Abstract
Familial hypercholesterolemia (FH) is an autosomal dominant disorder of lipoprotein metabolism caused mainly by mutations in the low-density lipoprotein receptor (LDLR) and apolipoprotein B 100 (APOB) genes. Until now, the molecular basis of FH has been demonstrated in detail in many populations, but there is still very limited Molecular data concerning FH in Iran. The aim of this study was to characterize the LDLR and APOB gene mutations in an Iranian population. A total of 30 non-related Iranian possible FH subjects were studied. Diagnosis of FH was based on the Dutch Lipid Clinic Network diagnostic criteria. All samples were initially tested for three common APOB gene mutations including R3500Q, R3500 W and R3531C using PCR-RFLP assay. Subsequently, promoter and coding region of the LDLR gene was screened by PCR-SSCP analysis and positive results were confirmed by DNA sequencing. Four previously reported polymorphisms 1413G > A, 1725C > T, 1773T > C and 2140 + 5G > A were found in ~17% (5/30) of population studied. Moreover, no variation was found in APOB gene. Our data indicated that LDLR and APOB gene mutations have not contribution to possible FH in Iranian population studied here. However, we examined three common APOB mutations and LDLR in only 30 patients, and to determine the role of these genes in developing FH in Iran, more FH samples and populations needed to be investigated for the mutations of the related genes.
Keywords: FH, APOB, LDLR, Iran
Introduction
Familial Hypercholesterolemia (FH; MIM# 143890) is a common autosomal dominant disorder of lipoprotein metabolism with a frequency of 1 in 500 of the heterozygous and 1 in a million of homozygous form. FH is characterized by elevations in LDL cholesterol, tendon xanthomata, arcus cornea and increased risk of coronary heart disease. The homozygous form of FH is much more severe and depending on the type of mutation may cause death in the early decades of life [1, 2]. Several different factors may affect the manifestation of FH such as age, gender, diet, type of LDLR mutations and other gene mutations [3–5].
Mutation in the gene encoding the low-density lipoprotein receptor (LDLR) is the most common genetic cause of FH (http://www.ucl.ac.uk/fh). The LDL receptor protein is a cell-surface protein which mediates specific uptake and degradation of LDL by its endocytosis, mainly in liver. A wide variety of mutations including insertions, deletions, nonsense and missense mutations has been described in patients with FH. LDLR gene (MIM# 606945) is located on chromosome 19 (19p13.3) and consists of 18 exons which spans ~45 kb with a mature protein of 860 amino acids [6, 7].
Mutation in apolipoprotein B-100 gene (APOB100; MIM 107730) is the second cause of the clinical FH phenotype. This gene is located on chromosome 2p24.1 that codes for the protein component of LDL particles [8, 9]. ApoB-100 is an integral component of LDL and functions as the ligand for the LDLR. Therefore mutations in the APOB 100 will drastically alter its functional activity leading to a decrease in its binding to LDLR, thereby delaying the clearance of LDL particles. The latter situation usually results in a mild or severe form of hypercholesterolemia together with an increased risk for early onset atherosclerosis [10, 11]. In contrast to LDLR, only a small number of functional mutations have been identified in APOB gene such as R3500Q [12], R3500 W [13], and R3531C [14]. There are very limited data regarding LDLR and APOB gene mutations in Iranian population [15, 16]. The aim of this study was to characterize the LDLR and three common apolipoprotein B 100 gene mutations in a group of 30 non-related Iranian possible FH patients.
Materials and Methods
Subjects
Thirty unrelated possible FH subjects were included based on the Dutch Lipid Clinic Network diagnostic criteria [17] from the 92 suspected FH individuals referred to our affiliated clinics in Shahrekord University of Medical Sciences in 2008. A total of 30 non-related Iranian possible FH subjects were studied. All patients were taking lipid-lowering drug. Subjects (21 males and 9 females) were from Chaharmahal va Bakhtiari province in southwest Iran with high serum cholesterol >240 mg/dl and LDL cholesterol >190 mg/dl aged between 29 and 73 years (mean: 55 years) (Table 1). All patients were clinically characterized and medical history were collected by a questionnaire. Secondary causes of hypercholesterolemia were excluded, such as diabetes mellitus (overnight fasting blood sugar >126 mg dl−1), hypertension (systolic blood pressure/diastolic blood pressure >140/90 mmHg), family history of premature coronary artery disease in first degree relatives and smoking habit. Blood samples were collected after obtaining informed consent from all patients. Glucose, total cholesterol (TC) and triglycerides (TG) were assayed using standard enzymatic procedures. HDL-C and LDL-C were measured with direct method. The study protocol was approved by the Review Board of the University.
Table 1.
Clinical and biochemical characteristics of the patients studied
| Parameters | Patients (N = 30) Mean ± SD | Men (N = 9) Mean ± SD | Women (N = 21) Mean ± SD |
|---|---|---|---|
| Age | 55.10 ± 13.16 | 57.33 ± 17.93 | 54.14 ± 10.93 |
| Sex | 30 | 9 | 21 |
| Total cholesterol (mg/dl) | 302.93 ± 34.72 | 304.22 ± 47.27 | 302.38 ± 29.21 |
| Triglycerides (mg/dl) | 229.96 ± 112.05 | 242.55 ± 196.58 | 224.57 ± 51.46 |
| HDL-cholesterol (mg/dl) | 42.52 ± 13.07 | 40.00 ± 13.36 | 43.78 ± 13.25 |
| LDL-cholesterol (mg/dl) | 208.47 ± 26.59 | 210.50 ± 33.79 | 207.53 ± 24.12 |
| Cholesterol/HDL ratio | 7.47 ± 1.98 | 8.02 ± 2.44 | 7.19 ± 1.74 |
Molecular Analyses
Genomic DNA was extracted from peripheral blood using phenol–chloroform procedure.
All of the Primers for the LDLR and APOB gene were designed using primer3 software. To exclude the APOB mutations, all samples were initially tested for three APOB gene mutations including R3500Q, R3500 W and R3531C using PCR-RFLP assay. The Primer sequences and restriction enzymes used for each mutation are shown in Table 2.
Table 2.
PCR primers and restriction enzymes used for the APOB gene mutations analysis
| Mutation | Forward primer 5′–3′ | Reverse primer 5′–3′ | Size (bp) | Res. enzyme |
|---|---|---|---|---|
| R3500Q | CTTACTTTTCCATTGAGTACTCTACC | AGTGCCCTGCAGCTTCACTGAGTAC | 143 | ScaI |
| R3500 W | CTTACTTTTCCATTGATGCATC | GTAAGTGGTTTTTCGTCATGTG | 251 | NlaIII |
| R3531C | CTTACTTTTCCATTGATGCATC | GTAAGTGGTTTTTCGTCATGTG | 251 | NsiI |
The underlined nucleotides showing the mismatch bases created to introduce a new recognition site for each desired mutation providing a within-assay control for the exclusion of false negative results
The PCR amplification was performed at annealing temperature of 64°C using primers described in Table 2 in a total volume of 25 μl. Ten micro liters of each PCR product were digested overnight with 5 U (for NlaIII) and 10 U (for ScaI and NsiI) of restriction enzyme in a volume of 20 μl. The digested products were separated on 6% polyacrylamide gel at 40 mA for 1.5 h and visualized by silver staining method.
To exclude the LDLR mutations, all of the 18 exons and promoter region of the LDLR gene were PCR amplified at annealing temperature of 51–67°C. The Primer sequences used for exons and promoter will be available in request.
The amplified products were then separated on 6% polyacrylamide gel and visualized by silver staining method. Following PCR amplification, 5 μl of PCR product was mixed with 5 μl SSCP loading solution and was made single stranded by heating at 95°C for 10 min followed by rapid chilling on ice. PCR-SSCP was carried out using different gel concentration, component, current and gel temperature (LDLR primers and PCR-SSCP conditions will be available in request). Afterwards, the gels were visualized by silver staining method. Fragments showing mobility shifts of single strands were sequenced directly using an ABI 3100 DNA Sequencer (Applied Biosystems).
Results
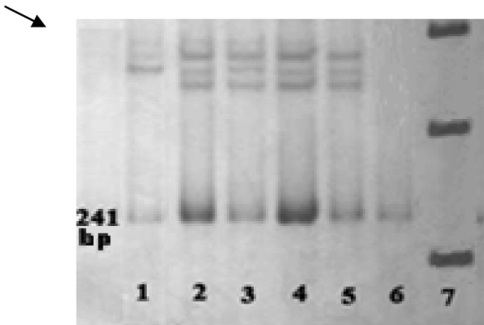
A total of 92 suspected FH individuals were investigated, from which 30 possible FH subjects met the Dutch Lipid Clinic Network diagnostic criteria (Table 1). All samples were initially tested for three APOB gene mutations including R3500Q, R3500 W and R3531C using PCR-RFLP assay. None of the three APOB gene mutations (R3500Q, R3500 W and R3531C) were detected in possible FH patients using PCR-RFLP assay. To exclude the LDLR mutations, all of the 18 exons and promoter region of the LDLR gene were examined using PCR-SSCP strategy. However, four previously reported LDLR polymorphisms including 1413G > A, 1725C > T, 1773T > C and 2140 + 5G > A were found in 2, 1, 1 and 1 affected individuals respectively. A gel picture of SSCP of exon 10 is showed in Fig. 1.
Fig. 1.
Polyacrylamide gel of SSCP for exon 10. Lane 1 mobility shifts of single strands, Lane2–5 normal samples, Lane 6 double strand PCR product, Lane 7 size marker
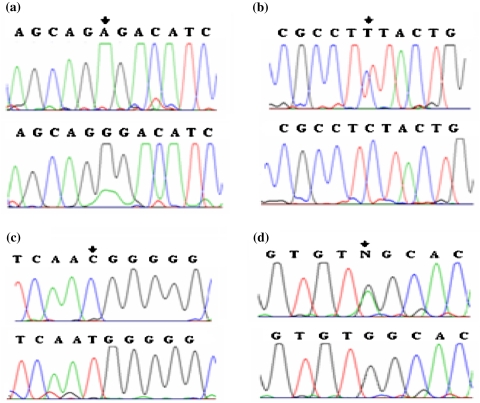
The polymorphisms 1413G > A, 1725C > T, 1773T > C and 2140 + 5G > A were found in 17% of patients from which 1413G > A and 1773T > C were found in both allele in 6 of 60 chromosomes (Fig. 2).
Fig. 2.
Electropherograms of LDLR gene polymorphisms a 1413G > A polymorphism, b 1725 C > T polymorphism, c 1773 T > C polymorphism, d 2140 + 5G > A polymorphism
Discussion
The present study revealed four previously reported LDLR gene polymorphisms including 1413G > A, 1725C > T, 1773T > C and 2140 + 5G > A among 30 Iranian possible FH patients. LDLR gene polymorphisms were found in 17% of individuals studied. But no mutation was detected in promoter and coding region of the LDLR gene studied. Also none of the three APOB gene mutations including R3500Q, R3500 W and R3531C were detected using PCR-RFLP assay. This is similar to the very low detection rate reported by other investigators that examined only one exon [16] of the LDLR gene and one common mutation of APOB (R3500Q) in 30 FH Iranian patients [15]. They found one allelic variation (445G > T) suggesting to be possible mutation.
The frequency and variants of LDLR and APOB gene mutations are different in populations.
The frequency of familial detective apolipoprotein apoB100 (FDB) is estimated as (1/200) in central Europe (e.g. Switzerland), decreasing gradually in Mediterranean or Northern European populations. In Germany, UK and USA, prevalence ranges from 1/700 to 1/500 [18, 19]. In most countries in Europe in 3–5% of patients the hypercholesterolemia is caused by a single mutation in the gene for ApoB [20]. The R3500Q mutation could not be found in Lebanon [21], Russia [22] and Turkey [23].
Mutations in the APOB gene were not detected in any of our patients. A similar result was also obtained in a previous study in Iran [15]. Our finding about R3500Q mutation is quite expected to the geographic distribution of this mutation. In our present study, not finding any mutation in APOB gene may be due to its low frequency rate.
A frequency of FH ranging from 1/411 (0.24%) for North Karelians of Finland [24] to 1/67 (1.5%) for Ashkenazi Jew in South Africa [25]. The frequency of FH is 1/900 (0.11%) for Japanese in Asia [26]. Except one report in Iranian HF patients, there is no report about the molecular basis of FH in Iran. In this report [16] from 30 clinically diagnosed heterozygous FH patients, one new variant (445G > T) was reported. In a case report, an Iranian boy with autosomal recessive hypercholesterolemia was described too [27].
In present study, except four polymorphisms such as 1413G > A, 1725C > T, 1773C > T and 2140 + 5G > A, no mutation was found in exons studied. In total LDLR gene alterations (polymorphisms) were found in 17% of population studied. In our present study, not finding any mutation maybe due to some factors:
In present study, no finding any mutation maybe due to small number of patient. As we examined small sample size of 30 patients, more samples must be tested to reveal the contribution of this gene in causing FH in Iranian families particularly in Chaharmahal va Bakhtiari province.
In some patients, mutations may not be present in the LDLR or APOB gene but occur in other genes such as PCSK9 or in the unidentified FH3 gene on chromosome 1 [28].
Acknowledgments
We would like to thank all the individuals in Chaharmahal va Bakhtiari province for their contribution to this study. This study was supported by Shahrekord University of Medical Sciences, Shahrekord, Iran.
References
- 1.Goldstein JL, Hobbs HH, Brown MS. Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle I, editors. The metabolic basis of inherited disease. 8. NewYork: McGraw-Hill; 2001. pp. 2863–2913. [Google Scholar]
- 2.Mahley RW, Weisgraber KH, Farese RV. Disorders of lipid metabolism. In: Wilson JD, Foster DW, Kronenberg HM, Larsen PR, editors. William’s textbook of endocrinology. 9. Philadelphia: WB Saunders Co; 1998. pp. 1125–1128. [Google Scholar]
- 3.Vohl MC, Gaudet D, Moorjani S, Tremblay G, Perron P, Gagné C, et al. Comparison of the effect of two low-density lipoprotein receptor class mutations on coronary heart disease among French-Canadian patients heterozygous for familial hypercholesterolaemia. Eur J Clin Invest. 1997;27(5):366–373. doi: 10.1046/j.1365-2362.1997.1250669.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoeg JM. Homozygous familial hypercholesterolemia: a paradigm for phenotypic variation. Am J Cardiol. 1993;72:11D–14D. doi: 10.1016/0002-9149(93)90004-V. [DOI] [PubMed] [Google Scholar]
- 5.Kotze MJ, Villiers WJ, Steyn K, Kriek JA, Marais AD, Langenhoven E, et al. Phenotypic variation among familial hypercholesterolemics heterozygous for either one of two Afrikaner founder LDL receptor mutations. Arterioscler Thromb. 1993;13(10):1460–1468. doi: 10.1161/01.ATV.13.10.1460. [DOI] [PubMed] [Google Scholar]
- 6.Lindgren V, Luskey KL, Russell DW, Francke U. Human genes involved in cholesterol metabolism: chromosomal mapping of the loci for the low density lipoprotein receptor and 3-hydroxy-3-methylglutaryl-coenzyme A reductase with cDNA probes. Proc Natl Acad Sci USA. 1985;82:8567–8571. doi: 10.1073/pnas.82.24.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 8.Knott TJ, Rall SC, Jr, Innerarity TL, Jacobson SF, Urdea MS, Levy-Wilson B, et al. Human apolipoprotein B: structure of carboxyl terminal domains, sites of gene expression, and chromosomal localization. Science. 1985;230:37–43. doi: 10.1126/science.2994225. [DOI] [PubMed] [Google Scholar]
- 9.Law SW, Lackner KJ, Hospattankar AV, Anchors JM, Sakaguchi AY, Naylor SL, et al. Human apolipoprotein B-100: cloning, analysis of liver mRNA, and assignment of the gene to chromosome 2. Proc Natl Acad Sci USA. 1985;82:8340–8344. doi: 10.1073/pnas.82.24.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 11.Boren J, Ekstrom U, Agren B, Nilsson-Ehle P, Innerarity TL. The molecular mechanism for the genetic disorder familial defective apolipoprotein B100. J Biol Chem. 2001;276:9214–9218. doi: 10.1074/jbc.M008890200. [DOI] [PubMed] [Google Scholar]
- 12.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86(2):587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaffney D, Reid JM, Cameron IM, Vass K, Caslake MJ, Shepherd J, et al. Independent mutations at codon 3500 of the apolipoprotein B gene are associated with hyperlipidemia. Arterioscler Thromb Vasc Biol. 1995;15(8):1025–1029. doi: 10.1161/01.ATV.15.8.1025. [DOI] [PubMed] [Google Scholar]
- 14.Pullinger CR, Hennessy LK, Chatterton JE, Liu W, Love JA, Mendel CM, et al. Familial ligand-defective apolipoprotein B. Identification of a new mutation that decreases LDL receptor binding affinity. J Clin Invest. 1995;95(3):1225–1234. doi: 10.1172/JCI117772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fard-Esfahani P, Mohammadi-Torabi P, Khatami S, Zeinali S, Taghikhani M, Allahyari M. Familial detective apolipoprotein B 100 : frequency of R3500Q mutation of apolipoprotein B gene in Iranian hypercholesterolemic patients. Acta Medica Iranica. 2005;43:193–196. [Google Scholar]
- 16.Fard-Esfahani P, Zeinali C, Rouhi Dehboneh S, Taghikhani M, Khatami SA. Novel mutation in exon 4 of the low density lipoprotein (LDL) receptor gene in an Iranian familial hypercholesterolemia patient. Iran Biomed J. 2005;9(3):139–142. [Google Scholar]
- 17.World Health Organization. Familial hypercholesterolemia report of a second WHO consultation. Geneva: World Health Organization; 1999. (WHO publication no. WHO/HGN/FH/CONS/99.2).
- 18.Brosseau T, Arveiler D, Cambou JP, Evans AE, Luc G, Fruchart JC, et al. Familial detective apolipoprotein B-100 and myocardial infarction. Atherosclerosis. 1995;116(2):269–271. doi: 10.1016/0021-9150(95)05579-L. [DOI] [PubMed] [Google Scholar]
- 19.Horvath A, Ganev V. The mutation APOB-100 in Eastern Europe. Atherosclerosis. 2001;156(1):241–242. doi: 10.1016/S0021-9150(01)00482-8. [DOI] [PubMed] [Google Scholar]
- 20.Tybjaerg-Hansen A, Humphries S. Familial defective apolipoprotein B-100: a single mutation that causes hypercholesterolaemia and premature coronary artery disease. Atherosclerosis. 1992;96:91–107. doi: 10.1016/0021-9150(92)90056-M. [DOI] [PubMed] [Google Scholar]
- 21.Sabbagh AS, Daher RT, Otrock ZK, Abdel Khalek RN, Zaatari GS, Mahfouz RAR. ApoB-100 R3500Q mutation in the Lebanese population: prevalence and historical review of the literature. Mol Biol Rep. 2007;34(4):267–270. doi: 10.1007/s11033-006-9041-7. [DOI] [PubMed] [Google Scholar]
- 22.Zakharova FM, Damgaard D, Mandelshtam MY, Golubkov VI, Nissen PH, Nilsen GG, et al. Familial hypercholesterolemia in St.-Petersburg: the known and novel mutations found in the low density lipoprotein receptor gene in Russia. BMC Med Genet. 2005;6(6):1–10. doi: 10.1186/1471-2350-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sozen MM, Whittall R, Oner C, Tokatli A, Kalkanoglu HS, Dursun A, Humphries SE, et al. The molecular basis of familial hypercholesterolaemia in Turkish patients. Atherosclerosis. 2005;180(1):63–71. doi: 10.1016/j.atherosclerosis.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 24.Vuorio AF, Turtola H, Piilahti KM, Repo P, Kanninen T, Kontula K. Familial hypercholesterolemia in the Finnish north Karelia. A molecular, clinical, and genealogical study. Arterioscler Thromb Vasc Biol. 1997;17:3127–3138. doi: 10.1161/01.ATV.17.11.3127. [DOI] [PubMed] [Google Scholar]
- 25.Seftel HC, Baker SG, Jenkins T, Mendelsohn D. Prevalence of familial hypercholesterolemia in Johannesburg Jews. Am J Med Genet. 1989;34:545–547. doi: 10.1002/ajmg.1320340418. [DOI] [PubMed] [Google Scholar]
- 26.Mabuchi H, Haba T, Ueda K, Ueda R, Tatami R, Ito S, et al. Serum lipids and coronary heart disease in heterozygous familial hypercholesterolemia in the Hokuriku district of Japan. Atherosclerosis. 1977;28:417–423. doi: 10.1016/0021-9150(77)90068-5. [DOI] [PubMed] [Google Scholar]
- 27.Rodenburg J, Wiegman A, Vissers MN, Kastelein JJP, Stalenhoef AFH. A boy with autosomal recessive hypercholesterolaemia. Neth J Med. 2004;62(3):89–93. [PubMed] [Google Scholar]
- 28.Hunt SC, Hopkins PN, Bulka K, McDermott MT, Thorne TL, Wardell BB, et al. Genetic localization to chromosome 1p32 of the third locus for familial hypercholesterolemia in a Utah kindred. Arterioscler Thromb Vasc Biol. 2000;20:1089–1093. doi: 10.1161/01.ATV.20.4.1089. [DOI] [PubMed] [Google Scholar]