Abstract
As a step in developing an understanding of plant adaptation to low atmospheric pressures, we have identified genes central to the initial response of Arabidopsis to hypobaria. Exposure of plants to an atmosphere of 10 kPa compared with the sea-level pressure of 101 kPa resulted in the significant differential expression of more than 200 genes between the two treatments. Less than one-half of the genes induced by hypobaria are similarly affected by hypoxia, suggesting that response to hypobaria is unique and is more complex than an adaptation to the reduced partial pressure of oxygen inherent to hypobaric environments. In addition, the suites of genes induced by hypobaria confirm that water movement is a paramount issue at low atmospheric pressures, because many of gene products intersect abscisic acid-related, drought-induced pathways. A motivational constituent of these experiments is the need to address the National Aeronautics and Space Administration's plans to include plants as integral components of advanced life support systems. The design of bioregenerative life support systems seeks to maximize productivity within structures engineered to minimize mass and resource consumption. Currently, there are severe limitations to producing Earth-orbital, lunar, or Martian plant growth facilities that contain Earth-normal atmospheric pressures within light, transparent structures. However, some engineering limitations can be offset by growing plants in reduced atmospheric pressures. Characterization of the hypobaric response can therefore provide data to guide systems engineering development for bioregenerative life support, as well as lead to fundamental insights into aspects of desiccation metabolism and the means by which plants monitor water relations.
Interest in the exploration of environments beyond Earth's atmosphere has brought unique challenges to bear on the understanding of the biological systems that will inhabit those environments. Among these challenges are alterations in atmospheric pressure, which are known to have effects on plant physiology and development (Mansell et al., 1968; Gale, 1973; Andre and Richaux, 1986; McKay and Toon, 1991; Andre and Massimino, 1992; Wheeler, 2000; He et al., 2003). Concepts for greenhouses on Mars, on the moon, and in Earth orbit incorporate low atmospheric pressures to address engineering and systems limitations (Boston, 1981; Drysdale, 2001).
Historically, low-pressure environments have been used throughout the U.S. human space exploration programs to reduce the masses of structural and consumable components of space vehicles. Such reductions have resulted in increased mission lengths and/or increased masses of launched payloads. For example, the Mercury, Gemini, and Apollo environments were designed to operate at 34 kPa with a pure oxygen environment to simplify support of humans in space (Baker, 1981; Martin and McCormick, 1992). Skylab was also operated at 34 kPa (with a 70% O2/30% N2 gas mixture), and that pressure was further reduced during periods without human crew habitation. It was not until the era of the Space Shuttle, Mir, and the International Space Station that spaceflight environments have operated at Earth-normal pressures near 101 kPa, although the pressure in the shuttle is lowered to approximately 70 kPa for extended periods (up to 24 h) during extravehicular activities (Winkler, 1992; Wieland, 1998). Although the internal pressures for future transit vehicles and surface missions to the moon and Mars have not been established, it seems reasonable to expect that reduced-pressure atmospheres will again be used to decrease the lift costs of structural components and consumables.
The consideration of plants for atmospheric regeneration and food production for long duration human space missions began in the 1960s, and the earliest tests on plant growth at reduced atmospheric pressure were conducted at Brooks Air Force Base (Mansell et al., 1968). Follow-on work further characterized physiological changes that occur at low atmospheric pressure (Lind, 1971; Gale, 1972, 1973). Growing plants in reduced pressures could result in both enhancement or repression of growth depending on the species under study and the composition of the atmosphere with respect to O2 and CO2 during the experiment (Rule and Staby, 1981; Andre and Richaux, 1986; Musgrave et al., 1988). These findings led to the general conclusion that plants appear to tolerate low atmospheric pressures (Corey et al., 2002). In all cases, however, significant adaptations must take place because, at a minimum, plants must cope with hypoxia induced by reduced partial pressure of O2 (Siegel, 1961; Siegel et al., 1963; Ferl et al., 2002). Hypoxic conditions at Earth-normal pressures have been widely used to study plant adaptation to hypoxia (Ramonell et al., 2001; Klok et al., 2002) and as a partial mimic for hypobaria.
Low atmospheric pressure environments place demands on water movement in addition to presenting de facto hypoxia. Transpiration rate increases as atmospheric pressure is reduced, even at high relative humidity (Gale, 1973; Corey et al., 2002) and influences stomatal aperture independent of relative humidity (Mott and Parkhurst, 1991). Aside from these growth and physiology observations, however, little is known about the mechanisms necessary to allow plant adaptation to low atmospheric pressure, and virtually no information exists on the patterns of gene expression that are influenced by low atmospheric pressure (Ferl et al., 2002).
To assess directly the molecular events that characterize plant adaptation to low atmospheric pressures, we used the 8K Affymetrix GeneChip arrays to characterize the effects of 24 h of exposure of Arabidopsis to 10 kPa and compared those results with 24 h of exposure to Earth-normal air at 101 kPa and 2% O2 at 101 kPa.
RESULTS
Test Conditions and Experimental Design
Environmental parameters within the low-pressure growth chamber (LPGC) were closely monitored and controlled to ensure that the lighting-, temperature-, and humidity-defined conditions that were non-stressful to Arabidopsis seedlings (Fig. 1A). Young Arabidopsis plants were grown on the vertical surface of nutrient agar plates. At 9 to 10 d old, the aerial portion consisted of two to three pairs of true leaves, and the roots had not yet reached the bottom of a 100-mm culture plate. Because the plants grew along the face of the agar (and did not tunnel into the agar), the entire surface of the plant received the impact of the environment in which it was placed (Fig. 1B). The plants demonstrated consistent growth during each of the 24-h experimental and control treatments within the LPGC and showed no visible signs of drying or distress (Fig. 1B). Quantitative measurements of plant biomass (Fig. 1C) suggested that neither hypoxic (2% O2) nor hypobaric (10 kPa) treatments caused any overt dehydration stress for plate-grown plants; the plants appeared turgid, and the before and after measurements of mass reflected similar changes in all treatments. The relative humidity inside a representative vertical plate containing 9-d-old plants was monitored with a small HOBO data logger during a 10-kPa run in the LPGC. The relative humidity inside the plate was consistently 95% or greater.
Figure 1.
The LPGC and the plants used for hypobaria-related experiments. A, A prototype LPGC was designed to control total pressure, gas composition, temperature, and light. B, The plants were grown on vertically oriented agar plates and, as such, had ample water supply, and the entire mass of roots and shoots was exposed to the atmospheric environment (Paul et al., 2001), before harvesting the shoots for RNA isolation and array analyses. For hypobaria, the total pressure was held at 10 kPa of Earth-normal air, whereas for hypoxia, the pressure was held at 101 kPa with flow of 98% N2:2% O2 (v/v). In all cases, the partial pressure of CO2 was held constant at 0.1 kPa, and chamber temperature was controlled to 22 ± 0.5°C. Although the relative humidity of the chamber averaged 85%, the relative humidity inside the growth plates containing the plants ranged between 95% and 100%. C, Measurements of plant biomass revealed no evidence of desiccation or any overt dehydration stress for all treatments. Seedling groups were weighed before and after 24-h experimental treatments in the LPGC. The relative change in biomass was determined for three groups of plants in each treatment set.
Differential Expression of Genes on the Array
More than 200 genes were differentially expressed in the shoots of plants exposed to the 10 kPa environment for 24 h, yet the response to 10 kPa was markedly different from the response to 24 h at 2% O2 (Fig. 2A). ANOVA and variance normalization analyses of GeneChip expression levels were used to identify those genes showing a significant treatment effect relative to the 101 kPa control samples, and the results from biological replicates were clustered according to their expression behavior. As summarized in the Venn diagrams (Fig. 2A), the genes induced by 10 kPa were markedly diverged from those induced by 2% O2, although the genes repressed by both treatments largely overlapped. Figure 2A is expanded in Figure 2B to provide identities and brief annotations of the genes induced by hypobaria and/or hypoxia, along with scaled ANOVA probabilities and average -fold change values of expression. These data revealed that the metabolic pathways being activated for adaptation to hypobaria included, and extended well beyond, those being activated for adaptation to hypoxia.
Figure 2.
Gene expression profiles from plants treated with hypoxia and low atmospheric pressure compared with Earth-normal controls. A, The induction and repression of gene activity is represented by the variance normalized score (Vnorm) for each sample against the average of the two control samples. Only those genes demonstrating significant treatment effects are shown. The Venn diagrams show the overlap between the hypoxia and low-pressure treatments. B, The A and C clusters from the indicated regions in A are shown, with the ANOVA value for the data and the average -fold increase or decrease in gene activity, together with the gene identifiers and brief annotations. Genes directly involved in dehydration responses are indicated by blue background in the annotations. Cluster B (genes coordinately down regulated in both treatments) annotations can be found in the supplemental data.
Exposure to 10 kPa engaged desiccation-related metabolic pathways, as revealed by the genes most highly induced by hypobaria (Fig. 2, A and B, clusters A1 and A2). These genes were repressed or unaffected by hypoxia and showed as much as a 30-fold difference in expression between 2% O2 and 10 kPa. By far the most numerous, most differentially induced genes included dehydrins, type-1 late embryogenesis abundant D113 (LEA)-like proteins, RD29B, cold-responsive (Cor)-related proteins, and Gly-rich proteins (Fig. 2B; e.g. Yamaguchi-Shinozaki and Shinozaki, 1993a; Siddiqui et al., 1998; Thomashow, 1999; Finkelstein et al., 2002; Soulages et al., 2003). In addition to the dehydration-associated genes, there were also examples of genes associated with stomatal distribution and regulation (Berger and Altmann, 2000) and signal transduction pathways induced by hypobaria that included kinases and kinase-related proteins (Guo et al., 2002; Yoshida et al., 2002), calcium-binding proteins (Takahashi et al., 2000; Sadiqov et al., 2002) and a cytochrome p450 (Reddy et al., 2002).
The genes induced by both hypobaria and hypoxia (Fig. 2, A and B, clusters C3, C2, and A3) were consistent with previous studies, which indicated that genes for fermentative pathways and those involving oxygen transport are required under low-oxygen conditions (e.g. Klok et al., 2002). The genes repressed by both hypoxia and hypobaria (Fig. 2A, cluster B) included representatives from a wide range of metabolic pathways and were also largely consistent with previous studies of hypoxia responses in Arabidopsis (an expanded, annotated view of cluster B is provided in the supplemental data, available in the online version of this article at http://www.plantphysiol.org). The hypobaria response therefore included, but was not limited to, a typical response to hypoxia.
There were a few genes that were down-regulated in response to hypoxia but remained unchanged in response to hypobaria (Fig. 2, A and B, cluster A1). Genes in this cluster encode proteins that may be involved in metabolic processes related to oxygen sensing that are mediated through a heme- or iron-based sensor (Aravind and Koonin, 2001; Zhu and Bunn, 2001; Quinn et al., 2002). There were also genes that were induced by hypoxia but not hypobaria (Fig. 2, A and B, cluster C1). This cluster contained additional heme-related proteins, as well as other genes typical of a hypoxic stress response.
Corroboration and Quantification of Microarray Data with RT-PCR
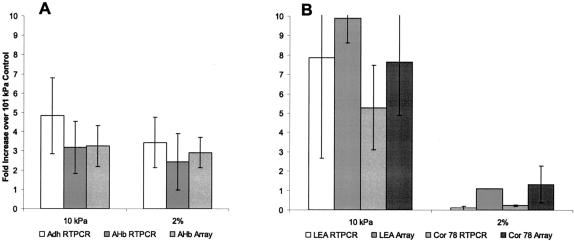
The degree of induction of representative genes in the hypobaria- and hypoxia-induced clusters was confirmed by real-time RT PCR (Taqman, Applied Biosystems, Foster City, CA). Figure 3 shows the -fold increase in response to environments of 10 kPa and 2% O2 over the 101 kPa control for representative genes. These RT-PCR data are derived from the RNA used for the arrays, replicate RNA isolations from the same biological samples used for the arrays, as well as RNA isolated from a third, independent biological replicate. Where available, the relative value from the array data is shown just to the right of the value from RT-PCR analyses. Figure 3A provides the -fold induction values for two genes associated with hypoxic stress response: alcohol dehydrogenase (Adh) and non-symbiotic hemoglobin (AHb). Adh, although not represented in the Affymetrix array used in these analyses, is a well-characterized hypoxia-induced gene (e.g. Paul et al., 2001; Klok et al., 2002). Adh was induced by hypobaria and hypoxia to similar levels. AHb was induced by both stresses to a similar degree, and there was a close agreement between values obtained from the quantitative RT-PCR and the relative induction values from the micoarrays.
Figure 3.
Real-time RT-PCR confirmation of expression profiles. RNA samples from the same tissues used for Gene Chip arrays, as well as additional experiment replications, were subjected to real-time RT-PCR using primers directed against key genes in the hypoxia and hypobaria adaptation pathways. A, Analysis of alcohol dehydrogenase (Adh) and non-symbiotic hemoglobin (AHb). Adh and AHb represent typical hypoxia-induced genes and are consistently induced approximately 3-fold in shoots from the 10 kPa and 2% O2 treatments. B, Analysis of LEA113 and Cor78. LEA and Cor78 represent typical desiccation-related genes and are induced only in shoots from 10 kPa treatment. Treatment with 2% O2 resulted in repression of LEA and Cor78 when examined by real-time RT PCR, as indicated by values of less than one on the graphs. Error bars extending the axis are truncated, and the replicate values for the 2% O2 LEA arrays were essentially identical.
Figure 3B provides a similar comparison for two genes typical of a desiccation response: LEA and Cor78. Both genes were strongly induced by hypobaria and not by hypoxia. Although the induction values derived from the array data for LEA and Cor78 were relatively higher than those obtained from quantitative RT-PCR, RT-PCR indicated that these desiccation-related genes were actually repressed by hypoxia.
Specificity of the Hypobaric Response in Transgenic Plants
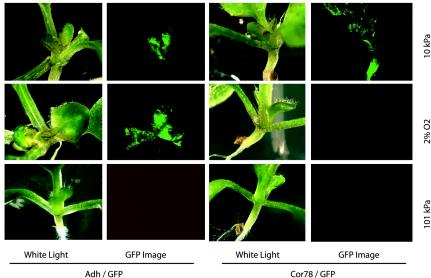
Arabidopsis plants carrying Adh/green fluorescent protein (GFP) or Cor78/GFP transcriptional promoter fusions were subjected to 24 h of 10 kPa or 2% O2 and then imaged for GFP expression in the shoot region (Fig. 4). The Adh/GFP construct was induced identically in the meristematic regions of the shoots of both hypobaric (10 kPa, top row) and hypoxic (2% O2, middle row) plants. The freshly emerging leaf pairs showed intense expression, whereas the fully emerged leaves showed little or no expression. The Cor78/GFP construct was induced in the shoots of hypobaric plants but was not apparently expressed in the shoots of hypoxic plants. In addition, Cor78/GFP expression was apparent in the central stem near the root-shoot junction and in the more fully expanded leaves. The hypobaria response was propagated over a wider distribution of shoot tissues than the hypoxia response, suggesting that the response to hypobaria is not limited to those tissues responding to hypoxia, and that the signaling mechanisms for the hypobaria-specific response are fundamentally different from those of the hypoxia response. Neither Adh/GFP nor Cor78/GFP was expressed in the corresponding control plants (101 kPa, bottom row).
Figure 4.
Specificity of the hypobaria response. Transgenic plants containing Adh/GFP and Cor78/GFP promoter fusions confirmed the selectivity of the hypobaria and hypoxia responses and highlight the tissue specificity of the gene activations revealed by the array and RT-PCR analyses. The response of Adh/GFP was essentially identical in plants exposed to 10 kPa or 2% O2 with the most responsive tissues being the young leaves surrounding the meristem. Fully expanded leaves showed a much reduced response. In contrast, the response of Cor78/GFP was markedly different between plants exposed to 10 kPa or 2% O2. Cor78/GFP was strongly induced in the central stem regions at 10 kPa, but plants at 2% O2 showed essentially no response. Plants maintained at 101 kPa showed no induction of either Adh/GFP or Cor78/GFP.
DISCUSSION
Arabidopsis plants respond to hypobaria with extensive changes in gene expression patterns. So even though plants have been known to survive exposure to hypobaria, clearly survival in hypobaric conditions requires a dynamic adaptive response. Some of the altered gene expression patterns are similar to those involved in the hypoxic stress response, as would be expected from the low-oxygen partial pressures present in hypobaria; yet some changes in gene expression patterns are unique to hypobaric conditions. These unique differential gene expression patterns demonstrate that hypobaria is not equivalent to hypoxia as an abiotic stress and suggest that, for plants, the response to hypobaria is more complex than the acclimation to the reduced partial pressures of oxygen inherent to low atmospheric pressures. By extension, environmental compensation for hypobaric stress in plants is not as simple as increasing the oxygen content as was done for humans in the low-pressure environments of the early space age.
Hypobaria Does Not Equal Hypoxia
Although hypobaria engages pathways distinct from those seen in response to hypoxia, there is significant overlap, indicating that the hypobaria response includes, but is not limited to, a typical response to hypoxia. A relatively small number of genes are induced by both hypobaria and hypoxia (Fig. 2, A and B, clusters C3, C2, and A3), primarily genes involved in fermentative pathways and oxygen transport (Klok et al., 2002). A much larger set of genes is repressed in response to both conditions (Fig. 2A, cluster B) and includes representatives from a wide range of metabolic pathways (see supplemental data). This wide-spread repression is a well-characterized phenomenon of hypoxic stress (e.g. Sachs et al., 1996; Vartapetian and Jackson, 1997; Klok et al., 2002). A much smaller cluster includes genes that were repressed in hypoxic conditions but were virtually unchanged in hypobaric conditions (Fig. 2B, cluster A1). The genes representing this group encode several putative proteins that may represent metabolic processes related to oxygen sensing that are mediated through a heme- or iron-based sensor (Quinn et al., 2002). Iron-based sensors have been implicated for direct oxygen or indirect redox sensing in various hypoxia-responsive pathways in eukaryotes (Bunn et al., 1998; Zhu and Bunn, 2001; Mecklenburgh et al., 2002), relying in part on subtilisin-catalyzed proteolytic inactivation of p450 that functions in the reduction of Fe(III) in the signal transduction cascade (Aravind and Koonin, 2001). Another small cluster is represented by genes that were induced in hypoxic conditions and repressed in hypobaric conditions (Fig. 2B, cluster C1). This group includes additional heme-related proteins, kinases, and phosphatases typical of a hypoxic response. The disparity in gene expression patterns displayed in each of these clusters suggests that that hypobaria and hypoxia may differentially activate certain alternative oxygen sensing and transport pathways.
Desiccation-Associated Pathways Are Induced by Hypobaria
Cluster A2 (Fig. 2B) defines a large group of genes showing increased expression in hypobaria while being unaffected or repressed in hypoxia, a group largely composed of genes associated with desiccation and abscisic acid signaling-related processes (e.g. Ozturk et al., 2002). Examples from cluster A2 include ABI2 (Chak et al., 2000), the APL3 subunit of ADP-Glc pyrophosphorylase (Weber et al., 1995; Rook et al., 2001), LEAs (e.g. Siddiqui et al., 1998; Kim et al., 2002), dehydrins and rab-like proteins (e.g. Mantyla et al., 1995; Nylander et al., 2001; Kizis and Pages, 2002), rd29B and cor15B (e.g. Wilhelm and Thomashow, 1993; Yamaguchi-Shinozaki and Shinozaki, 1993a), and aldehyde dehydrogenase (Kirch et al., 2001). Thus, it appears that the adaptation response of Arabidopsis to 10 kPa is, at a minimum, a specific adaptation to perceived desiccation that is overlaid upon an adaptation to hypoxia. However, the fact that the plants were grown in a humid environment (>95% relative humidity within the plates), showed no loss of fresh weight or turgor, and yet still responded to 10 kPa as if they were dehydrated indicates that the desiccation response is likely due to the perception of increased water flux caused by the low-pressure environment (Corey et al., 2002) rather than absolute loss of water in the plants (Figs. 1C, 2B, and 4).
Arabidopsis plants at 10 kPa presented no wilting, browning, or other hallmarks of desiccation. This observation begs the question as to whether successful adaptation to a 10 kPa environment actually requires the activation of desiccation-related pathways. It is possible that as long as there is sufficient environmental water available in the growth medium, an induction of desiccation-related metabolism may not be necessary for survival. With regard to bioregenerative life support for space, if the desiccation response is not necessary for adaptation to low pressure, then the induction of these pathways could represent a drain on metabolism with a resultant cost to production. Under this scenario, metabolic engineering designed to divert the inappropriate desiccation response could reduce the drain of these pathways on metabolism. If, however, the desiccation response is required, then enhancing desiccation tolerance may increase production at low pressure. Given the specificity and detail of the 10 kPa response indicated by the present analyses, studies at intermediate and extended low pressures and mutations in the desiccation and abscisic acid response pathways should clarify the role of desiccation responses to low atmospheric pressures. The present data do suggest that the desiccation stress response is tied as much to the flux of water through stomata as to the physical dehydration of tissue.
CONCLUSIONS
The data presented here address future needs to drive efficient plant growth at low pressures in extraterrestrial habitats and also address current issues involved in plant growth experiments in spaceflight environments where atmospheric pressure may be a variable. In the long term, reduced pressure environments may permit the use of life support systems that otherwise would be impractical or impossible under higher atmospheric pressures. For example, estimates indicate that a transparent greenhouse structure using currently available materials on Mars could be constructed only if the internal pressure of the greenhouse could be maintained below 7.5 kPa (Boston, 1981). Even if newer materials were developed to withstand higher pressure differentials, it is clear that leak rates are always higher with increased pressure differentials, and any reduction of internal pressure would have engineering benefits. In the near term, there are several mission-relevant issues regarding reduced atmospheric pressure. These include the effects of the transitory reduced Space Shuttle cabin pressures routinely experienced during extravehicular activities, and if emergency transition to space suits becomes a mission safety scenario, then the standard human mission pressures could potentially return to the 30 to 40 kPa range used during the Apollo and Skylab era to eliminate the time it takes to pre-adapt the crew to space suit pressures (Waligora et al., 1991).
MATERIALS AND METHODS
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
The LPGC
The LPGC was constructed by modifying a commercial 45-L vacuum oven (Vacuum Oven 285, Fisher Scientific, Fair Lawn, NJ; Fig. 1A). Sealed high-vacuum manifolds (not shown) facilitated the insertion of sensors to monitor and control environmental conditions. Temperature was maintained at 22°C ± 0.5°C by circulating chilled water through copper tubing lining the inner walls of the LPGC. Pressure sensing, coupled with automated vacuum pumping, maintained the total atmospheric pressure to within 1% of the set point. Hypoxic conditions (2% O2) were created by flowing a humidified 98% N2:2% O2 (v/v) gas mix into the LPGC. CO2 levels were maintained at a partial pressure of 0.1 kPa (± 5%) referencing a pressure-calibrated non-dispersive infrared CO2 probe. An externally mounted 400-W metal halide lamp supplied 150 μmol m-2 s-1 photosynthetically active radiation (400–700 nm). A PC-driven data acquisition system monitored the sensors and initiated appropriate compensations.
Plant Culture and Experiment Design
Arabidopsis (ecotype Wassilewskija [WS]) plants were grown on nutrient media agar plates held in the vertical position to encourage root growth along the aerated and exposed surface of the agar (Paul et al., 2001; Fig. 1B). The culture conditions preceding the LPGC environmental treatments consisted of 9 d of growth in continuous light (70–80 μmol m-2 s-1) at 22°C to 24°C. These growth conditions produce no overt or incipient hypoxia due to roots penetrating the agar substrate (Paul et al., 2001). The experimental treatments were conducted on two separate occasions. On each occasion, multiple plates were treated as a group at 10 kPa with normal air, with a 2% O2:98% N2 (v/v) gas mixture at 101 kPa, or with normal air at normal pressure of 101 kPa. Subsequent to the chamber treatments, the plants were pulled off the plates with forceps and immersed immediately in RNAlater (Ambion, Austin, TX). Tissues from several plates for each treatment were pooled, and the total time from chamber venting to immersion of tissue into RNAlater was less than 10 min. In a separate experiment, replicate plates of plants grown for the chamber analyses were assayed for possible water loss on the basis of fresh weight as compared with plants grown outside the chamber for the same time period. Representative plants were harvested and weighed before and after treatments (Fig. 1C). The environment inside a representative vertical plate containing 9-d-old plants was monitored for temperature and relative humidity with a HOBO data logger during a 10 kPa run in the LPGC.
Microarray Sample Preparation
Total RNA from each pooled shoot sample was extracted using RNAeasy kits from Qiagen (Valencia, CA): Tissue (150 mg) was ground in a solution composed of 100 μL of RNAlater and 450 μL of the Qiagen RLT buffer. All subsequent steps were as described by the Qiagen kit instructions, including a DNase I treatment interposed in the wash steps. The target RNAs were labeled and prepared for hybridization according to the protocols outlined in the GeneChip Expression Analysis Technical Manual (Revision 1, 2001, Affymetrix, Santa Clara, CA). In brief, 8 μg of total RNA was used as template for first-strand cDNA synthesis (Superscript, Invitrogen, Carlsbad, CA), which was primed with a T7-(dT)24 primer containing a T7 RNA polymerase promoter sequence (Genset Oligos, La Jolla, CA). In vitro transcription was performed on the second-strand product (Bioarray High Yield RNA Transcript labeling kit, Enzo Diagnostics, New York), and the resulting biotinylated cRNA was heated at 94°C for 35 min in a magnesium and potassium acetate-containing buffer (Affymetrix), which produces fragments 35 to 200 bases in length. Arabidopsis genome arrays (Affymetrix 900292) were hybridized for 16 h at 45°C with 15 μg of fragmented cRNA, stained with a streptavidin-phycoerythrin conjugate (Molecular Probes, Eugene, OR), and scanned with an Agilent argon-ion laser with a 488-nm emission and a 570-nm detection (GeneArray scanner).
Quantitative RT-PCR
Selected genes identified from the microarrays were quantified with Taqman real-time RT-PCR from Applied Biosystems (Bustin, 2000). The Applied Biosystems Prism 7700 Sequence Detection System was used for the analyses. Forward and reverse primers flanking a 60- to 100-bp section of the gene of interest were ordered from Applied Biosystems along with the fluorescently tagged probe sequence that lies between the primer pairs. The amount of targeted message in each sample was determined by relating the Taqman results for the gene of interest to a standard curve.
Experiment Analysis
Affymetrix cel files were analyzed with Probe Profiler software (Corimbia, Berkeley, CA) to generate quantitative estimates of gene expression. Probes that were not considered present (P = 0.05) in any of the samples were eliminated from further analyses. A one-way ANOVA for three treatments and two replications was performed on the expression values. ANOVA probabilities were then scaled for color. Expression values of genes for which there was a significant treatment effect were then normalized by first subtracting the mean of the control values for that gene. This difference was then divided by the sd of the six expression values for that gene. K-Means clustering was performed on the normalized expression values using Cluster (Eisen et al., 1998). Examination of the different K-Means clusters was the fundamental way different patterns of gene expression were identified. Vizard v1.2 (Moseyko and Feldman, 2002) was used to generate -fold change data on the non-normalized expression values. The K-Means cluster data, together with the ANOVA values and the derived fold-change data, were visualized using Treeview (Eisen et al., 1998).
Promoter/GFP Transgenic Plants
The Cor78 promoter/sGFP(S65T) construct was developed through PCR amplification from Arabidopsis genomic DNA using primers 5′-CCCAAGCTTAAGTTTGAAAGAAAATTTATTTCTTC and 3′-GCTCTAGAAAAAAATATAAGTTTCTGAAATTAGAA (where underlined sequence represent HindIII and XbaI restriction sites, respectively) that were designed to amplify 961 bp up-stream of the Cor78 (RD29A) gene start site. A β-glucuronidase reporter gene study demonstrated that this segment of the Cor78 promoter contains regulatory sequences that are activated upon exposing Arabidopsis to cold, drought, high salt, and abscisic acid (Yamaguchi-Shinozaki and Shinozaki, 1993b). The sGFP(S65T) gene was cloned into the pCAMBIA1300 vector via XbaI and SacI restriction sites before the addition of the Cor78 promoter via HindIII and XbaI restriction sites. Of the more than 15 Cor78/GFP transgenic lines (produced in ecotype WS), four were extensively characterized for consistent response to dehydration before choosing the line for low pressure analysis in Figure 4. The Adh/GFP transgenic line (also in WS) were extensively characterized for response to hypoxia as previously described (Manak et al., 2002).
Supplementary Material
Acknowledgments
We thank V. Rygalov and P. Fowler for initial chamber design considerations and discourse, M.W. Meisel for gas law discussions, and R. Wheeler for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.032607.
This work was supported by the National Aeronautics and Space Administration (grant nos. NAG 10–316, NAG 10–291, and AO–99–HEDS–01–032). This is manuscript R-09787 of the Florida Agricultural Experiment Station.
The online version of this article contains Web-only data.
References
- Andre M, Massimino D (1992) Growth of plants at reduced pressures. Experiments in wheat: technological advantages and constraints. Adv Space Res 12: 97-106 [DOI] [PubMed] [Google Scholar]
- Andre M, Richaux C (1986) Can plants grow in quasi-vacuum? In CELSS 1985 Workshop, Ames Research Center. NASA Publication TM 88215: 395-404 [Google Scholar]
- Aravind L, Koonin EV (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol 2: 007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D (1981) The History of Spaceflight. Crown Publishers, Inc., New York
- Berger D, Altmann T (2000) A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14: 1119-1131 [PMC free article] [PubMed] [Google Scholar]
- Boston PJ (1981) Low-pressure greenhouses and plants for a manned research station on Mars. J Br Interplanetary Soc 54: 189-192 [Google Scholar]
- Bunn HF, Gu J, Huang LE, Park JW, Zhu H (1998) Erythropoietin: a model system for studying oxygen-dependent gene regulation. J Exp Biol 201: 1197-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169-193 [DOI] [PubMed] [Google Scholar]
- Chak RK, Thomas TL, Quatrano RS, Rock CD (2000) The genes ABI1 and ABI2 are involved in abscisic acid- and drought-inducible expression of the Daucus carota L. Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta 210: 875-883 [DOI] [PubMed] [Google Scholar]
- Corey KA, Barta DJ, Wheeler RM (2002) Toward Martian agriculture: responses of plants to hypobaria. Life Support Biosph Sci 8: 103-114 [PubMed] [Google Scholar]
- Drysdale AE (2001) Life Support Trade Studies Involving Plants. SAE Technical Paper Series 2001-01-2362
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ, Schuerger AC, Paul A-L, Gurley WB, Corey K, Bucklin R (2002) Plant adaptation to low atmospheric pressures: potential molecular responses. Life Support Biosph Sci 8: 93-101 [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell Suppl 14: S15-S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale J (1972) Availability of carbon dioxide for photosynthesis at high altitudes: theoretical considerations. Ecology 53: 494-497 [Google Scholar]
- Gale J (1973) Experimental evidence for the effect of barometric pressure on photosynthesis and transpiration. In Plant Responses to Climatic Factors. Proceedings of the Uppsala Symposium. UNESCO, Paris, pp 289-294
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233-244 [DOI] [PubMed] [Google Scholar]
- He C, Davies FT, Lacey RE, Drew MC, Brown DL (2003) Effect of hypobaric conditions on ethylene evolution and growth of lettuce and wheat. J Plant Physiol 160: 1341-1350 [DOI] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL (2002) Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol 130: 688-697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch H-H, Nair A, Bartels D (2001) Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. Plant J 28: 555-567 [DOI] [PubMed] [Google Scholar]
- Kizis D, Pages M (2002) Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J 30: 679-689 [DOI] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481-2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind CT (1971) Germination and Growth of Selected Higher Plants in a Simulated Space Cabin Environment. Wright-Patterson AFB, Ohio AMRL-TR-70-121
- Manak MS, Paul A-L, Sehnke PC, Ferl RJ (2002) Remote sensing of gene expression in planta: transgenic plants as monitors of exogenous stress perception in extraterrestrial environments. Life Support Biosph Sci 8: 83-91 [PubMed] [Google Scholar]
- Mansell RL, Rose GW, Richardson B, Miller RL (1968) Effects of Prolonged Reduced Pressure on the Growth and Nitrogen Content of Turnip (Brassica rapa L.). SAM-TR-68–100. School of Aerospace Medicine Technical Report, pp 1-13 [PubMed]
- Mantyla E, Lang V, Palva E (1995) Role of abscisic acid in drought-Induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol 107: 141-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CE, McCormick AK (1992) Air Handling and Atmosphere Conditioning Systems for Manned Spacecraft. ICES Paper No. 921350. SAE International, Warrendale, PA
- McKay CP, Toon OB (1991) Making Mars habitable. Nature 352: 489-496 [DOI] [PubMed] [Google Scholar]
- Mecklenburgh KI, Walmsley SR, Cowburn AS, Wiesener M, Reed BJ, Upton PD, Deighton J, Greening AP, Chilvers ER (2002) Involvement of a ferroprotein sensor in hypoxia-mediated inhibition of neutrophil apoptosis. Blood 100: 3008-3016 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Feldman LJ (2002) VIZARD: analysis of Affymetrix Arabidopsis GeneChip data. Bioinformatics 18: 1264-1265 [DOI] [PubMed] [Google Scholar]
- Mott KA, Parkhurst DF (1991) Stomatal responses to humidity in air and helox. Plant Cell Environ 14: 509-515 [Google Scholar]
- Musgrave ME, Gerth WA, Scheld HW, Strain BR (1988) Growth and mitochondrial respiration of mungbeans (Phaseolus aureus Roxb.) germinated at low pressure. Plant Physiol 86: 19-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263-279 [DOI] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48: 551-573 [DOI] [PubMed] [Google Scholar]
- Paul A-L, Daugherty CJ, Bihn EA, Chapman DK, Norwood KL, Ferl RJ (2001) Transgene expression patterns indicate that spaceflight affects stress signal perception and transduction in Arabidopsis. Plant Physiol 126: 613-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Eriksson M, Moseley JL, Merchant S (2002) Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol 128: 463-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell KM, Kuang A, Porterfield DM, Crispi ML, Xiao Y, McClure G, Musgrave ME (2001) Influence of atmospheric oxygen on leaf structure and starch deposition in Arabidopsis thaliana. Plant Cell Environ 24: 419-428 [DOI] [PubMed] [Google Scholar]
- Reddy AR, Ramakrishna W, Sekhar AC, Ithal N, Babu PR, Bonaldo MF, Soares MB, Bennetzen JL (2002) Novel genes are enriched in normalized cDNA libraries from drought-stressed seedlings of rice (Oryza sativa L. subsp. indica cv. Nagina 22). Genome 45: 204-211 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421-433 [DOI] [PubMed] [Google Scholar]
- Rule DE, Staby GL (1981) Growth of tomato seedlings at sub-atmospheric pressures. HortScience 16: 331-332 [Google Scholar]
- Sachs MM, Subbaiah CC, Saab IN (1996) Anaerobic gene expression and flooding tolerance in maize. J Exp Bot 47: 1-15 [Google Scholar]
- Sadiqov ST, Akbulut M, Ehmedov V (2002) Role of Ca2+ in drought stress signaling in wheat seedlings. Biochemistry 67: 491-497 [DOI] [PubMed] [Google Scholar]
- Siddiqui NU, Chung HJ, Thomas TL, Drew MC (1998) Abscisic acid-dependent and -independent expression of the carrot late-embryogenesis-abundant-class gene Dc3 in transgenic tobacco seedlings. Plant Physiol 118: 1181-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SM (1961) Effects of reduced oxygen tension on vascular plants. Physiol Plant 14: 554-557 [Google Scholar]
- Siegel SM, Rosen LA, Giumarro C (1963) Plants at sub-atmospheric oxygen levels. Nature 198: 1288-1289 [Google Scholar]
- Soulages JL, Kim K, Arrese EL, Walters C, Cushman JC (2003) Conformation of a group 2 late embryogenesis abundant protein from soybean: evidence of poly (l-proline)-type II structure. Plant Physiol 131: 963-975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K (2000) An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol 41: 898-903 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Mol Biol 50: 571-599 [DOI] [PubMed] [Google Scholar]
- Vartapetian BB, Jackson MB (1997) Plant adaptations to anaerobic stress. Ann Bot 79: 3-20 [Google Scholar]
- Waligora JM, Horrigan DJ, Nicogossian A (1991) The physiology of spacecraft and space suit atmosphere selection. Acta Astronaut 23: 171-177 [DOI] [PubMed] [Google Scholar]
- Weber H, Heim U, Borisjuk L, Wobus U (1995) Cell-type specific, coordinate expression of two ADP-glucose pyrophosphorylase genes in relation to starch biosynthesis during seed development of Vicia faba L. Planta 195: 352-361 [DOI] [PubMed] [Google Scholar]
- Wheeler RM (2000) Can plants grow at high carbon dioxide partial pressures? Inflatable Greenhouse Workshop NASA TM 2000-2 08577: 58-63
- Wieland PO (1998) Living together in space: the design and operation of the life support systems on the International Space Station. NASA TM 1998-2 06956: 1
- Wilhelm KS, Thomashow MF (1993) Arabidopsis thaliana cor15b, an apparent homologue of cor15a, is strongly responsive to cold and ABA, but not drought. Plant Mol Biol 23: 1073-1077 [DOI] [PubMed] [Google Scholar]
- Winkler HE (1992) Shuttle Orbiter ECLSS Flight Experience. ICES Paper No. 921348. SAE International, Warrendale, PA
- Yamaguchi-Shinozaki K, Shinozaki K (1993a) Arabidopsis DNA encoding two desiccation-responsive rd29 genes. Plant Physiol 101: 1119-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993b) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236: 331-340 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473-1483 [DOI] [PubMed] [Google Scholar]
- Zhu H, Bunn HF (2001) Signal transduction: How do cells sense oxygen? Science 292: 449-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.