Abstract
The pathogenesis of idiopathic nephrotic syndrome is not completely understood. We postulate that cytokine gene polymorphisms may influence susceptibility or clinical course in Idiopathic Nephrotic Syndrome. Polymorphisms of IL-4, IL-6, and TNF-α cytokines were investigated in 150 children with Idiopathic Nephrotic Syndrome and 569 healthy controls by using polymerase chain reaction and restriction fragment length polymorphism. On comparing patient with controls strong association were found for IL-6, TNF-α and IL-4 at allelic level (IL-6-G174C (G vs. C): P = <0.001; OR = 6.33, TNF-α-G308A (G vs. A): P = <0.001; OR = 1.99, IL-4-C590T (C vs. T): P = 0.048; OR = 1.38). Further when SR group was compared with SS group significant association was found at genotypic level in all the studied genetic polymorphisms. Studied cytokine gene polymorphisms may influence susceptibility to idiopathic nephrotic syndrome and might affect steroid response in INS patients.
Keywords: Idiopathic nephrotic syndrome, IL-6, IL-4, TNF-α, PCR, RFLP
Introduction
Nephrotic syndrome (NS) is a glomerular disease that is characterized by heavy proteinuria, hypoalbuminemia, hypercholesterolemia, generalized edema, and a relapse/remission course, without histological evidence of classical inflammatory immune-mediated injury. Although its pathogenesis remains to be elucidated, there are some evidences which suggest that Idiopathic Nephrotic Syndrome (INS) is primary immune disease associated with immuno-regulatory imbalance between T helper subtype 1 (Th1) and T helper subtype 2 (Th2) cytokines [1]. Patients with MCNS often display a defect in delayed-type hypersensitivity (DTH) response, suggesting an abnormal Th1-dependent cellular immunity [2]. Th1 cells produce IL-2, IFN-γ and tumor necrosis factor-beta (TNF-β), and promote both macrophage activation resulting into DTH, and production of complement-fixing and opsonizing antibodies. Th2 cells synthesize IL-4, IL-5, IL-6, IL-10 and IL-13 provide optimal help for antibody production, and promote both mast cell growth and eosinophil differentiation and activation causing humoral responses [2]. INS is considered to be an immune mediated disease still the contributions of Th1 and Th2 cytokines is a matter of debate. The close relationship between atopy and MCNS suggests a common immune pathway. Stimuli, such as allergens, may activate common immune mechanisms, which subsequently result in proteinuria in children with MCNS. Several investigators have demonstrated the potential role of Th2 cytokines in this disease [3–6].
Cytokines play a critical role as mediators of inflammation and as progressive factors in INS, various cytokines are considered as prime candidates for mediating INS progression [7–9]. Recent studies on several cytokine gene polymorphisms like, interleukin-1b (IL-1b), interleukin-1 receptor antagonist (IL-1ra), and tumor necrosis factor-a (TNF-α), have shown an association with various inflammatory diseases, including glomerulonephritis, ankylosing spondylitis, and multiple sclerosis [10–13].
The cytokines selected in the present study were IL-6-G174C (db SNP ID rs1800795), IL-4-C590T (db SNP ID rs2243250) and TNF-α-G308A (db SNP ID rs1800629). The reason for selecting these genes was that the single nucleotide polymorphism of these genes may influence susceptibility or clinical course of the disease. This study was conducted on 150 INS patients and 569 controls.
Materials and Methods
150 children with NS (Male = 106, Female = 44) who attended the nephrology clinic from January 2006 to March 2009; and 569 age and sex matched healthy children as controls were included in this study. The mean age at diagnosis of NS was 4.8 ± 3.4 years (range: 1–18 years). An informed written consent was obtained from both patients and controls as per the institute guidelines. If the children were less than 15 years of age, informed written consent was obtained from the parents or their guardian. The study was approved by the Ethical committee of the Institute.
Nephrotic syndrome was defined as per the diagnostic criteria laid by International Study of Kidney Disease in Children (ISKDC) as mentioned in our earlier study [14].The NS in children was defined as proteinuria of 40 mg/m2/h or spot urine protein (mg)/creatinine (mg) ratio of 2 in first morning urine sample. The remission of NS was defined by urinary protein excretion <4 mg/m2/h or urine dipstix nil/trace for three consecutive days. The relapse was defined by urinary protein excretion >40 mg/m2/h or urine dipstix ++ or more for three consecutive days. Frequent relapses were defined by 2 or more relapses within 6 months of initial response or four or more relapses within any 12 months period. The steroid dependence was defined by two consecutive relapses occurring during the period of steroid taper or within 14 days of its cessation. Patients with poor compliance and not on regular follow-up were excluded from the study. Renal biopsies were carried out in all steroid resistant cases. The NS patients were categorized in two 2 groups: Group 1: Steroid responsive group (Infrequent relapser’s (IFR), Frequent relapser’s (FR) and Steroid Dependent (SD)) and Group 2: Steroid resistant group. The genotyping was performed in all cases and controls selected for the study.
All children were subjected to a detailed history and physical examination. In addition, the following biochemical tests were done to confirm the diagnosis of nephrotic syndrome—serum creatinine, total protein, albumin, cholesterol, triglycerides and urinary routine and microscopy and urine protein to creatinine ratio in a spot sample.
Blood Collection and DNA Extraction
Blood samples for measuring serum biochemical parameters were obtained in the morning after 8 h of fasting. 3 ml of venous blood was collected in EDTA vials. Genomic DNA was extracted from the whole blood using a commercially available genomic DNA purification kit (Qiagen kit).
Analysis of the IL-6, IL-4 and TNF-α Genotype
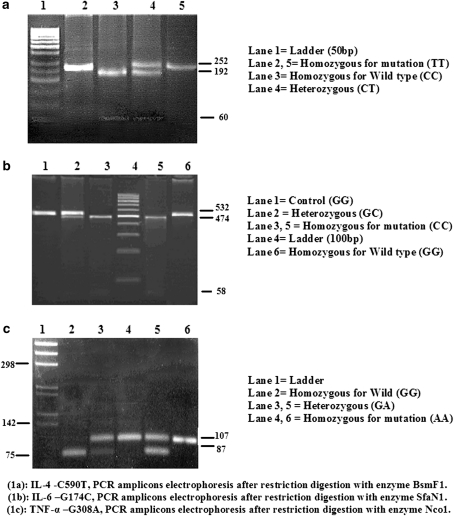
IL-6, IL-4 and TNF-α were genotyped by PCR-RFLP method (Fig. 1a, b and c) in accordance with Tischendorf et al. [15], Kobayashi et al. [16] and Wilson et al. [17]. To improve the genotyping quality and validation, Random samples were reanalyzed, in a double blind manner to confirm the RFLP results.
Fig. 1.
Gel photograph of IL-4, IL-6 and TNF-α after restriction digestion
Statistical Analysis
Statistical analysis was carried out using the GraphPad Prism program (version 2.01, USA). Quantitative variables are presented as the mean ± standard deviation. Comparisons of Quantitative variables between two independent groups were done by Student unpaired t test. All statistical tests were two-tailed, and P < 0.05 was considered as the level of significance. The χ2 test was employed to perform univariate analysis of the association of each polymorphism with NS and categorical clinical features singly and in combinations. Allelic and genotypic frequencies were estimated by genotype count. The association between genotypes and clinical characteristics was expressed as odds ratio (OR) with 95% confidence interval (95% CI). Yate’s corrections were applied wherever, required.
Results
Biochemical and Demographic Profile
The demographic and biochemical profiles of both INS patient and control groups are shown in Table 1. Table 1 showed that there is a significant difference between the INS patients and controls at all the biochemical parameters except age, serum creatinine and male to female ratio.
Table 1.
Biochemical and demographic characteristics of nephrotic patients and controls
| Parameters (SI) | Patients = 150 (mean ± SD) | Controls = 569 (mean ± SD) | P value |
|---|---|---|---|
| Age | 11.0 ± 6.6 years | 12.0 ± 3.5 years | 0.075 |
| S. Alb. (mg/dl) | 2.7 ± 0.7 | 3.9 ± 0.2 | <0.001* |
| S. Prot. | 6.5 ± 1.1 | 6.9 ± 0.3 | <0.001* |
| S. Calc. (mmol/l) | 8.2 ± 0.8 | 9.6 ± 0.4 | <0.001* |
| Hb | 11.2 ± 1.4 | 13.5 ± 1.1 | <0.001* |
| S. URIC (mg/dl) | 5.1 ± 0.8 | 4.9 ± 1.0 | 0.010* |
| S. BUN (mg/dl) | 16.2 ± 7.7 | 9.8 ± 2.7 | <0.001* |
| S. ALKP (U/l) | 239.3 ± 92.8 | 87.8 ± 16.7 | <0.001* |
| S. Na (mmol/l) | 135.7 ± 8.5 | 138.4 ± 2.6 | 0.002* |
| S. K (mmol/l) | 4.7 ± 0.6 | 3.7 ± 0.24 | <0.001* |
| S. PHOS (mg/dl) | 4.4 ± 1.0 | 3.7 ± 0.3 | <0.001* |
| S. Creatinine (mg/dl) | 0.81 ± 0.7 | 0.81 ± 0.2 | 1.00 |
| Proteinuria | 2.9 ± 1.1 | 0.33 ± 0.6 | <0.001* |
| Gender % (male/female) | 106 (70.7%)/44 (29.3) | 399 (70.2)/170 (29.8) | Ns |
* Significant value (P < 0.05), Ns not significant
S. Alb serum albumin, S. Prot. serum protein, S. Calc. serum calcium, Hb Hemoglobin, S. URIC serum uric acid, S. BUN serum blood urea nitrogen, S. ALKP serum alkaline phosphatase, S. Na serum sodium, S. K serum potassium, S. PHOS serum phosphorus, S. Creatinine serum creatinine
Distribution of IL-6-G174C, IL-4-C590T and TNF-α-G308A Genotypes and Alleles in Patient and Control Group
The frequency distribution of genotypes and alleles of IL-6-G174C, IL-4-C590T and TNF-α-G308A in patient and control groups are summarized in Table 2. Table 2 showed that IL-6 and TNF-α exhibited significantly (P < 0.05) different genotype distribution (IL-6-G174C (GG vs. CC): P = <0.001; OR = 31.40, TNF-α-G308A (GG vs. AA): P = 0.006; OR = 10.55), as well as alleles distribution (IL-6-G174C (G vs. C): P = <0.001; OR = 6.33, TNF-α-G308A (G vs. A): P = <0.001; OR = 1.99) whereas IL-4 exhibit significant difference in distribution of alleles only (IL-4-C590T (C vs. T): P = 0.048; OR = 1.38) when patient group was compared with control group.
Table 2.
Distribution of IL-6-G174C, IL-4-C590T and TNF-α-G308A genotypes and alleles in patient and control group
| Genotype | Patient’s N = 150 (%) | Controls N = 569 (%) | P value | OR (95% CI) |
|---|---|---|---|---|
| IL-6-G174C (db SNP ID rs1800795) polymorphism | ||||
| Genotype frequency | ||||
| GG | 82 (54.7) | 515 (90.5) | Reference genotype | |
| GC | 63 (42.0) | 53 (9.3) | <0.001* | 7.46 (4.84–11.52) |
| CC | 05 (3.3) | 01 (0.2) | <0.001* | 31.40 (3.62–272.3) |
| Allele frequency | ||||
| G | 227 (75.7) | 1083 (95.2) | Reference allele | |
| C | 73 (24.3) | 55 (4.8) | <0.001* | 6.33 (4.33–9.24) |
| IL-4-C590T (db SNP ID rs2243250) polymorphism | ||||
| Genotype frequency | ||||
| CC | 93 (62.0) | 397 (69.8) | Reference genotype | |
| CT | 51 (34.0) | 160 (28.1) | 0.118 | 1.36 (0.92–2.01) |
| TT | 06 (4.0) | 12 (2.1) | 0.131 | 2.13 (0.78–5.84) |
| Allele frequency | ||||
| C | 237 (79.0) | 954 (83.8) | Reference allele | |
| T | 63 (21.0) | 184 (16.2) | 0.048* | 1.38 (1.01–1.90) |
| TNF-α-G308A (db SNP ID rs1800629) polymorphism | ||||
| Genotype frequency | ||||
| GG | 118 (78.7) | 498 (87.5) | Reference genotype | |
| GA | 27 (18.0) | 69 (12.1) | 0.042* | 1.65 (1.01–2.69) |
| AA | 05 (3.3) | 02 (0.4) | 0.006* | 10.55 (2.02–55.07) |
| Allele frequency | ||||
| G | 264 (88.0) | 1065 (93.6) | Reference allele | |
| A | 36 (12.0) | 73 (6.4) | <0.001* | 1.99 (1.31–3.03) |
* Significant value (P < 0.05)
Further synergistic effects of IL-6, IL-4 and TNF-α gene polymorphisms were evaluated as shown in Table 3. Our results indicated that individuals with mutant alleles of IL-6 (C), IL-4 (T), and TNF-α (A) in different combinations were at high risk of disease progression as compared to other combinations (Table 3).
Table 3.
Combined analysis of IL-6, IL-4 and TNF-α genotypes among INS patients and controls
| Genotype | Patient’s N = 150 (%) | Controls N = 569 (%) | P value | OR (95% CI) |
|---|---|---|---|---|
| Double: IL-4-C590T + IL-6-G174C | ||||
| IL-4 (1) & IL-6 (1) | 48 (32.0) | 361 (63.4) | Reference genotype | |
| IL-4 (1) & IL-6 (0) | 45 (30.0) | 36 (6.3) | <0.001* | 9.40 (5.52–16.00) |
| IL-4 (0) & IL-6 (1) | 34 (22.7) | 154 (27.1) | 0.036* | 1.66 (1.03–2.60) |
| IL-4 (0) & IL-6 (0) | 23 (15.3) | 18 (3.2) | <0.001* | 9.61 (4.84–19.09) |
| Double: IL-4-C590T + TNF-α-G308A | ||||
| IL-4 (1) & TNF-α (1) | 70 (46.7) | 345 (60.6) | Reference genotype | |
| IL-4 (1) & TNF-α (0) | 23 (15.3) | 52 (9.1) | 0.005* | 2.18 (1.25–3.79) |
| IL-4 (0) & TNF-α (1) | 48 (32.0) | 153 (26.9) | 0.038* | 1.55 (1.02–2.34) |
| IL-4 (0) & TNF-α (0) | 09 (6.0) | 19 (3.3) | 0.041* | 2.34 (1.01–5.38) |
| Double: IL-6-G174C + TNF-α-G308A | ||||
| IL-6 (1) & TNF-α (1) | 64 (42.7) | 456 (80.1) | Reference genotype | |
| IL-6 (1) & TNF-α (0) | 18 (12.0) | 59 (10.4) | 0.008* | 2.17 (1.21–3.92) |
| IL-6 (0) & TNF-α (1) | 54 (36.0) | 42 (7.4) | <0.001* | 9.16 (5.66–14.82) |
| IL-6 (0) & TNF-α (0) | 14 (9.3) | 12 (2.1) | <0.001* | 8.31 (3.68–18.77) |
| Triple: IL-4-C590T + IL-6-G174C + TNF-α-G308A | ||||
| IL-4 (1) + IL-6 (1) + TNF-α (1) | 35 (23.3) | 320 (56.2) | Reference genotype | |
| IL-4 (0) + IL-6 (0) + TNF-α (1) | 19 (12.7) | 18 (3.2) | <0.001* | 9.65 (4.64–20.09) |
| IL-4 (0) + IL-6 (1) + TNF-α (0) | 05 (3.3) | 19 (3.3) | 0.090 | 2.40 (0.85–6.84) |
| IL-4 (0) + IL-6 (1) + TNF-α (1) | 29 (19.3) | 135 (23.7) | 0.011* | 1.96 (1.15–3.34) |
| IL-4 (1) + IL-6 (0) + TNF-α (0) | 10 (6.7) | 12 (2.1) | <0.001* | 7.62 (3.07–18.91) |
| IL-4 (1) + IL-6 (0) + TNF-α (1) | 35 (23.3) | 24 (4.2) | <0.001* | 13.33 (7.13–24.93) |
| IL-4 (1) + IL-6 (1) + TNF-α (0) | 13 (8.7) | 40 (7.1) | 0.002* | 2.97 (1.45–6.08) |
| IL-4 (0) + IL-6 (0) + TNF-α (0) | 04 (2.7) | 01 (0.2) | <0.001* | 36.57 (3.97–336.5) |
0 mutant + heterozygous, 1 wild type genotype
* Significant value (P < 0.05)
Distribution of IL-6-G174C, IL-4-C590T and TNF-α-G308A Genotypes and Alleles among Idiopathic Nephrotic Syndrome Patients
Out of 150 nephrotic patients 115 (76.7%) were steroid responsive (SS) (28 IFR + 42 FR + 45SD), and 35 (23.3%) were steroid resistant (SR). When SR group was compared with SS group significant association was found at genotypic level in case of IL-6, and IL-4 (IL-6-G174C (GG vs. CC): P = 0.002; OR = 15.29, IL-4-C590T (CC vs. TT): P = 0.020; OR = 6.46) respectively. Where as in case of TNF-α significant association was found both at genotypic (TNF-α-G308A (GG vs. AA): P = 0.002; OR = 14.88) as well as allelic level (TNF-α-G308A (G vs. A): P = 0.025; OR = 2.25) (Table 4).
Table 4.
Distribution of IL-6-G174C, IL-4-C590T and TNF-α-G308A genotypes and alleles among idiopathic nephrotic syndrome patients
| Genotype | Steroid resistant | Steroid sensitive | P value | OR (95% CI) |
|---|---|---|---|---|
| N = 35 (%) | N = 115 (%) | |||
| IL-6-G174C (db SNP ID rs1800795) polymorphism | ||||
| Genotype frequency | ||||
| GG | 17 (48.6) | 65 (56.5) | Reference genotype | |
| GC | 14 (40.0) | 49 (42.6) | 0.828 | 1.09 (0.49–2.43) |
| CC | 04 (11.4) | 01 (0.9) | 0.002* | 15.29 (1.60–146.0) |
| Allele frequency | ||||
| Allele G | 48 (68.6) | 179 (77.8) | Reference allele | |
| Allele C | 22 (31.4) | 51 (22.2) | 0.114 | 1.61 (0.89–2.91) |
| IL-4-C590T (db SNP ID rs2243250) polymorphism | ||||
| Genotype frequency | ||||
| CC | 22 (62.8) | 71 (61.7) | Reference genotype | |
| CT | 09 (25.8) | 42 (36.6) | 0.401 | 0.69 (0.29–1.64) |
| TT | 04 (11.4) | 02 (1.7) | 0.020* | 6.46 (1.11–37.66) |
| Allele frequency | ||||
| Allele C | 53 (75.7) | 184 (80.0) | Reference allele | |
| Allele T | 17 (24.3) | 46 (20.0) | 0.440 | 1.28 (0.68–2.42) |
| TNF-α-G308A (db SNP ID rs1800629) polymorphism | ||||
| Genotype frequency | ||||
| GG | 25 (71.5) | 93 (80.9) | Reference genotype | |
| GA | 06 (17.1) | 21 (18.2) | 0.905 | 1.06 (0.39–2.92) |
| AA | 04 (11.4) | 01 (0.9) | 0.002* | 14.88 (1.59–139.2) |
| Allele frequency | ||||
| Allele G | 56 (80.0) | 207 (90) | Reference allele | |
| Allele A | 14 (20.0) | 23 (10) | 0.025* | 2.25 (1.09–4.66) |
* Significant value (P < 0.05)
Discussion
Recently, several studies have reported that polymorphisms of cytokine genes were associated with the development and severity of inflammatory diseases. Many researchers have evaluated the association with genetic polymorphisms in the IL-4, IL-1ra, IL-1β, IL-6, and TNF-α genes in patients with various inflammatory diseases, including IgA nephropathy, ankylosing spondylitis, and multiple sclerosis [10–13]. TNF-α is pro-inflammatory cytokine, and IL-4 and IL-6 are anti-inflammatory cytokines. Pro-inflammatory cytokines are of central importance in T and B-cell help and initiation of the immune response [18]. Minimal change nephrotic syndrome (MCNS) in children is frequently associated with allergy and immunoglobulin E production. Th2 cytokines, such as interleukin-4 (IL-4) [19], may have an important role in the development of atopy.
In the present study we observed that when patient group was compared with control group IL-6 and TNF-α exhibited significantly different genotype distribution as well as allele distribution whereas IL-4 exhibit significant difference in distribution of alleles only. Further When SR group was compared with SS group significant association was found at genotypic level in case of IL-6, and IL-4 respectively. Where as in case of TNF-α significant association was found both at genotypic as well as allelic level which indicated that genetic polymorphism at IL-6, IL-4 or TNF-α genes may be one of the genetic risk factor for progression of INS and this may affect the steroid response.
Some of the previous studies have suggested that genetic variations in IL-4 may be associated with predisposition to MCNS, and partially to the clinical course of MCNS [16, 20, 21]. IL-4 production by peripheral Th2 cells is up-regulated in patients with MCNS and correlated with the severity of proteinuria [22]. Our results demonstrated, that IL-4-C590T polymorphism may influence the prognosis as well as clinical course of the disease.
Role of IL-6-G174C is not well studied in nephrotic syndrome however, its role has been found in mesangial proliferative glomerulonephritis and end-stage renal disease [23]. There are few studies on recurrent infections, thrombophilia and immunological alterations, such as celiac disease, breast cancer, systemic lupus erythromatosus and leishmaniasis [15, 24–26]. Our study revealed that CC genotype of IL-6-G174C is more prevalent in the INS patient, and steroid resistant group, hence may be considered responsible for progression as well as for steroid resistance.
Tumour necrosis factor alpha (TNF-α) is a potent immunomodulator and pro-inflammatory Th1 cytokine [19] and has been implicated in many pathological processes. The gene coding for this polymorphism is located on the chromosome six in class III region of major histocompatibility complex. Several biallelic polymorphism of this gene are known, including the TNF-α-G308A gene polymorphism, which is the first discovered gene polymorphism. TNF-α is pro-inflammatory cytokine, especially TNF-α carries a central importance in T and B-cell help and initiation of the immune response. Polymorphism at position 308 of the TNF-α promoter, representing G to A base transitions, has been linked to increased TNF transcription [17, 27]. Elevation of TNF-α has been found in the plasma and urine of patients with INS [28]. Earlier studies have shown increase of TNF-α synthesis and gene expression in patients with idiopathic nephrotic syndrome and focal glomerular sclerosis [29, 30]. In our study strong association of TNF-α gene polymorphism was observed in patients with INS and steroid resistance.
We may conclude that studied genetic polymorphisms may influence susceptibility to idiopathic nephrotic syndrome and might affect steroid response in INS patients. However, the role of other genetic and environmental factors cannot be ruled out. The findings of the present study may be a small step put forward. Further studies are required to confirm these findings and to elucidate the role of the IL-6, IL-4 and TNF-α gene polymorphism in development of INS and steroid resistance among Indians.
Acknowledgment
We are indebted to SGPGIMS, Lucknow for financial support.
References
- 1.Mathieson PW. Cytokine polymorphisms and nephrotic syndrome. Clin Sci. 2002;102:513–514. doi: 10.1042/CS20020025. [DOI] [PubMed] [Google Scholar]
- 2.Fodor P, Saitua MT, Rodriguez E, Gonzalez B, Schlesinger L. T-cell dysfunction in minimal-change nephrotic syndrome of childhood. Am J Dis Child. 1982;136:713–717. doi: 10.1001/archpedi.1982.03970440057016. [DOI] [PubMed] [Google Scholar]
- 3.Kimata H, Fujimoto M, Furusho K. Involvement of interleukin (IL)-13, but not IL-4, in spontaneous IgE and IgG4 production in nephrotic syndrome. Eur J Immunol. 1995;25:1497–1501. doi: 10.1002/eji.1830250604. [DOI] [PubMed] [Google Scholar]
- 4.Neuhaus TJ, Wadhwa M, Callard R, Barratt TM. Increased IL-2, IL-4 and interferon-gamma (IFN-gamma) in steroid sensitive nephrotic syndrome. Clin Exp Immunol. 1995;100:475–479. doi: 10.1111/j.1365-2249.1995.tb03725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yap HK, Cheung W, Murugasu B, et al. Th1 and Th2 cytokine mRNA profiles in childhood nephrotic syndrome: evidence for increased IL-13 mRNA expression in relapse. J Am Soc Nephrol. 1999;10:529–537. doi: 10.1681/ASN.V103529. [DOI] [PubMed] [Google Scholar]
- 6.Punnonen J, Aversa G, Cocks BG, et al. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90:3730–3734. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho BS, Yoon SR, Jang JY, Pyun KH, Lee CE. Upregulation of interleukin-4 and CD23/Fcepsilon RII in minimal change nephrotic syndrome. Pediatr Nephrol. 1999;13:199–204. doi: 10.1007/s004670050592. [DOI] [PubMed] [Google Scholar]
- 8.Noronha IL, Niemir Z, Stein H, Waldherr R. Cytokines and growth factors in renal disease. Nephrol Dial Transplant. 1995;10:775–786. [PubMed] [Google Scholar]
- 9.Tenbrock K, Schubert A, Stapenhorst L, Kemper MJ, Gellermann J, Timmermann K, Müller-Wiefel DE, Querfeld U, Hoppe B, Michalk D. Type I IgE receptor, interleukin 4 receptor and interleukin 13 polymorphisms in children with nephrotic syndrome. Clin Sci. 2002;102:507–512. doi: 10.1042/CS20010229. [DOI] [PubMed] [Google Scholar]
- 10.Liu ZH, Cheng ZH, Yu YS, Tang Z, Li LS. Interleukin-1 receptor antagonist allele: Is it a genetic link between Henoch-Schonlein nephritis and IgA nephropathy? Kidney Int. 1997;51:1938–1942. doi: 10.1038/ki.1997.264. [DOI] [PubMed] [Google Scholar]
- 11.Shu KH, Lee SH, Cheng CH, Wu MJ, Lian JD. Impact of interleukin-1 receptor antagonist and tumor necrosis factor alpha gene polymorphism on IgA nephropathy. Kidney Int. 2000;58:783–789. doi: 10.1046/j.1523-1755.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- 12.Paardt M, Crusius JB, Garcia-Gonzalez MA, Baudoin P, Kostense PJ, Alizadeh BZ, Dijkmans BA, Pena AS, Horst-Bruinsma IE. Interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms in ankylosing spondylitis. Rheumatology. 2002;41:1419–1423. doi: 10.1093/rheumatology/41.12.1419. [DOI] [PubMed] [Google Scholar]
- 13.Mann CL, Davies MB, Stevenson VL, Leary SM, Boggild MD, Ko Ko C, Jones PW, Fryer AA, Strange RC, Thompson AJ, Hawkins CP. Interleukin 1 genotypes in multiple sclerosis and relationship to disease severity. J Neuroimmunol. 2002;129:197–204. doi: 10.1016/S0165-5728(02)00181-9. [DOI] [PubMed] [Google Scholar]
- 14.Gulati S, Godbole M, Singh U, Gulati K, Srivastava A. Are children with idiopathic nephrotic syndrome at risk for metabolic bone disease? Am J Kidney Dis. 2003;41:1163–1169. doi: 10.1016/S0272-6386(03)00348-2. [DOI] [PubMed] [Google Scholar]
- 15.Tischendorf JJ, Yagmur E, Scholten D, Vidacek D, Koch A, Winograd R. The interleukin-6 (IL6)-174 G/C promoter genotype is associated with the presence of septic shock and the ex vivo secretion of IL6. Int J Immunogenet. 2007;34:413–418. doi: 10.1111/j.1744-313X.2007.00712.x. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y, Arakawa H, Suzuki M, Takizawa T, Tokuyama K, Morikawa A. Polymorphisms of interleukin-4-related genes in Japanese children with minimal change nephrotic syndrome. Am J Kidney Dis. 2003;42:271–276. doi: 10.1016/S0272-6386(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 17.Wilson AG, di Giovine FS, Blakemore AI, Duff GW. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by NcoI restriction of PCR product. Hum Mol Genet. 1992;1(5):353–6. [DOI] [PubMed]
- 18.Carter DB, Deibel MR, Jr, Dunn CJ, Tomich CS, Laborde AL, Slightom JL, Berger AE, Bienkowski MJ, Sun FF, McEwan RN. Purification, cloning, expression and biological characterization of an interleukin-1 receptor antagonist protein. Nature. 1990;344:633–638. doi: 10.1038/344633a0. [DOI] [PubMed] [Google Scholar]
- 19.Mosmann TR, Cherwinski H, Bond MW. Two types of murine helper T cell clone. 1. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 20.Liu HM, Shen Q, Xu H, Yang Y. Significance of polymorphisms in variable number of tandem repeat region of interleukin-4 gene in recurrence of childhood steroid sensitive nephrotic syndrome. Zhonghua Er Ke Za Zhi. 2005;43:431–433. [PubMed] [Google Scholar]
- 21.Acharya B, Shirakawa T, Pungky A, Damanik P, Massi MN, Miyata M. Polymorphism of the interleukin-4, interleukin-13, and signal transducer and activator of transcription 6 genes in Indonesian children with minimal change nephrotic syndrome. Am J Nephrol. 2005;25:30–35. doi: 10.1159/000083729. [DOI] [PubMed] [Google Scholar]
- 22.Masutani K, Taniguchi M, Nakashima H, Yotsueda H, Kudoh Y, Tsuruya K. Up-regulated interleukin-4 production by peripheral T-helper cells in idiopathic membranous nephropathy. Nephrol Dial Transplant. 2004;19:580–586. doi: 10.1093/ndt/gfg572. [DOI] [PubMed] [Google Scholar]
- 23.Mittal RD, Manchanda PK. Association of interleukin (IL)-4 intron-3 and IL-6-174 G/C gene polymorphism with susceptibility to end-stage renal disease. Immunogenetics. 2007;59:159–165. doi: 10.1007/s00251-006-0182-6. [DOI] [PubMed] [Google Scholar]
- 24.DeMichele A, Martin AM, Mick R, Gor P, Wray L, Klein-Cabral M. Interleukin-6-174G3C polymorphism is associated with improved outcome in high-risk breast cancer. Cancer Res. 2003;63:8051–8056. [PubMed] [Google Scholar]
- 25.Castellucci L, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M. IL6-174 G/C promoter polymorphism influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. J Infect Dis. 2006;194:519–527. doi: 10.1086/505504. [DOI] [PubMed] [Google Scholar]
- 26.Schotte H, Schlüter B, Rust S, Assmann G, Domschke W, Gaubitz M. Interleukin-6 promoter polymorphism (-174 G/C) in Caucasian German patients with systemic lupus erythematosus. Rheumatology. 2001;40:393–400. doi: 10.1093/rheumatology/40.4.393. [DOI] [PubMed] [Google Scholar]
- 27.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;1:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suranyi MG, Guasch A, Hall BM, Myers BD. Elevated levels of tumor necrosis factor-α in the nephrotic syndrome in humans. Am J Kidney Dis. 1993;21:251–259. doi: 10.1016/s0272-6386(12)80742-6. [DOI] [PubMed] [Google Scholar]
- 29.Bustos C, González E, Muley R, Alonso JL, Egido J. Increase of tumor necrosis factor α synthesis and gene expression in peripheral blood mononuclear cells of children with idiopathic nephrotic syndrome. Eur J Clin Investig. 1994;24:799–805. doi: 10.1111/j.1365-2362.1994.tb02022.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto K. Spontaneous and LPS-stimulated release of tumor necrosis factor-alpha by peripheral blood monocytes in patients with focal glomerular sclerosis. Nephron. 1995;70:118–119. doi: 10.1159/000188559. [DOI] [PubMed] [Google Scholar]