piqueras P, Micol JL (2000) Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156: 1363-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena J, Robles P, Ponce MR, Micol JL (1999) Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol Gen Genet 261: 725-739 [DOI] [PubMed] [Google Scholar]
- Sieberer T, Seifert GJ, Hauser MT, Grisafi P, Fink GR, Luschnig C (2000) Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr Biol 10: 1595-1598 [DOI] [PubMed] [Google Scholar]
- Sieburth LE (1999) Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol 121: 1179-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171-182 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH (1995) The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev 9: 97-107 [DOI] [PubMed] [Google Scholar]
- Terol J, Bargues M, Carrasco P, Pérez-Alonso M, Paricio N (2002) Molecular characterization and evolution of the protein phosphatase 2A B′ regulatory subunit family in plants. Plant Physiol 129: 808-822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T (2002) Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193-196 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Brown MI, Skibinski G, Langdale JA (1999) Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol 121: 1163-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M, Langdale J (1998) The formation of leaves. Curr Opin Plant Biol 1: 43-48 [DOI] [PubMed] [Google Scholar]
- Ueda K, Okamura N, Hirai M, Tanigawara Y, Saeki T, Kioka N, Komano T, Hori R (1992) Human P-glycoprotein transports cortisol, aldosterone, and dexamethasone, but not progesterone. J Biol Chem 267: 24248-24252 [PubMed] [Google Scholar]
- Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C (1998) Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol Cell Biol 18: 3034-3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucich VA, Gasser CS (1996) Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol Gen Genet 252: 510-517 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505-513 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380-383 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377-383 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Zhu JK (2002) Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J Biol Chem 277: 8588-8596 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-García S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181-191 [DOI] [PubMed] [Google Scholar]
- Young JC, Obermann WM, Hartl FU (1998) Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem 273: 18007-18010 [DOI] [PubMed] [Google Scholar]