Abstract
Voltage-dependent anion channels (VDACs) are generally considered as the main pathway for metabolite transport across the mitochondrial outer membrane. Recent proteomic studies on isolated symbiosome membranes from legume nodules indicated that VDACs might also be involved in transport of nutrients between plants and rhizobia. In an attempt to substantiate this, we carried out a detailed molecular and cellular characterization of VDACs in Lotus japonicus and soybean (Glycine max). Database searches revealed at least five genes encoding putative VDACs in each of the legumes L. japonicus, Medicago truncatula, and soybean. We obtained and sequenced cDNA clones from L. japonicus encoding five full-length VDAC proteins (LjVDAC1.1–1.3, LjVDAC2.1, and LjVDAC3.1). Complementation of a yeast (Saccharomyces cerevisiae) mutant impaired in VDAC1, a porin of the mitochondrial outer membrane, showed that LjVDAC1.1, LjVDAC1.2, LjVDAC2.1, and LjVDAC3.1, but not LjVDAC1.3, are functional and targeted to the mitochondrial outer membrane in yeast. Studies of the expression pattern of the five L. japonicus VDAC genes revealed largely constitutive expression of each throughout the plant, including nodules. Antibodies to LjVDAC1.1 of L. japonicus and the related POM36 protein of potato (Solanum tuberosum) recognized several proteins between 30 and 36 kD on western blots, including LjVDAC1.1, LjVDAC1.2, LjVDAC1.3, and LjVDAC2.1. Immunolocalization of VDACs in L. japonicus and soybean root nodules demonstrated their presence on not only mitochondria but also on numerous, small vesicles at the cell periphery. No evidence was found for the presence of VDACs on the symbiosome membrane. Nonetheless, the data indicate that VDACs may play more diverse roles in plants than suspected previously.
Porins are a diverse group of β-barrel proteins that fulfill a variety of functions in prokaryotes and eukaryotes. They are located in the outer membranes of Gram-negative (Delcour, 2002) and -positive bacteria (Riess et al., 1999, 2001; Lichtinger et al., 1998), mitochondria of eukaryotes (Benz, 1994), and plastids of plants (Fischer et al., 1994; for review, see Bolter and Soll, 2001). In addition to the common β-barrel fold and perhaps related to this pore-forming structure, porins share the electrophysiological property of symmetric voltage gating (Bainbridge et al., 1998). Porins can adopt two conformational states: an open state, which is selective for small anions, and a closed state, in which conductivity decreases and selectivity for cations increases. Eukaryotic porins show voltage gating at potentials of ±20 to 40 mV, whereas an electrical potential of ±100 mV is needed to close certain bacterial porins (Benz, 1994). The physiological significance of voltage gating remains unclear, however, given that porins are generally located in “nonenergized” membranes (Blachly-Dyson and Forte, 2001).
Despite uncertainty about the physiological significance of voltage gating in porins, this feature has been used to name one important family of these proteins, the voltage-dependent anion channel (VDAC) family in eukaryotes. VDAC protein sequence and function have been conserved during evolution. Thus, homologs have been found in yeast (Saccharomyces cerevisiae), animals, and plants, and VDACs from animals (Blachly-Dyson et al., 1993; Xu et al., 1999) can suppress mutations in yeast VDAC1. Genetic studies have shown that yeast VDAC1, which is located in the outer mitochondrial membrane (OMM), is essential for mitochondrial respiration (Dihanich et al., 1987; Lee et al., 1998). Similar data is lacking for plants. Evolution of multicellularity was accompanied by an expansion of the number of VDAC genes in eukaryotic genomes. This makes genetic studies of the physiological role(s) of VDACs in higher eukaryotes intrinsically more difficult than in yeast, which has only two VDAC genes, both encoding proteins of the OMM. Adding to the complexity in higher eukaryotes is the observation that VDAC porins may not be confined to the OMM. In animals, VDACs also appear to be located in the caveolae domains of the plasma membrane (Bathori et al., 1999; Buettner et al., 2000). Caveolae are microdomains that form a unique endocytotic and exocytotic compartment at the cell surface, which is capable of importing molecules and delivering them to specific locations within the cell and exporting molecules to the extracellular space (Anderson, 1998). In plants, VDAC proteins may be located in the boundary membrane of glyoxisomes, in addition to the OMM (Corpas et al., 2000).
We were drawn to the study of VDACs in legumes after learning that they might be located in the peribacteroid or symbiosome membrane (SM) of pea (Pisum sativum) and Lotus japonicus nodules (Saalbach et al., 2002; Wienkoop and Saalbach, 2003). Given the transport properties of VDAC porins and the strategic role of the SM in controlling nutrient exchange between the plant and nitrogen-fixing bacteroids in legume root nodules (Udvardi and Day, 1997), we sought to verify these results, using a method that leaves cell structures intact. Thus, in addition to presenting the molecular and functional characterization of a family of five new legume VDACs, we present data here on the immunolocalization of VDAC proteins in root nodule cells.
RESULTS
Isolation and Sequence Analysis of Full-Length cDNAs Encoding Five Different VDACs in L. japonicus
Amino acid sequence data obtained from proteins of isolated SM from soybean (Glycine max; M. Wandrey, B. Trevaskis, and M.K. Udvardi, unpublished data), pea (Saalbach et al., 2002), and L. japonicus (Wienkoop and Saalbach, 2003) indicated the possible presence of VDACs in this membrane of legume root nodules. Given the broad substrate range of porins that includes dicarboxylic acids (Reumann et al., 1998), which are a primary source of carbon for nitrogen-fixing bacteroids (Udvardi et al., 1988; Udvardi and Day, 1997), we sought to confirm the SM location of these proteins. To facilitate the production of antibodies to VDACs for immunolocalization, we began by isolating partial and then full-length VDAC cDNA from soybean and L. japonicus, respectively. A 400-bp VDAC gene fragment was PCR amplified from soybean nodule cDNA using degenerate primers (forward, 5′-CARTAYYTICAYGARTAYGCNGG-3′; and reverse, 5′-TTIGGICKCCAYTCRTGYTG-3′), designed from conserved regions of plant porins (ZmPor1, GenBank accession no. X73429; PsPor1, GenBank accession no. Z25540; POM34, GenBank accession no. X80386; POM36I, GenBank accession no. X80388; POM36II, GenBank accession no. X80387; and SVDAC1, GenBank accession no. U50900). This gene fragment, which showed 83% identity to PsPor1, was used to screen a nodule cDNA library from L. japonicus at low stringency. Six positive clones were selected and sequenced. All six clones were derived from the same gene, which we named LjVDAC1.1. Subsequent in silico screening of the growing L. japonicus expressed sequence tag (EST) databases (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov; Kazusa DNA Research Institute, Chiba, Japan, http://www.kazusa.or.jp; and The Institute for Genomic Research, http://www.tigr.org) identified potential full-length cDNA clones encoding a further four porins. These were obtained, fully sequenced, and named LjVDAC1.2, LjVDAC1.3, LjVDAC2.1, and LjVDAC3.1. These VDACs correspond to EST clones MWM207c01 (GenBank accession no. AV411556), MWM211g08 (GenBank accession no. AV411841), MWM066g10 (GenBank accession no. AV426384), and MWM239d07 (GenBank accession no. AV414031), respectively. The GenBank accession numbers for the full-length sequences are AY316737 to AY31641, respectively.
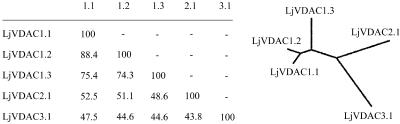
The five L. japonicus VDACs encode proteins that are either 276 or 277 amino acids in length with calculated molecular masses in the range from 29 to 30 kD. They show between 44% to 88% sequence identity and form three main evolutionary branches (Fig. 1).
Figure 1.
Homology of L. japonicus VDACs. Amino acid identities are given in percentages. The multiple sequence alignment and unrooted phylogenetic tree were carried out using ClustalW (Jeanmougin et al., 1998; http://clustalw.genome.ad.jp).
To identify conserved or variable domains that may be involved in secondary structure formation, targeting, or regulation of channel activity, deduced protein sequences of all five clones were aligned. In addition, secondary structure prediction using PROFsec (Rost, 1996), and a pattern search using the PROSITE database (Hofmann et al., 1999) were performed (Fig. 2). Alignment of L. japonicus VDAC proteins revealed highly conserved regions at the N terminus, including a loop (light-gray box under the aligned sequences) and an α-helical region (boxed H). The positions of β-sheets (dark gray) among the different VDACs are also highly conserved, although sequence conservation is not greater than in the rest of the protein. The PROSITE database pattern search revealed a 23-amino acid-long eukaryotic mitochondrial porin pattern (Pattern-ID: PS00558), beginning at position 219, for LjVDAC1.1 to 1.3 (Fig. 2, underlined). This pattern was not identified in LjVDAC2.1 or LjVDAC3.1.
Figure 2.
Comparison of primary and secondary structure of L. japonicus VDACs. The multiple sequence alignment was carried out using ClustalW (Jeanmougin et al., 1998; http://clustalw.genome.ad.jp). Black shading indicates similar amino acids in at least three VDAC sequences. The graphic below the protein sequence alignment displays a comparison of the secondary structure of L. japonicus VDACs in the same order as above. Secondary structure prediction and protein pattern search were performed at the “PredictProtein server” (http://cubic.bioc.columbia.edu/predictprotein) with PROFsec (Rost, 1996) and PROSITE (Hofmann et al., 1999), respectively. Dark-gray boxes, β-Sheets; boxed “H,” α-helical protein structures; light-gray boxes, likely formation of a loop, predicted with an average accuracy of 82%. The underlined sequences show eukaryotic mitochondrial porin signatures (PROSITE Pattern-ID: PS00558) of the LjVDAC1.1, LjVDAC1.2 and LjVDAC1.3 proteins following the pattern [YH]-x(2)-D-[SPCAD]-x-[STA]-x(3)-[TAG]-[KR]-[LIVMF]-[DNSTA]-[DNS]x(4)-[GSTAN]-[LIVMA]-x-[LIVMY].
Phylogenetic Analysis of Plant VDACs
BLAST searches (Altschul et al., 1990) of public databases identified numerous homologs of the L. japonicus VDACs in other higher plants, with sequence identities ranging from 58% to 81%. Searches of the Plant Gene Index at TIGR (http://www.tigr.org) revealed at least five different Tentative Consensus sequences corresponding to VDACs in each of soybean, Medicago truncatula, and L. japonicus. The Arabidopsis genome contains five VDAC genes. Significant homology was also found between L. japonicus VDACs and proteins of other eukaryotes, including mammals, insects, nematodes, amoebae, and fungi. For instance, the VDAC from yeast (VDAC1, GenBank accession no. NP 014343) shares at least 18% amino acid sequence identity with L. japonicus porins. No sequence homology was found with prokaryotic proteins, which is typical for VDAC porins.
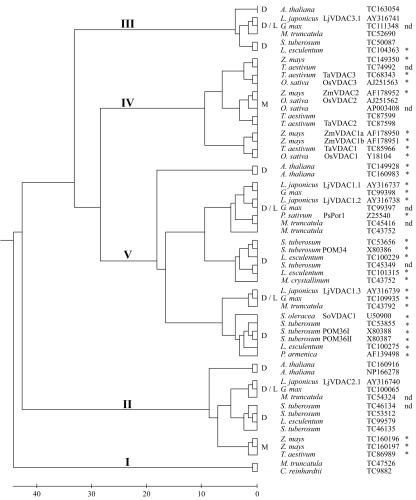
The phylogenetic relationships between VDACs from legumes and other plants are depicted in Figure 3. The dendrogram splits into five main branches, some of which contain monocot or dicot VDACs exclusively. Branch I contains only two plant VDACs, one from the single-celled organism Chlamydomonas reinhardtii and one from the legume M. truncatula. BLAST searches of these proteins revealed that they are more closely related to eukaryotic VDACs of fungi, mammals, and insects. Branch II contains VDACs from both dicotyledonous and monocotyledonous plants. Branches III and IV contain VDACs from dicots and monocots, respectively. Branch V is the largest branch with multiple VDAC isoforms of the dicots L. japonicus, soybean, M. truncatula, and potato (Solanum tuberosum). Some of these may have evolved by gene duplication after the monocot/dicot split, such as the L. japonicus VDACs LjVDAC1.1 and LjVDAC1.2, which exhibit high sequence identity (88%).
Figure 3.
Phylogenetic tree of plant VDACs. Multiple alignment of plant VDAC protein sequences was performed as described in the legend of Figure 1. The phylogenetic tree was calculated applying the neighbor-joining method (Saitou and Nei, 1987; http://clustalw.genome.ad.jp). The scale below indicates the number of amino acid differences calculated with the open source software MEGA2 (http://www.megasoftware.net). VDACs of legumes (L), other dicots (D), and monocots (M) are shown. GenBank accession numbers or tentative consensus sequence (TC) numbers from the TIGR Gene Index (http://www.tigr.org/tdb/tgi/plant.shtml) are given. Proteins marked with an asterisk contain the eukaryotic mitochondrial porin signature (PROSITE ID: PS00558). Cases where incomplete N-terminal sequence precluded identification of the porin signature are also indicated (nd).
VDAC Expression Analysis
Division of labor between different proteins of the same family is often indicated by distinct patterns of gene expression in time and/or space within an organism. To determine whether specialization of L. japonicus VDAC function could be programmed at the level of transcription, we used RNA gel-blot analysis and real-time reverse transcriptase (RT)-PCR to monitor expression in different organs. Northernblot analysis was performed with total RNA from leaves, roots, flowers, and nodules of 8-week-old L. japonicus plants and showed constitutive expression of VDAC genes throughout the plant (data not shown). However, dot-blot analysis with in vitrotranscribed RNA revealed cross-hybridization among cloned LjVDAC genes. Therefore, we employed real-time RT-PCR to increase specificity and sensitivity of transcript detection. Leaf, root, and nodule poly(A+) RNA from 8-week-old plants was reverse transcribed and the resulting cDNA subjected to PCR analysis. Primers were designed to bind to the 3′-untranslated regions of LjVDAC1.1 to 1.3, LjVDAC2.1, and LjVDAC3.1 and tested for their specificity before use. Transcript levels of a polyubiquitin gene, which is expressed constitutively in L. japonicus (Colebatch et al., 2002), were measured to facilitate normalization of data. Real-time RT-PCR revealed only slight differences in transcript levels for the various VDAC genes in different organs (Table 1). LjVDAC2.1 was the most highly expressed and LjVDAC3.1 the least highly expressed of the VDAC genes in different organs. However, transcript levels of LjVDAC2.1 were never more than 10 times higher than those of LjVDAC3.1, and transcripts of each VDAC gene were as abundant as those of the highly expressed ubiquitin gene.
Table I.
Threshold cycle (CT) values and relative abundance of LjVDAC transcripts in different organs
Transcript levels for each gene were measured by real-time RT-PCR, normalized to the level of ubiquitin transcript in the same sample, and then expressed relative to the level of LjVDAC1.1 transcript in leaves (set at 1). CT values (mean ± sd, n = 3) are presented above the relative gene expression level in each case.
| Transcript | Leaf | Root | Nodule |
|---|---|---|---|
| Ubiquitin | 22.40 ± 0.19 | 23.73 ± 0.73 | 22.99 ± 0.07 |
| LjVDAC1.1 | 19.75 ± 0.18 | 20.09 ± 0.25 | 20.92 ± 0.39 |
| Ratio | 1 | 1.99 | 0.67 |
| LjVDAC1.2 | 19.04 ± 0.08 | 20.49 ± 0.12 | 20.66 ± 0.25 |
| Ratio | 1.64 | 1.51 | 0.80 |
| LjVDAC1.3 | 19.28 ± 0.06 | 20.05 ± 0.17 | 19.93 ± 0.19 |
| Ratio | 1.39 | 2.04 | 1.33 |
| LjVDAC2.1 | 19.12 ± 0.11 | 19.20 ± 0.09 | 19.09 ± 0.08 |
| Ratio | 1.55 | 3.69 | 2.37 |
| LjVDAC3.1 | 21.77 ± 0.07 | 22.36 ± 0.12 | 21.43 ± 0.08 |
| Ratio | 0.25 | 0.41 | 0.47 |
Functional Complementation of a Yeast VDAC1 Mutant
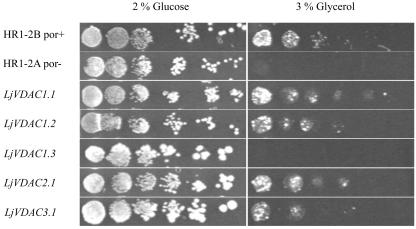
The functionality of LjVDAC1.1 to 1.3, LjVDAC2.1, and LjVDAC3.1 was assessed by complementation of yeast mutant strain HR1-2A (Dihanich et al., 1987), which lacks VDAC1, the VDAC of the OMM. This mutant is impaired in respiration and relies mainly on fermentation for energy production. The metabolic defect results in an inability to grow on non-fermentable carbon sources like glycerol. Transformation of the yeast mutant with L. japonicus porins LjVDAC1.1 and 1.2, LjVDAC2.1, and LjVDAC3.1, cloned in the expression vector p112A1NE (Riesmeier et al., 1992), suppressed the growth defect (Fig. 4). In contrast, LjVDAC1.3 was unable to complement the mutation, although the LjVDAC1.3 protein was expressed in the yeast mutant, as shown by western analysis (Fig. 5, A and B). The results show that all of the L. japonicus VDACs except LjVDAC1.3 are functional in yeast and indicate that four of the five proteins are targeted to the OMM.
Figure 4.
Functional complementation of a yeast VDAC1 mutant with L. japonicus VDACs. The VDAC1-deficient yeast mutant strain HR1-2A was transformed with cDNAs encoding LjVDAC1.1, LjVDAC1.2, LjVDAC1.3, LjVDAC2.1, and LjVDAC3.1, cloned in yeast expression vector p112A1NE. As controls, wild-type yeast strain (HR1-2B) and the VDAC1-deficient mutant (HR1-2A) were transformed with the cloning vector. A series of 10-fold dilutions of overnight cultured cells, starting with an OD600 of 1.0, was made, and 3 μL of cells was dropped onto plates containing minimal medium and 2% (w/v) Glc or 3% (v/v) glycerol as sole carbon sources. Plates containing Glc were incubated for 3 d at 30°C, whereas glycerol plates were incubated for 6 d at 30°C.
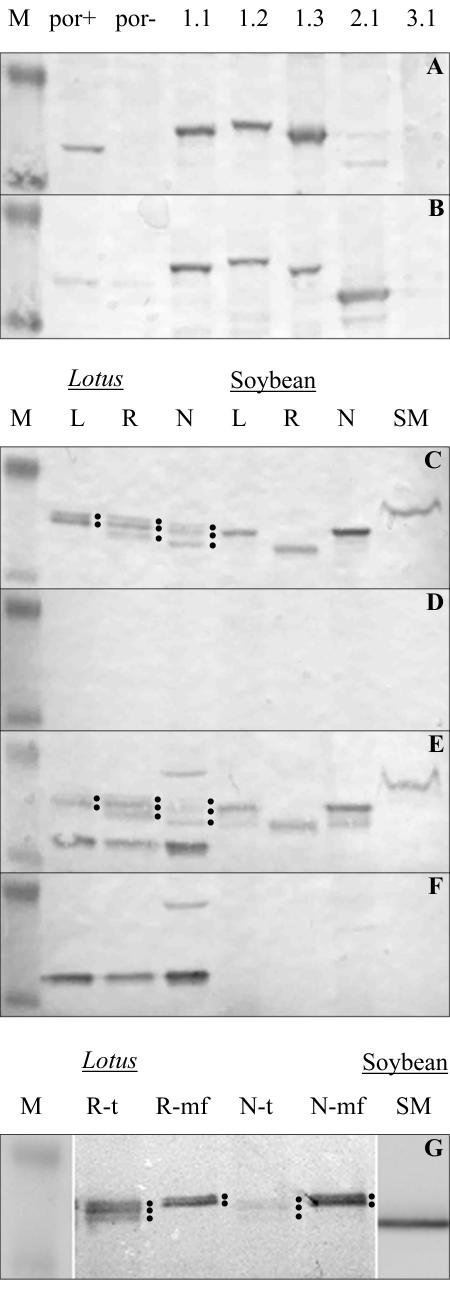
Figure 5.
Western-blot analysis of L. japonicus VDAC expressing yeast and VDACs of L. japonicus and soybean. A and B, L. japonicus VDACs were expressed in a VDAC1-deficient yeast mutant (por-). Approximately 20 μg of total protein was loaded per lane. por+, Strain HR1-2B; por-, strain HR1-2A; 1.1, LjVDAC1.1; 1.2, LjVDAC1.2; 1.3, LjVDAC1.3; 2.1, LjVDAC2.1; 3.1, LjVDAC 3.1. Western blots were incubated with anti-LjVDAC1.1 (A) and anti-POM36 (B) antibody. C to F, Total protein extracts from leaves, roots, and nodules from L. japonicus and soybean and SMs purified via aqueous two-phase partitioning were prepared from 8-week-old plants. Around 20 μg of protein was loaded per lane. L, Leaf; R, root; N, nodule. Western blots were incubated with anti-LjVDAC1.1 (C) and anti-POM36 (E) antibody or with the same antibodies blocked with purified LjVDAC1.1 proteins (D and F, respectively). G, Microsomal fractions of roots and nodules from L. japonicus and SMs from soybean prepared via Percoll density centrifugation were obtained from 8-week-old plants. Around 20 μg of microsomal protein extract and 5 μg of symbiotic membranes were loaded. For comparison, total protein extract of L. japonicus roots and nodules was loaded. VDAC proteins were detected with anti-LjVDAC1.1 antibody. R, Root; N, nodule; t, total protein extract; mf, microsomal fraction. Protein bands representing L. japonicus VDACs are marked with dots. Molecular masses of protein standard are 28 and 39 kD.
Expression of VDAC Proteins in L. japonicus
The analysis of VDAC expression in L. japonicus was extended to the protein level. Western blots were performed with affinity-purified anti-LjVDAC1.1 and anti-POM36 antibodies (Heins et al., 1994). In preliminary experiments with proteins expressed in yeast, it was shown that both antibodies cross-react with LjVDAC1.1, LjVDAC1.2, LjVDAC1.3, and LjVDAC2.1. However, cross-reaction of the anti-LjVDAC1.1 antibody with LjVDAC2.1 was weak. Neither of the two antibodies recognized LjVDAC3.1 (Fig. 5, A and B). Despite the similarities in predicted Mr of the L. japonicus VDACs (29–30 kD), they were separated and distinguishable on western blots. Interestingly, both antibodies recognized yeast VDAC1. Blocking of both antibodies with purified LjVDAC1.1 protein resulted in disappearance of bands corresponding to LjVDAC1.1 to 1.3 and LjVDAC2.1 (data not shown).
The two antibodies were used to detect VDAC proteins in crude extracts and microsomal fractions from different organs of L. japonicus and soybean. Western-blot analysis using anti-LjVDAC1.1 antibody revealed two or three distinct VDAC bands around 32 to 36 kD in extracts of leaves, roots, and nodules (Fig. 5C). In L. japonicus extracts, these bands may correspond to LjVDAC1.1, LjVDAC1.2, and LjVDAC1.3. The anti-POM36 antibody recognized these and two additional bands at 30 and 38 kD (Fig. 5E). Both antibodies also cross-reacted with soybean VDACs. Specificity of the antibodies was tested using antibodies that were blocked with purified LjVDAC1.1 protein. Blocked anti-LjVDAC1.1 antibody detected no proteins on western blots of L. japonicus or soybean extracts (Fig. 5D), whereas blocked anti-POM36 antibody still cross-reacted with the 30- and 38-kD bands, which, therefore, might not be VDACs (Fig. 5F).
The presence of VDACs in microsomal fractions of L. japonicus roots and nodules was also investigated. Anti-LjVDAC1.1 antibody detected only two protein bands in microsomal fractions from L. japonicus roots and nodules, instead of the three bands detected in total protein extracts (Fig. 5G). This indicated that one of the LjVDAC isoforms, possibly LjVDAC1.3, was located on a membrane that was removed by centrifugation during preparation of microsomal fractions.
Subcellular Localization of VDAC Proteins
The subcellular location of VDACs in L. japonicus and soybean nodules was investigated further using western blot and immunolocalization methods. SM was purified from soybean nodules via Percoll gradients (Panter et al., 2000) and aqueous two-phase partitioning (Christiansen et al., 1995). A protein was detected using anti-LjVDAC1.1 and anti-POM36 antibodies on western blots of SM prepared using both Percoll gradients (Fig. 5G) and aqueous two-phase partitioning (Fig. 5, C and E).
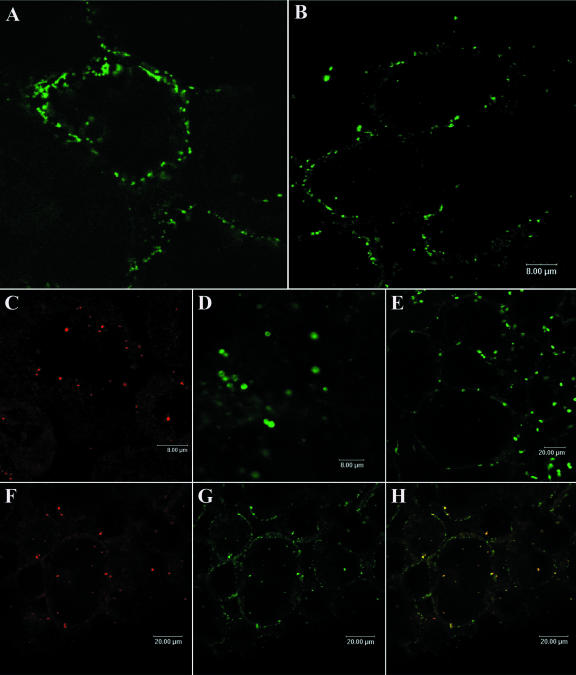
To establish more clearly the subcellular location of VDACs in L. japonicus and soybean nodules, we carried out immunofluorescence and immunogold labeling of tissue sections, using affinity-purified anti-LjVDAC1.1 antibody. Anti-LjVDAC1.1 antibody bound to punctate structures in the cell periphery close to, or associated with, the plasma membrane in L. japonicus and soybean nodules (Fig. 6, A and B). Control labeling with antibodies against the mitochondrial protein, Cyt c (Fig. 6C); the peroxisomal protein, uricase (Fig. 6D); and the plastidial protein, ferritin (Fig. 6E) were carried out on soybean nodule sections because of lack of cross-reactivity of some of the antibodies with the corresponding L. japonicus proteins. Peroxisomes and plastids were clearly larger (2–4 μm) than the structures labeled with anti-LjVDAC1.1. Thus, peroxisomes and plastids were not labeled by anti-LjVDAC1.1 antibody. An overlay of Cyt c labeling (Fig. 6F) and LjVDAC labeling (Fig. 6G) showed that a subset of LjVDAC-labeled structures were mitochondria (Fig. 6H, yellow). However, the anti-LjVDAC1.1 antibody also labeled vesicular, non-mitochondrial structures close to, or associated with, the plasma membrane that were not labeled with anti-Cyt c antibody (Fig. 6H, green).
Figure 6.
Immunolocalization of VDAC proteins in root nodules of soybean and L. japonicus. A and B, Single labeling studies with affinity-purified anti-LjVDAC1.1 antibody of L. japonicus and soybean nodule sections, respectively. C to E, Labeling of soybean mitochondria, peroxisomes, and plastids with anti-cytochrome c (Cyt c), anti-uricase, and anti-ferritin antibody, respectively. Colocalization of mitochondrial VDACs and Cyt c in soybean sections, using anti-Cyt c antibody (F) and LjVDAC1.1 antibody (G), is shown by image overlay (H, yellow). Note the non-mitochondrial signal in green in the overlay.
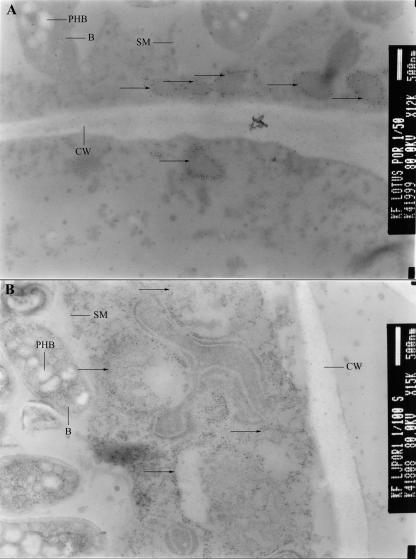
Further characterization of VDAC-containing structures was done using immunogold labeling of L. japonicus nodule sections with the anti-LjVDAC1.1 antibody. Once again, two types of structures were labeled: abundant 200- to 500-nm vesicular structures near the plasma membrane (Fig. 7A) and less abundant but larger (typically >1 μm) mitochondria (Fig. 7B). Mitochondria exhibited internal cristae typical of this organelle, whereas the small vesicles did not.
Figure 7.
Immunogold labeling of L. japonicus VDACs on root nodule sections. Nodules from 8-week-old L. japonicus nodules were used for immunogold labeling. Ultrathin sections (90 nm) were incubated with affinity-purified LjVDAC1.1 antibody. Secondary antibody was conjugated with 15-nm gold particles. Immunogold-labeled structures are marked with arrows. A, Small peripheral vesicles; B, mitochondria. SMs, bacteroids (B), polyhydroxybutyrate (PHB), and cell walls (CW) are also indicated.
Symbiosomes in nodule sections were not labeled with anti-LjVDAC1.1 antibody (Figs. 6 and 7), even though a VDAC antigen band was detected in SM preparations on western blots (Fig. 5). This apparent contradiction suggests that the SM preparations illustrated in Figure 5 were actually contaminated with another membrane type.
DISCUSSION
VDAC genes are present in fungi, animals, and plants. However, the numbers of homologs in different plant species exceed those in other eukaryotes. For example, Arabidopsis has five VDAC genes, whereas humans have three and yeast only two. At least five VDAC genes have been uncovered in each of L. japonicus, M. truncatula, and soybean by EST sequencing projects. Plant VDAC genes fall into five distinct subfamilies; the largest of these appear to have arisen since the divergence of monocots and dicots (Fig. 3). Three of the five L. japonicus VDAC genes characterized here fall into this category. Two of these, LjVDAC1.1 and LjVDAC1.2, are very similar in sequence and have a matching pair of genes in soybean, which may reflect a gene duplication event before the divergence of the Glycine and Lotus genera. Relatively recent gene duplication events in several other lineages are revealed by phylogenetic analysis (Fig. 3). Presumably, such events have provided the plant kingdom with an opportunity to diversify the physiological roles of VDAC proteins.
One way in which specialization of gene/protein function may come about during evolution is through a change in the pattern of gene expression. To determine whether any of the L. japonicus VDAC genes are expressed in an organ-specific manner, we carried out real-time RT-PCR analysis of leaves, roots, and nodules from mature L. japonicus plants. All five L. japonicus VDAC genes were expressed at high levels, although there were slight differences in the level of expression between genes and organs. Differential expression of VDAC genes during development has been reported for wheat (Triticum aestivum) and rice (Oryza sativa; Elkeles et al., 1995; Al Bitar et al., 2003).
Protein specialization can arise from changes in protein sequence and structure, which can alter the biochemical properties of a protein and/or its sub-cellular location. Clear differences in primary structure were revealed by comparisons of the deduced amino acid sequences of the five L. japonicus VDACs (Fig. 2). However, the predicted secondary structural elements, including an α-helical domain and numerous β-sheets, are conserved among all five proteins (Fig. 2). Thus, these structural elements may be essential for channel formation and functionality A 23-amino acid-long eukaryotic mitochondrial porin pattern (PROSITE Pattern-ID PS00558), which is conserved among plant, animal, and fungal VDACs, is present in LjVDAC1.1 to 1.3 but not in LjVDAC2.1 or LjVDAC3.1. This difference, together with sequence divergence, contributed to our naming of three separate VDAC subfamilies in L. japonicus. With the identification of an increasing number of VDAC sequences, the 23-amino acid porin pattern is losing its generality. Thus, most plant VDACs of branches I to III of the phylogenetic tree (Fig. 3) do not posses this porin signature. Surprisingly, the plant VDACs that posses the eukaryotic mitochondrial porin signature (branches IV and V of the tree; Fig. 3) are most distantly related to yeast and animal VDACs, which also posses the signature. Thus, although VDACs of yeast, animals, and plants have diverged during evolution, the porin signature has been maintained in some plant porin families, suggesting a common function of the motif in the different kingdoms. The functional significance of this pattern remains unknown, however.
The N-terminal α-helix is conserved among eukaryotic VDAC porins. Although positively charged N-terminal α-helices often function as mitochondrial signal sequences, and VDACs are imported via proteins of the Translocator of outer membrane (TOM) complex (Krimmer et al., 2001), it has been found that this domain is not involved in mitochondrial targeting (Court et al., 1996). On the other hand, circular dicroism, crystal structure analysis of yeast VDAC1 (Mannella, 1998), and point mutation analysis (Thomas et al., 1993) have shown that the N-terminal helix forms a mobile domain, which is involved in voltage sensing and opening and closing of the channel. Thus, the conserved N-terminal region appears to be involved in basic mechanisms of channel function, like voltage sensing and channel gating, rather than in specific functions like targeting or selectivity. This is supported to some extent by our observation that LjVDAC1.3, which like all other L. japonicus VDACs contains the conserved N-terminal α-helix, is apparently not targeted to the OMM (see below).
Functionality and intracellular location of L. japonicus VDAC proteins was examined by expressing them in a VDAC1-deficient yeast mutant. Complementation or suppression of the por- mutation in yeast strain HR1-2A by LjVDAC1.1, LjVDAC1.2, LjVDAC2.1, and LjVDAC3.1 demonstrated not only that these proteins function in yeast but also that they are targeted to the OMM (Fig. 4). This is the first time, to our knowledge, that plant VDACs have been shown to function in yeast, which opens up the possibility to carry out more detailed biochemical characterization of these proteins in the future. LjVDAC1.3 was unable to complement the yeast mutant, although the protein was expressed in transformed cells (Fig. 5, A and B). Immunolocalization showed that LjVDAC1.3 was not targeted to the OMM in yeast (data not shown), which provides an explanation for its inability to complement the mutant. Interestingly, LjVDAC1.3 is the least related of all the L. japonicus VDACs to animal and yeast VDACs. Sequence differences must account for the altered intracellular targeting of this protein in yeast, and this result raises the possibility that the protein is not located on the OMM in L. japonicus.
The anti-LjVDAC1.1 antibody that we produced is highly specific for VDACs. It binds strongly to LjVDAC1.1, LjVDAC1.2, and LjVDAC1.3, weakly to LjVDAC2.1, and not at all to LjVDAC3.1 (Fig. 5A) or other L. japonicus proteins (Fig. 5, C and D). The anti-LjVDAC1.1 antibody also recognizes VDAC proteins in soybean (Fig. 5, C and D). In contrast, the anti-POM36 antibody cross-reacted strongly with two proteins in L. japonicus that could not be confirmed as VDACs (Fig. 5, E and F). Localization studies, using affinity-purified LjVDAC1.1 antibody together with Cyt c antibody, confirmed the mitochondrial location of some VDACs in root nodules (Fig. 6, F–H). However, anti-LjVDAC1.1 antibody also labeled distinct vesicular structures close to the plasma membrane, which were not labeled with anti-Cyt c antibody (Fig. 6, F–H). Because the anti-LjVDAC1.1 antibody cross-reacts with LjVDAC1.1 to 1.3, and LjVDAC1.1 and LjVDAC1.2 are targeted to the OMM in yeast, these proteins are likely to be located in the OMM in root nodules as well. In contrast, the LjVDAC1.3 protein, which was not targeted to the OMM of yeast and was probably absent from microsomal fractions (Fig. 5G), may be located in the non-mitochondrial, vesicular structures observed in root nodules. These structures were apparently not peroxisomes or plastids (Fig. 6). We can only speculate on the identity of these vesicles at this stage. VDAC proteins have been found in extra-mitochondrial membranes in animals, in the caveolea, or Tritoninsoluble fraction domains of plasma membranes (Bathori et al., 1999; Bathori et al., 2000). Such domains have not been described in plants. However, because the ultrastructure of caveolea varies and can take on a vesicular form also (Anderson, 1998), the VDAC-decorated vesicles in nodules (Fig. 7A) could represent caveolae-like domains. Alternatively, they could represent vesicles involved in the secretory pathway. Salient in this regard is the observation that VDAC fusion proteins are targeted to secretory pathway vesicles in animals (Buettner et al., 2000). Previous work on plant VDACs localized an anonymous protein in the boundary membrane of glyoxisomes in cucumber (Cucumis sativus) plants, using the anti-POM36 antibody (Corpas et al., 2000). However, the same antibody cross-reacted strongly with two L. japonicus proteins that may not be VDACs (Fig. 5E). Therefore, results obtained from the use of the anti-POM36 antibody in immunolocalization studies should be interpreted with caution.
Results from proteomics studies have indicated the presence of VDACs in SM preparations from pea and L. japonicus nodules (Saalbach et al., 2002; Wienkoop and Saalbach, 2003). The VDAC sequenced from L. japonicus SM preparations is identical to LjVDAC1.1 described here. However, using cytological methods and the affinity-purified LjVDAC1.1 antibody, we were unable to detect the presence of VDAC proteins on the SM in sections of soybean or L. japonicus nodules. Therefore, we conclude that VDAC proteins are probably not located on the SM in intact legume root nodules. Although we cannot exclude the possibility that the VDAC antigens recognized by the LjVDAC1.1 antibody are somehow masked and not detectable in the SM in situ, our results urge caution in interpreting the results of proteomics experiments with isolated, potentially contaminated organelles, such as symbiosomes. Ideally, the results of subcellular proteomics should be confirmed by in situ methods, such as immunolocalization.
To summarize, we have characterized five VDAC porins from the model legume, L. japonicus, four of which are functional at the OMM when expressed in yeast. Transcripts of all five genes accumulate to similar, high levels throughout the plant. Immunolocalization of VDACs in L. japonicus and soybean root nodules revealed their presence not only on mitochondria but also on distinct, small vesicles, which remain to be identified. Our results indicate that the biological roles of VDAC proteins have diversified during the evolution of plants.
MATERIALS AND METHODS
Plant Culture
Lotus japonicus GIFU B-129-S9 and soybean (Glycine max cv Stevens) plants were grown in quartz sand-filled pots in a greenhouse under artificial light in a 16-h-light/8-h-dark rhythm. Inoculation of L. japonicus with Mesorhizobium loti strain R7A and of soybean with Bradyrhizobium japonicum USDA110 was done at the time of sowing and repeated 4 to 7 d later.
DNA Manipulation
DNA manipulations such as plasmid purification, restriction digests, agarose gel electrophoresis, ligations, and screening of cDNA libraries were performed using standard protocols (Sambrook et al., 1989). Synthesis of cDNA was carried out with the Superscript Lambda cDNA synthesis kit (Gibco BRL, Eggenstein, Germany).
RNA Gel-Blot Analysis
Leaves, stems, roots, flowers, and nodules from 6-week-old L. japonicus plants were harvested and ground to a fine powder in liquid nitrogen, using mortar and pestle, and RNA was extracted using the detergent-based method of Jacobsen-Lyon et al. (1995). After separation of RNA by gel electrophoresis under denaturing conditions (Lehrach et al., 1977), RNA was transferred to nylon membranes (Porablot NY amp plus, Macherey & Nagel, Düren, Germany) and fixed using a UV crosslinker (Stratagene, Heidelberg). Blots were prehybridized for 4 h at 65°C and hybridized overnight at 65°C in 250 mm sodium phosphate buffer (pH 7.2) containing 7% (w/v) SDS, 1% (w/v) bovine serum albumin (BSA), and 1 mm EDTA. DNA probes were 32P-labeled using the RediPrime random primer labeling kit (Amersham, Braunschweig, Germany) and purified with NAP5 columns (Amersham), which removed unincorporated nucleotides. Membranes were washed twice at 65°C in 2× SSC (1× SSC contains 0.15 m NaCl and 0.015 m sodium citrate) plus 0.1% (w/v) SDS, and once in 0.2× SSC and 0.1% (w/v) SDS. Blots were subjected to autoradiography overnight at -80°C, using Kodak X-Omat AR Films (Sigma, Deisenhofen, Germany).
Real-Time RT-PCR
Poly(A+) RNA was purified from 10 μg of DNase-treated total RNA, using the Qiagen Oligotex mRNA kit (Qiagen, Hilden, Germany). Approximately 10 ng of mRNA was then reverse transcribed with Superscript II RT (Invitrogen, Karlsruhe, Germany) using an oligo(dT) primer, to generate 50 μL of first strand cDNA. Real-time RT-PCR was performed using 1 μL of a 1/10 (v/v) dilution of the first strand cDNA reaction and the SYBRGreen reagent (Eurogentec, Seraing, Belgium) in a reaction volume of 25 μL on a GeneAmp 5700 Sequence Detection System (PE-Applied Biosystems, Foster City, CA). Primers were designed to the 3′-untranslated region of LjVDAC1.1 (forward, 5′-GATAATTT TGATCCTTGGCAAGAC-3′; and reverse, 5′-GAACACTTCTTAGCCAAGAAGAG-3′), LjVDAC1.2 (forward, 5′-ATGA TTTTGAATTGATAGGCTGCG-3′; and reverse, 5′-ACCCAGC AAGAAATTAACAGCTC-3′), LjVDAC1.3 (forward, 5′-TTTTGAGCCATTATGGCAAGA GC-3′; and reverse, 5′-AACACTT ATGTCCAATAGAACCAG-3′), LjPor2.1 (forward, 5′-TTG AAAGGAGGGCAGAATAA TTAG-3′; and reverse, 5′-GTCAAACAGAAGCCATTGGTGAT-3′), and LjPor3.1 (forward, 5′-GCCTTGTCATAAAGGTAAAACCAA-3′; and reverse, 5′-CTCTAGCCATTATGTAAGTATT TC-3′). Each primer pair amplified a single product, as shown by the melting temperature of the amplicons and agarose gel electrophoresis. Likewise, ubiquitin primers (forward, 5′-TTCACCTTGTGCTCCGTCTTC-3′; and reverse, 5′-AACAACAGCACACACAGACAATCC-3′) were designed against the L. japonicus polyubiquitin gene (GenBank accession no. AW720576). Samples without template were used as negative controls. Expression data were normalized to ubiquitin and then compared using the formulae:
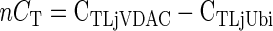 |
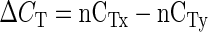 |
 |
where CT is the PCR cycle number at which a set threshold value (usually 0.1) of SYBR Green fluorescence is reached, nCT is the normalized CT value, ΔCT is the difference in normalized CT values in tissues x and y, and Ration is the ratio of gene transcript level in tissue x compared with y.
Yeast (Saccharomyces cerevisiae) Complementation
Yeast strains HR1-2B (por+) and HR1-2A (por-) were a kind gift of Dr. Carla Koehler (University of California, Los Angeles; Dihanich et al., 1987). Rich-medium yeast peptone dextrose for yeast growth contained 1% (w/v) yeast extract, 1% (w/v) peptone, and 2% (w/v) Glc. Full-length VDAC cDNAs were cloned into yeast expression vector p112A1NE (Riesmeier et al., 1992) and transformed into the por- strain, using the lithium acetate method (Gietz et al., 1995). As controls, por+ and por- strains were transformed with the non-recombinant vector. Transformants were selected for Trp prototrophy on solid minimal medium containing 0.67% (w/v) yeast nitrogen base (Difco, Augsburg, Germany), 2% (w/v) Glc, 0.14% (w/v) yeast synthetic drop-out mix (Sigma), and 2% (w/v) Bacto-Agar (Gibco BRL) supplemented with 20 mg L-1 uracil, 30 mg L-1 Leu, and 20 mg L-1 His.
Protein Extraction and Western-Blot Analysis
All protein extraction methods were carried out at 4°C, unless otherwise stated. Total protein extracts from L. japonicus and soybean organs were prepared from 8-week-old plants by harvesting fresh tissue into liquid nitrogen and grinding the frozen tissue with mortar and pestle to a fine powder. The powder was resuspended in an equal volume of extraction buffer (10 mm Tris [pH 7.5], 140 mm NaCl, 5 mm EDTA, 1% [v/v] Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 1 mm dithiothreitol). Cell debris and insoluble components were pelleted twice by centrifugation at 14,000 rpm for 10 min. Protein concentration of supernatants was determined according to Bradford using the Bio-Rad Protein Assay Kit I (Bio-Rad, Munich). Approximately 20 μg of protein was loaded per lane.
SMs from soybean root nodules of 8-week-old plants were prepared as described by Panter et al. (2000) and by aqueous polymer two-phase partitioning after Christiansen et al. (1995). Membrane pellets were resuspended in 20 mm Tris (pH 7.2) and 1% (w/v) SDS and subjected to SDS-PAGE. Approximately 20 μg of protein was loaded per lane.
Crude yeast protein extracts were prepared by first pelleting 5 mL of cells of an overnight culture and resuspending them in 150 μL of LDS-Sample buffer (Invitrogen). After heating for 10 min at 95°C, a half volume of glass beads was added, and the cells were vortexed for 10 min before being subjected to SDS-PAGE. Approximately 20 μg of protein, in 10 μL of extract, was loaded per lane.
To obtain polyclonal anti-LjVDAC1.1 antiserum, the open reading frame of the LjVDAC1.1 cDNA was amplified with primers containing either BamHI or HindIII restriction sites (forward, 5′-GGATCCATGGCTAAGGGTCCTGGTCTC-3′; and reverse, 5′-AAGCTTCC CAATGTCTTGCCAAGGATC-3′). After subcloning the 871-bp PCR fragment into pGEMTEasy (Promega, Mannheim, Germany), the BamHI-HindIII fragment was isolated and ligated to a poly-His-encoding sequence in vector pQE30 (Qiagen). Protein expression in Escherichia coli and protein purification using nickel affinity columns was performed as per the manufacturer's instructions (QIAexpressionist, Qiagen). Purified protein was then injected into rabbits (Pineda Antikörper Service, Berlin), and polyclonal antisera were collected 61 and 90 d after immunization. Antiserum was then affinity purified using Sepharose columns with immobilized LjVDAC1.1 protein (Pineda Antikörper Service).
After separation on NuPage precast gels (Invitrogen), proteins were electroblotted onto nitrocellulose membranes (Schleicher & Schuell, Ein-beck, Germany) overnight at 70 mA or for 150 min at 400 mA with an XCell II Blot Module (Invitrogen) in 25 mm Tris-Base, 190 mm Gly, and 20% (v/v) methanol. Protein detection on western blots using 1:1,000 (v/v) diluted primary antibody, alkaline phosphatase-conjugated anti-rabbit secondary antibody (1:1,500 [v/v] dilution, Promega), and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate tablets (Roche Diagnostics, Mannheim, Germany) was performed as described by Blake et al. (1984).
Immunolocalization by Laser-Scanning Confocal Microscopy
Root nodules of around 8-week-old L. japonicus and soybean plants were fixed overnight in PEM (50 mm 1,4-Piperazinediethanesulfonic acid (PIPES) [pH 6.8], 5 mm EDTA, and 5 mm MgSO4), containing 3.7% (w/v) paraformaldehyde, under a slight vacuum. Using a VT1000S Vibratome (Leica, Bensheim, Germany), 100-μm sections were taken and mounted onto poly-l-Lys-coated slides (Poly-Prep-Slides, Sigma). Sections were treated for 10 min with 0.5% (w/v) Cellulase R10 in PEM (Yakult, Tokyo) before blocking with PEM, containing 3% (w/v) BSA, 0.1% (v/v) Tween20, and 20 mm Lys for 2 h at room temperature. Affinity-purified anti-LjVDAC1.1 and anti-Cyt c antibodies (BD Biosciences, Heidelberg) were applied in a 1:10 (v/v) dilution, anti-uricase antibody (VandenBosch and Newcomb, 1986) was applied in a 1:250 (v/v) dilution, and anti-ferritin antibody (Lucas et al., 1998) in a 1:50 (v/v) dilution in blocking solution overnight at 4°C. Sections were then washed in 200 mL of blocking solution for 1 h at room temperature with gently stirring. As fluorescent conjugates, anti-Rabbit antibody labeled with Alexa Fluor 488 and anti-mouse antibody labeled with Alexa Fluor 568 (Molecular Probes Europe, Leiden, Netherlands) were used and applied at 1:50 (v/v) dilution in blocking solution for 4 h at RT. After washing in blocking solution for 2 to 4 h, the sections were viewed on a laser-scanning confocal microscope (Leica DM IRBE microscope, TCS SPII confocal scanner). Images were generated using processing software from Leica.
Protein Localization by Immunogold Labeling and Electron Microscopy
Root nodule pieces from 8-week-old plants were fixed overnight at 4°C in 0.1 m sodium phosphate buffer (pH 7.3), containing 4% (w/v) paraformaldehyde. Samples were then dehydrated for 6 h in 30% (v/v) ethanol, 3 h in 50% (v/v) ethanol at room temperature, and overnight in 70% (v/v) ethanol at 4°C. Afterward samples were embedded in LR-White (LRW) Resin Medium Grade (Plano, Wetzlar, Germany) by infiltration for 6 h in a 1:1 (v/v) LRW:ethanol mixture and for 24 h in pure LRW at 4°C. Polymerization was carried out for 2 d at 50°C. Ultrathin sections (90 nm) were taken with an Ultracat E ultramicrotome (Leica) and mounted onto pyroxylin- and carboncoated 200-mesh gold grids (Plano). For immunogold staining, the sections were blocked for 1 h at RT in phosphate-buffered saline (PBS; 10 mm sodium phosphate buffer [pH 7.5] and 130 mm NaCl), containing 1% (w/v) Aurion-BSA (Science-Services, Munich) and 0.1% (v/v) Tween 20. Affinity-purified LjVDAC1.1 antibody was diluted 1:10 (v/v) in incubation solution (PBS, 0.1% [w/v] Aurion-BSA, and 0.01% [v/v] Tween20) and applied overnight at 4°C. After three washes for 15 min in wash buffer (PBS and 0.1% [v/v] Tween20), the gold-conjugated secondary antibody (10 nm of gold, British BioCell International, Cardiff, UK) diluted 1:30 (v/v) in incubation solution was applied for 3 to 4 h at room temperature. After washing three times for 15 min in (successively) wash buffer, PBS, and distilled water, the grids were contrast stained with uranyl acetate and lead citrate. The labeled sections were monitored with a Joel Transmission electron microscope (Tokyo, Japan) at a voltage of 80 mV and photographed.
Acknowledgments
We would like to thank the Kazusa DNA Research Institute (Chiba, Japan) for VDAC cDNA clones and Drs. Carla Koehler (University of California, Los Angeles), Udo Schmitz (Universität Hannover, Germany), Jean-Francois Briat (University of Montpellier, France), and Desh Pal Verma (Ohio State University, Columbus) for the VDAC1-deficient yeast mutant and antibodies to yeast VDAC1, POM36, ferritin, and uricase, respectively. We also thank Kim Findlay (John Innes Centre, Norwich, UK) for help with electron microscopy.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.031484.
This work was supported by the Deutsche Forschungsgemeinschaft and by the Max Planck Society.
References
- Al Bitar F, Roosens N, Smeyers M, Vauterin M, Van Boxtel J, Jacobs M, Homble F (2003) Sequence analysis, transcriptional and posttranscriptional regulation of the rice vdac family. Biochim Biophys Acta 1625: 43-51 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410 [DOI] [PubMed] [Google Scholar]
- Anderson RG (1998) The caveolae membrane system. Annu Rev Biochem 67: 199-225 [DOI] [PubMed] [Google Scholar]
- Bainbridge G, Gokce I, Lakey JH (1998) Voltage gating is a fundamental feature of porin and toxin beta-barrel membrane channels. FEBS Lett 431: 305-308 [DOI] [PubMed] [Google Scholar]
- Bathori G, Parolini I, Szabo I, Tombola F, Messina A, Oliva M, Sargiacomo M, De Pinto V, Zoratti M (2000) Extramitochondrial porin: facts and hypotheses. J Bioenerg Biomembr 32: 79-89 [DOI] [PubMed] [Google Scholar]
- Bathori G, Parolini I, Tombola F, Szabo I, Messina A, Oliva M, De Pinto V, Lisanti M, Sargiacomo M, Zoratti M (1999) Porin is present in the plasma membrane where it is concentrated in caveolae and caveolae-related domains. J Biol Chem 274: 29607-29612 [DOI] [PubMed] [Google Scholar]
- Benz R (1994) Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. Biochim Biophys Acta 1197: 167-196 [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E, Forte M (2001) VDAC channels. Int Union Biochem Mol Bio Life 52: 113-118 [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E, Zambronicz EB, Yu WH, Adams V, McCabe ER, Adelman J, Colombini M, Forte M (1993) Cloning and functional expression in yeast of two human isoforms of the outer mitochondrial membrane channel, the voltage-dependent anion channel. J Biol Chem 268: 1835-1841 [PubMed] [Google Scholar]
- Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC (1984) A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on western blots. Anal Biochem 136: 175-179 [DOI] [PubMed] [Google Scholar]
- Bolter B, Soll J (2001) Ion channels in the outer membranes of chloroplasts and mitochondria: open doors or regulated gates? EMBO J 20: 935-940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner R, Papoutsoglou G, Scemes E, Spray DC, Dermietzel R (2000) Evidence for secretory pathway localization of a voltage-dependent anion channel isoform. Proc Natl Acad Sci USA 97: 3201-3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen JH, Rosendahl L, Widell S (1995) Preparation and characterization of sealed inside-out peribacteroid membrane vesicles from Pisum Sativum L. and Glycine Max L. root nodules by aqueous polymer two-phase partitioning. J Plant Physiol 147: 175-181 [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant-Microbe Interact 15: 411-420 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Sandalio LM, Brown MJ, del Rio LA, Trelease RN (2000) Identification of porin-like polypeptide(s) in the boundary membrane of oilseed glyoxysomes. Plant Cell Physiol 41: 1218-1228 [DOI] [PubMed] [Google Scholar]
- Court DA, Kleene R, Neupert W, Lill R (1996) Role of the N- and C-termini of porin in import into the outer membrane of Neurospora mitochondria. FEBS Lett 390: 73-77 [DOI] [PubMed] [Google Scholar]
- Delcour AH (2002) Structure and function of pore-forming beta-barrels from bacteria. J Mol Microbiol Biotechnol 4: 1-10 [PubMed] [Google Scholar]
- Dihanich M, Suda K, Schatz G (1987) A yeast mutant lacking mitochondrial porin is respiratory-deficient, but can recover respiration with simultaneous accumulation of an 86-kd extramitochondrial protein. EMBO J 6: 723-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkeles A, Devos KM, Graur D, Zizi M, Breiman A (1995) Multiple cDNAs of wheat voltage-dependent anion channels (VDAC): isolation, differential expression, mapping and evolution. Plant Mol Biol 29: 109-124 [DOI] [PubMed] [Google Scholar]
- Fischer K, Weber A, Brink S, Arbinger B, Schunemann D, Borchert S, Heldt HW, Popp B, Benz R, Link TA et al. (1994) Porins from plants: molecular cloning and functional characterization of two new members of the porin family. J Biol Chem 269: 25754-25760 [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355-360 [DOI] [PubMed] [Google Scholar]
- Heins L, Mentzel H, Schmid A, Benz R, Schmitz UK (1994) Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J Biol Chem 269: 26402-26410 [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A (1999) The PROSITE database, its status in 1999. Nucleic Acids Res 27: 215-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen-Lyon K, Jensen EO, Jorgensen JE, Marcker KA, Peacock WJ, Dennis ES (1995) Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca. Plant Cell 7: 213-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23: 403-405 [DOI] [PubMed] [Google Scholar]
- Krimmer T, Rapaport D, Ryan MT, Meisinger C, Kassenbrock CK, Blachly-Dyson E, Forte M, Douglas MG, Neupert W, Nargang FE et al. (2001) Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J Cell Biol 152: 289-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, Xu X, Blachly-Dyson E, Forte M, Colombini M (1998) The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J Membr Biol 161: 173-181 [DOI] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boetcker H (1977) RNA molecular weight determination by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 16: 4743-4751 [DOI] [PubMed] [Google Scholar]
- Lichtinger T, Burkovski A, Niederweis M, Kramer R, Benz R (1998) Biochemical and biophysical characterization of the cell wall porin of Corynebacterium glutamicum: the channel is formed by a low molecular mass polypeptide. Biochemistry 37: 15024-15032 [DOI] [PubMed] [Google Scholar]
- Lucas MM, Van de Sype G, Hérouart D, Hernández MJ, Puppo A, de Felipe MR (1998) Immunolocalization of ferritin in determinate and indeterminate legume root nodules. Protoplasma 204: 61-70 [Google Scholar]
- Mannella CA (1998) Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications. J Struct Biol 121: 207-218 [DOI] [PubMed] [Google Scholar]
- Panter S, Thomson R, de Bruxelles G, Laver D, Trevaskis B, Udvardi M (2000) Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol Plant-Microbe Interact 13: 325-333 [DOI] [PubMed] [Google Scholar]
- Reumann S, Maier E, Heldt HW, Benz R (1998) Permeability properties of the porin of spinach leaf peroxisomes. Eur J Biochem 251: 359-366 [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1992) Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J 11: 4705-4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess FG, Dorner U, Schiffler B, Benz R (2001) Study of the properties of a channel-forming protein of the cell wall of the gram-positive bacterium Mycobacterium phlei. J Membr Biol 182: 147-157 [DOI] [PubMed] [Google Scholar]
- Riess FG, Lichtinger T, Yassin AF, Schaal KP, Benz R (1999) The cell wall porin of the gram-positive bacterium Nocardia asteroides forms cation-selective channels that exhibit asymmetric voltage dependence. Arch Microbiol 171: 173-182 [DOI] [PubMed] [Google Scholar]
- Rost B (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol 266: 525-539 [DOI] [PubMed] [Google Scholar]
- Saalbach G, Erik P, Wienkoop S (2002) Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2: 325-337 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Thomas L, Blachly-Dyson E, Colombini M, Forte M (1993) Mapping of residues forming the voltage sensor of the voltage-dependent anion-selective channel. Proc Natl Acad Sci USA 90: 5446-5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493-523 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett 231: 36-40 [Google Scholar]
- VandenBosch KA, Newcomb EH (1986) Immunogold localization of nodule-specific uricase in developing soybean root nodules. Planta 167: 425-436 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Saalbach G (2003) Proteome analysis: novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Decker W, Sampson MJ, Craigen WJ, Colombini M (1999) Mouse VDAC isoforms expressed in yeast: channel properties and their roles in mitochondrial outer membrane permeability. J Membr Biol 170: 89-102 [DOI] [PubMed] [Google Scholar]