SUMMARY
Recently a new definition of surrogate endpoint, the ‘principal surrogate’, was proposed based on causal associations between treatment effects on the biomarker and on the clinical endpoint. Despite its appealing interpretation, limited research has been conducted to evaluate principal surrogates, and existing methods focus on risk models that consider a single biomarker. How to compare principal surrogate value of biomarkers or general risk models that consider multiple biomarkers remains an open research question. We propose to characterize a marker or risk model’s principal surrogate value based on the distribution of risk difference between interventions. In addition, we propose a novel summary measure (the standardized total gain) that can be used to compare markers and to assess the incremental value of a new marker. We develop a semiparametric estimated-likelihood method to estimate the joint surrogate value of multiple biomarkers. This method accommodates two-phase sampling of biomarkers and ismore widely applicable than existing nonparametric methods by incorporating continuous baseline covariates to predict the biomarker(s), and is more robust than existing parametric methods by leaving the error distribution of markers unspecified. The methodology is illustrated using a simulated example set and a real data set in the context of HIV vaccine trials.
Keywords: Estimated likelihood, Predictiveness curve, Principal stratification, Semiparametric, Surrogate marker, Total gain
1. Background
In a randomized trial, substituting a surrogate endpoint(s) for the primary clinical endpoint when evaluating the intervention effect on the clinical outcome can be appealing. If the effect of treatment on the short-term biomarkers reliably predicts its effect on the long-term clinical endpoint, then a much shorter and smaller trial can be conducted on the basis of the biomarker with enormous savings of time and cost. Moreover, surrogate markers might help elucidate the mechanism of intervention effect on the clinical endpoint.
We consider a two-arm randomized trial in this manuscript. Let Z be the binary treatment indicator, 0 for placebo and 1 for active treatment. Let S be the candidate surrogate, possibly multivariate, with each component measured on the continuous scale at fixed time τ after randomization. Let Y denote the binary clinical endpoint, 0 for nondiseased and 1 for diseased. Acknowledging the possibility that Y occurs before S is measured, let Yτ be the indicator of whether disease develops before τ; S is only measured if Yτ = 0. Let W be baseline covariates such as demographics and laboratory measurements. We allow for a two-phase sampling design wherein S and perhaps components of W are only measured in a subcohort. When missingness only occurs in S, let δ be the indicator of whether S is measured. The observed data are n iid copies . Let S(z), Yτ (z), Y (z) be the corresponding potential outcomes under treatment assignment z, for z = 0, 1. If Yτ (z) = 1, S(z) is undefined and we set S(z) = *.
Joffe and Greene (2009) summarized four different frameworks that have been used for evaluating a surrogate endpoint. The greatest amount of research has been done within the framework stimulated by the Prentice (1989) definition and operational criteria for a surrogate endpoint. This framework defines a surrogate as a replacement endpoint that provides a valid test of the null hypothesis of no clinical treatment effect. Regression methods have been used to evaluate Prentice’s operational criteria (e.g., Freedman et al. (1992); Lin et al. (1997); Burzykowski et al. (2005)). This framework seems most advantageous when there is substantial variability of the biomarker in both treatment groups, and the trial collects data on all of the important clinical risk factors, which facilitate achieving a valid check of the Prentice definition (Joffe and Greene, 2009; Wolfson and Gilbert, 2010). A second framework supposes the investigator can assign subjects to joint levels of the treatment and the biomarker, and evaluates direct and indirect effects (Robins and Greenland, 1992). This framework seems most advantageous for contexts where it is possible to reliably manipulate the biomarker in most subjects. The third and fourth frameworks are closely related, which evaluate the causal association of treatment effects on the biomarker and on the clinical endpoint, either based on many trials via a meta-analysis (e.g., Daniels and Hughes (1997); Buyse et al. (2000)) or based on a single large trial (Frangakis and Rubin, 2002). Here we develop new methods for evaluating a principal surrogate within the fourth framework; it is not our goal to compare methods across the different frameworks, and we concur with Joffe and Greene (2009) that the frameworks are all useful and complementary, with their relative suitability depending on the problem context.
After pointing out that evaluation of a surrogate endpoint based on the Prentice criteria can be misleading if simultaneous predictors of S and Y are not accounted for, Frangakis and Rubin (2002) proposed a new definition of surrogate endpoint that avoids this potential pitfall, the ‘principal surrogate’, based on principal stratification. By construction, this approach cannot give misleading results due to unmeasured simultaneous predictors, because the causal estimand conditions on principal strata (S(0) and S(1)), which can be treated as baseline covariates. Criteria for defining a univariate principal surrogate have been proposed based on comparing the risk of Y under assignment Z = 0 versus under assignment Z = 1, i.e., risk(0){s1, s0} = P{Y (0) = 1|S(1) = s1, S(0) = s0, Yτ (1) = Yτ (0) = 0} versus risk(1){s1, s0} = P{Y (1) = 1|S(1) = s1, S(0) = s0, Yτ (1) = Yτ (0) = 0} (Gilbert and Hudgens (2008), henceforth GH). The first criterion, Average Causal Necessity, states that if Z has no effect on S, then Z has no average effect on Y, i.e., risk(1){S(1), S(0)} = risk(0){S(1), S(0)} for S(1) = S(0) (i.e., no ‘dissociative effects’). The second criterion, Average Causal Sufficiency, states that if Z has a large enough effect on S, then Z has an average effect on Y, i.e., risk(1){S(1), S(0)} ≠ risk(0){S(1), S(0)} for all S(1) ≠ S(0) with S(1) − S(0) ⩾ c > 0 (i.e., complete ‘associative effects’ when S(1) − S(0) ⩾ c). Frangakis and Rubin (2002) only required Average Causal Necessity to define a principal surrogate. This seems appropriate, as Average Causal Sufficiency is ‘less fundamental’ in a sense. Specifically, consider the setting where either no clinical events occur before S is measured or there is no treatment effect before S is measured (P{Yτ (1) ≠ Yτ (0)} = 0). For this setting, Average Causal Necessity plus some overall treatment effect P{Y (1) = 1} ≠ P{Y (0) = 1} imply the existence of some associative effects (values S(1) ≠ S(0) such that risk(1){S(1), S(0)} ≠ risk(0){S(1), S(0)}). Moreover, the existence of some associative effects (not requiring complete associative effects), provides guidance for how to reformulate a partially effective treatment or vaccine (Wolfson and Gilbert, 2010). Thus, Average Causal Necessity is the more essential condition to evaluate, which in mediation analysis is referred to as no direct effects (e.g., Gallop et al. (2009)).
Average Causal Necessity and Sufficiency can be assessed based on coefficients in risk models. GH proposed fully parametric and nonparametric methods for estimation. The search for a ‘totally valid’ or ‘perfect’ surrogate has usually failed (Fleming and DeMets, 1996; Weir and Walley, 2006; Burzykowski et al., 2005), and hence in practice the more germane objective is to compare the degree of surrogate value between different risk models. For example, two markers might both satisfy Average Causal Necessity and have some associative effects, and it is interesting to know which one has better surrogacy. Or, one biomarker included in a risk model may have mediocre surrogate value, and we want to know whether adding new markers to the risk model improves surrogate value. Comparisons of these kinds usually cannot be based on coefficients in risk models because the interpretation of coefficients depends on other components in the model as well as the model’s functional form. Moreover, to be useful in practice it is important that the comparison is based on a clinically meaningful measure.
We propose a graphical tool to characterize and compare the principal surrogate value between risk models. A clinically meaningful summary measure derived from this graphical tool– the standardized total gain (STG)– is proposed for making inferences. We develop a semiparametric estimated-likelihood method to estimate the STG, allowing for the joint effect of multiple biomarkers. This work is based on a similar set-up and set of identifiability assumptions as in GH, and, like their work, accommodates two-phase sampling of the biomarkers. In addition to considering more than one biomarker and the new graphical tool and the summary measure STG, its new developments are to relax the fully parametric assumptions in the estimated-likelihood technique and to incorporate the innovative close-out placebo vaccination augmented study design proposed by Follmann (2006). We illustrate the new methodology using simulation studies and an HIV vaccine trial example.
2. Methods
2.1 Identifiability Assumptions
Throughout we adopt identifiability assumptions A1–A4 made by GH.
(A1) SUTVA and Consistency: {S(1), S(0), Yτ (1), Yτ (0), Y (1), Y (0)} is independent of the treatment assignments of other subjects, and given the treatment a subject actually received, a subject’s potential outcomes equal the observed outcomes.
(A2) Ignorable Treatment Assignments: Z ⊥ W, S(1), S(0), Yτ (1), Yτ (0), Y (1), Y (0).
(A3) Equal Early Clinical Risk: Yτ (1) = Yτ (0).
(A4) Risk of Y (z) given S and W follows a generalized linear model for z = 0, 1.
GH and Wolfson and Gilbert (2010) discussed the role, plausibility, and testable consequences of these identifiability assumptions. Briefly, A1 is plausible in trials where participants do not interact with one another and A2 is plausible in randomized blinded trials. A3 will usually be implausible if the treatment is efficacious but violations of it will be inconsequential if few clinical events happen before the biomarker is measured. A4 extends GHs work (A4-P) to model multiple biomarkers, and we defer its specification to Section 2.3. The current work is quite different from Wolfson and Gilbert (2010), which restricted attention to a binary biomarker and nonparametric models, and focused on relaxing A3 and identifiability issues.
A1–A4 and the observed iid data identify risk(0) and risk(1) and our proposed summary measure STG, which we describe next. A3 is useful because it implies risk(z){S(1), S(0)} = P{Y (z) = 1|S(1), S(0), Yτ = 0} for each z = 0, 1, so that risk(z) can be identified based on the subset of subjects assigned Z = z who are observed to have the marker measured at time τ. Wolfson and Gilbert (2010) showed that relaxing A3 makes identification and estimation of the risk(z) considerably more difficult, and hence the methods described here are restricted to applications where A3 is reasonable. Henceforth we simplify the notation and drop the conditioning of all probabilities on Yτ (1) = Yτ (0) = 0; by A3 these probabilities all equivalently condition on Yτ = 0, such that subjects contribute to the analysis if and only if they are observed to be at-risk at time τ.
2.2 Characterizing the Principal Surrogate Value of Biomarkers
Without loss of generality, we introduce the idea behind the STG for a risk model conditional on the biomarkers only. The generalization to include baseline covariates W is straight-forward. Let D = Y (0) − Y (1), the individual treatment effect on the clinical endpoint. The risk difference between the two treatment arms, which we denote as Δ{S(1), S(0)} = risk(0){S(1), S(0)} − risk(1){S(1), S(0)} = E{D|S(1), S(0)}, is a measure of the treatment effect conditional on biomarker values. We propose to characterize the surrogacy of the candidate biomarker using the distribution of the risk difference Δ. The amount of variability in D explained by Δ indicates how reliably the biomarkers/risk model predicts the treatment effect, with larger variability explained implying superior surrogacy.
We use a quantile plot of Δ to demonstrate its distribution graphically. With υ a number taking value in (0, 1) and R(υ) the υth quantile of Δ{S(1), S(0)}, we plot a monotone increasing curve of R(υ) versus υ. Earlier a plot of similar flavor was proposed in a different setting where a marker’s predictive capacity for disease risk was assessed (Bura and Gastwirth, 2001; Pepe et al., 2008; Huang et al., 2007). It can be easily shown (GH) that the area under this curve is equal to the difference in prevalence of potential clinical outcomes between the two treatment arms, i.e. ρ0 − ρ1, where ρz = P{Y (z) = 1}, z = 0, 1. This facilitates visual comparison of two curves in terms of their steepness. A steeper curve corresponds to larger variability in Δ, and hence better prediction of D.
To formally compare risk models, we propose to use the total gain (TG) and its standardized version derived from the corresponding quantile curve. Here TG, originally proposed by Bura and Gastwirth (2001) in the risk prediction setting, is the area sandwiched between the quantile curve and the horizontal line ρ0 − ρ1, . Larger TGs indicate larger differences in risk model compared to a useless model that predicts the same D value for each subject, thus implying larger surrogate value. Moreover, under an additional assumption A5 below, the standardized TG, STG = TG/[2(ρ0 − ρ1){1 − (ρ0 − ρ1)}], has an appealing interpretation related to the accuracy of classifying a subject according to treatment-effectiveness.
(A5) No Harm by Active Treatment (Monotonicity): Y (1) ⩽ Y (0).
Theorem 1. Assuming Y (0) ⩾ Y (1), given a threshold c, suppose a subject is classified into the treatment-effective category D = Y (0)−Y (1) = 1 if his/her predicted risk difference, Δ{S(1), S(0)}, is greater than c, or classified into the treatment-ineffective category D = 0 if his/her Δ value is equal to or less than c. Then sensitivity and specificity at given c are Sensitivity(c) = P [Δ{S(1), S(0)} > c|D = 1]; Specificity(c) = P [Δ{S(1), S(0)} ⩽ c|D = 0], and STG = maxc{Sensitivity(c)+Specificity(c)} − 1, the Youden’s Index (Youden, 1950).
The proof of Theorem 1 is sketched in Web Supplementary Appendix A. Through its link with the classification accuracy measures, the STG is a clinically meaningful criterion that can be used to compare general risk models in terms of their principal surrogate value. Two hypothetical examples are presented in Web Supplementary Appendix B, where we demonstrate the comparison of principal surrogacy between two markers applied to the same population using the proposed graphical tool and the STG. Because of the standardization by ρ0 − ρ1, this measure is free of disease prevalence, which implies its potential utility for comparing surrogacy across different populations. Note that sensitivity and specificity in the principal surrogate marker setting is defined based on individual treatment-effectiveness status that is not observable. Hence we cannot compute these classification accuracy measure directly. Nevertheless, the STG can still be estimated by modeling the risks for each treatment arm, and serves as a bridge between the risk model and classification accuracy measures.
Previously proposed summary measures of principal surrogate value include the proportion associative (PA) (Taylor et al., 2005), the surrogate associative proportion (SAP), the surrogate dissociative proportion (SDP), the common associative proportion (CAP) (Li et al., 2010), the proportion associative effect (PAE), and the associative span (AS) (GH). Under A1–A3 and A5, PA = P{S(1) > S(0)|D = 1} is related to Sensitivity(c) above, although the PA does not involve modeling risk(z). Introduced in a binary marker setting, the SAP and SDP correspond to the positive and negative predictive value, respectively, while the CAP is a mixture of the sensitivity and the positive predictive value. The PAE and the AS measure a combination of the positive and negative predictive values of how well differences S(1) − S(0) predict clinical treatment effects. In contrast, the TG and STG measure a combination of how well the risk model reflects positive and absent clinical treatment effects. In Section 3 and 4, we assess PAE and AS together with TG and STG since none of them requires the monotonicity assumption A5 for estimation, whereas A5 is critical for defining and estimating the other measures mentioned above.
2.3 Semiparametric Method for Evaluating Principal Surrogacy of a General Risk Model
In this section, we propose a semiparametric method to estimate a risk model’s principal surrogate value. Without loss of generality, we assume W is measured for everyone.
Missing potential biomarkers pose a challenge to the evaluation of principal surrogate value. In some applications such as HIV vaccine trials where the biomarker of interest is the immune response to HIV targets, the problem is simplified because subjects receiving placebo have no HIV-specific immune response, i.e., S(0) equals the zero-vector 0 for every subject. This condition was named ‘Constant Biomarkers’ (Case CB) by GH. From now on, we assume Case CB for mathematical convenience. When S(0) = 0 for all subjects, only S(1) needs to be incorporated into the risk model. However, even outside of Case CB, conditioning on S(1) and omitting S(0) in the risk model still yields a causal estimand of interest: P{Y (0) = 1|S(1), Yτ (1) = 0} − P{Y (1) = 1|S(1), Yτ (1) = 0} averages Δ{S(1), S(0)} over the distribution of S(0). Moreover, if we are interested in risks conditional on the joint values {S(1), S(0)}, the method we propose in the next subsection can be easily extended to some settings where Case CB does not hold, given proper enforcement of study design, as will be explained in the discussion.
GH proposed a parametric and a nonparametric method for risk model estimation, both relying on W to predict the missing S(1)’s of subjects assigned placebo. The former models both the risks and the joint distribution of S(1) and W parametrically. The latter models both these components nonparametrically, but requires discretization of both W and S(1). Our semiparametric method allows continuous S(1) and either continuous or discrete W and models the distribution of S(1) conditional on W semiparametrically for flexibility. Note our method is not designed to deal with truly high-dimensional data wherein the number of biomarkers is greater than the sample size. That would require additional research beyond the scope of this work. But the method is designed to handle several markers. We consider a general risk model with J markers. With a slight abuse of notation, let Sj indicate the S(1) value for marker j, j = 1, …, J. Assumptions A1–A3 guarantee . The former distribution can be estimated from the subjects in the Z = 1 arm and applied to subjects in the Z = 0 arm. This baseline immunogenicity predictor (BIP) design for vaccine trials was suggested by Follmann (2006).
We partition W into W = (X, A), where X are covariates predicting risk of Y and A are auxiliaries not associated with Y with sole function to help predict S(1). Assumption A4 posits a generalized linear model for the structural risks , for some parametric link function g. We assume missingness of markers is determined by design and that S1, …, SJ are always missing together. This missingness at random assumption allows estimation of the risk model based on the observed conditional likelihood. The ith subject’s contribution to the likelihood of disease conditional on observed covariates is P(Yi|Zi, Wi, δiSi1, …, δiSiJ), i.e., P(Yi|Zi, Wi, Si1, …, SiJ) when δi = 1, and P(Yi|Zi, Wi) when δi = 0. The latter can be represented as ∫ P(Yi|Zi, s1, …, sJ, Wi)dF(s1, …, sJ|Wi), where F is the joint CDF for S1, …, SJ conditional on W. We propose to maximize an estimated version of the likelihood,
| (1) |
by plugging in an estimator for F(s1, …, sJ|W, Z), which equals F(s1, …, sJ|W) by A1–A3. Maximum estimated likelihood methods that use nonparametric or parametric approaches for estimating the distribution of missing covariates conditional on observed covariates have been developed (e.g., Pepe and Fleming (1991), GH). Here we employ a semiparametric approach by modeling each Sj given W (for j = 1, …, J) with a location-scale model. Assume is the baseline CDF for the univariate residual εj. Let be the joint CDF for (ε1, …, εJ)T. We have
| (2) |
Thus estimation of F(S1, …, SJ|W) can be achieved by estimating the location and scale parameters μj and σj for each marker and estimating .
Suppose a random sample of nV subjects with Z = 1 have S1, …, SJ measured, which we call the validation set. Assume μj(W), log{σj(W)} are parametric functions of . Then γ̂j, η̂j, the estimators of γj, ηj, can be obtained by solving estimating equations for the mean and variance (Heagerty and Pepe, 1999) for Sj based on the validation set:
| (3) |
Thus for the nV validation samples, we obtain a series of residuals (e1k, …, eJk)T, k = 1, …, nV, where . We estimate empirically based on these residuals. Entering and μ̂j, σ̂j into (2) we get F̂(s1, …, sJ|W) for any W of interest. Then for subject i with δi = 0, we compute P̂(Yi = 1|Zi, Wi)
| (4) |
where . Plugging these into (1) we obtain the estimated likelihood.
As shown in Web Appendix C an EM-algorithm can be employed to estimate the risk model parameters β: (I) Apply the semiparametric location-scale model to subjects in the validation set for each marker in the model and obtain corresponding estimates for model parameters and the baseline residuals distribution; (II) Start with an initial estimate of β; (III) For subjects i with δi = 1, use their observed data. For subjects i with δi = 0, construct a set of filled-in data, ; (IV) For each filled-in observation, calculate an associated weight,
(V) Fit a weighted GLM to the augmented dataset and obtain a new estimate of β; (VI) Repeat steps (IV) to (V) until convergence. After obtaining β̂, for a particular W (and X) of interest, we estimate disease prevalence with for z = 0, 1, and estimate TG(W) with , and estimate STG(W) with . We use the bootstrap to compute standard errors and confidence intervals; standard asymptotic-based inferences do not work because some subjects have zero probability that S1, …, SJ are sampled.
2.4 Alternative Sampling Designs and Closeout Placebo Vaccination
The estimation procedure we developed in Section 2.3 can easily be generalized to accommodate general missing at random two-phase sampling designs. For example, S(1) in the validation set may be randomly sampled within disease cases and controls separately, and/or within baseline covariate strata separately. The sampling design can be accommodated in our setting by: (I) Multiplying subject i’s contribution to the estimating equations (3) by a weight proportional to the inverse estimated probability that he/she is sampled, 1/P̂(δi = 1 | Yi, Wi); (II) Multiplying subject i’s contribution by the same weight in calculating the joint CDF of the baseline residuals. Typically, for subjects with Yi = 1, P(δi = 1) will be set to the observed fraction of disease cases for whom the biomarker(s) is measured (ideally near 1), and for subjects with Yi = 0, P(δi = 1) will be estimated by stratum-specific observed fractions of controls for whom the biomarker(s) is measured, or, if multiple auxiliary covariates are used, by fitted values of a binary model regressing δ on the auxiliaries. If in addition W are only measured on a subset of samples, a weighted likelihood technique (e.g., Breslow and Wellner (2008)) can be applied to the samples with W observed.
GH showed that, because S(1) is missing in all placebo subjects, it is not possible to identify risk(0) in a standard randomized trial design without imposing an untestable constraint on the risk(0) model. As such, the method described above uses an untestable modeling assumption A4 for risk(0). An alternative solution is to enhance the study design. Follmann (2006) proposed a closeout placebo vaccination (CPV) design where a portion of placebo subjects who are uninfected at the end of the trial receive vaccine at closeout and their immune response Sc is measured τ time-units after close-out. If A6 and A7 below hold, then for uninfected placebo subjects we can substitute Sc for S(1), and the methods developed in Section 2.3 apply directly. Again we use subjects in arm Z = 1 with S(1) measured as our validation set, but now δ is 1 if the marker is measured either during or after the trial.
(A6) No infectious during the closeout period. That is, no placebo subjects uninfected at close-out have a disease event over the next τ time-units.
(A7) Time Constancy of the immune response distribution: are iid vectors of random errors with mean zero and have the same distribution. A7 implies that Sc can be used in place of S(1) without changing the risk model. Under A1–A3 and A6–A7, the risk(0) model is fully testable, as addressed further in the Discussion. An advantage of the CPV design is that it applies in the same way if there are multiple markers, so that the methods in Section 2.3 apply directly. Next we evaluate in simulations the BIP and CPV designs used together.
3. Simulation Studies
We consider simulation studies where n subjects are 1:1 randomized to placebo and active treatment for n ranging from 500 to 3,000. Suppose Case CB holds, a continuous baseline covariate W = X is measured for everyone, and the biomarkers value S(1) or Sc is available for everyone in the active treatment arm and all uninfected placebo subjects. We consider different surrogate values with STG ranging from 0.2 to 0.6. For each simulation setting, 1,000 simulations are conducted; and for each simulated dataset, 250 bootstrap samples are generated for construction of confidence intervals (CIs). Risk model parameters are chosen such that ρ0 = 0.12 and ρ1 = 0.06.
We first study a one-marker setting where risk of Y given S(1), W, and Z follows a probit model, and S(1) conditional on W follows a location-scale model with ε ~ t5 (details are presented in Web Supplementary Appendix D). Our proposed semiparametric approach is evaluated together with GH’s parametric method assuming a joint normal distribution of W and S(1) and GH’s nonparametric method where S(1) and W are discretized by quintiles. For two different risk models with small-to-large STG and allowing surrogate value of the marker to vary with W, we compare performance of different estimation methods for estimating the risk model coefficients (Web Supplementary Table 1) and for estimating W –specific TG, STG, PAE and AS (Web Supplementary Tables 2 and 3). The semiparametric approach has satisfactory performance in terms of minimal bias and accurate coverage probabilities using bootstrap percentile intervals for estimation of model coefficients and all summary measures. The parametric method, on the other hand, has non-ignorable bias with less than nominal coverage in general, due to the deviation of the joint distribution of W and S(1) from bivariate normal. The nonparametric method is also biased with less than norminal coverage. Comparing Wald tests for any surrogate value based on different summary measures and the semiparametric method, their relative powers vary with the risk model and baseline W level. For example, for risk model I with true γ = (−0.41, −0.2, −0.5, −0.48, 0.2, 0.26)T and small-to-medium PAE, the power of PAE tends to be less than that of other summary measures (Web Supplementary Table 2), but exceeds that of AS in risk model II with true γ = (−1.73, −1.5, −1.0, −0.7, 1.0, 0.68)T and large PAE (Web Supplementary Table 3). However, in general we found the power of TG and STG either better than or comparable to that of other summary measures, with the power of TG slightly larger compared to STG.
For the two-marker setting, we focus on the semiparametric approach and study TG and STG. We assume (S1, S2, W) follows a multivariate normal distribution with correlation 0.7 between markers and 0.5 between each marker and W. We assume a probit two-marker risk model, which induces a probit risk model based only on S1 and W (Web Supplementary Appendix E). Suppose the base-model contains marker 1 and we are interested in whether adding marker 2 increases surrogate value. We model the location parameters for the distributions of S1 and S2 conditional on W as linear in W. Performances of estimators are evaluated in two different settings where STG ranges from 0.2 to 0.4 for the one-marker model and from 0.3 to 0.5 for the two-marker model.
Table 1 presents results for the risk model parameter estimators. For both the one-marker and two-marker models, the semiparametric estimators appear to be approximately unbiased, with coverage of percentile bootstrap confidence interval close to the nominal level at sample size 500 or greater. Precision of the estimators increases with sample size.
Table 1.
Performance of the estimators for risk model parameters. Two-marker model: risk(Z)(S1, S2, W) = Φ(β0 + β1Z + β2S1 + β3S1Z + β4S2 + β5S2Z + β6W + β7W Z). One-marker model: risk(Z)(S1,W) = Φ (γ0 + γ1Z + γ2S1 + γ3S1Z + γ4W + γ5W Z).
| Parameter | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| One-marker model | |||||||||
| n | γ0 2.12 |
γ1 −0.60 |
γ2 −0.80 |
γ3 −0.094 |
γ4 −0.49 |
γ5 0.093 |
|||
| Bias | 500 | 0.17 | −0.04 | −0.07 | 0.003 | −0.04 | 0.02 | ||
| 1000 | 0.07 | 0.003 | −0.03 | −0.01 | −0.02 | 0.01 | |||
| 3000 | 0.034 | −0.02 | −0.018 | 0.01 | −0.003 | −2e-4 | |||
| SE | 500 | 1.00 | 1.20 | 0.54 | 0.60 | 0.22 | 0.31 | ||
| 1000 | 0.62 | 0.75 | 0.33 | 0.37 | 0.14 | 0.21 | |||
| 3000 | 0.34 | 0.43 | 0.18 | 0.20 | 0.08 | 0.11 | |||
| Cover* | 500 | 92.37 | 93.95 | 92.87 | 93.86 | 92.77 | 95.24 | ||
| 1000 | 92.85 | 94.45 | 93.38 | 93.98 | 93.32 | 94.18 | |||
| 3000 | 93.60 | 93.52 | 93.77 | 94.75 | 93.85 | 94.59 | |||
| Two-marker model | |||||||||
| n | β0 3.04 |
β1 −0.40 |
β2 −0.40 |
β3 −0.05 |
β4 −0.90 |
β5 −0.25 |
β6 −0.40 |
β7 0.12 |
|
| bias | 500 | 0.42 | −0.03 | −0.06 | 0.01 | −0.14 | −0.04 | −0.06 | 0.03 |
| 1000 | 0.19 | −0.01 | −0.03 | −0.003 | −0.06 | −0.01 | −0.03 | 0.01 | |
| 3000 | 0.08 | −0.04 | −0.02 | 0.01 | −0.02 | 0.005 | −0.004 | 0.001 | |
| SE | 500 | 1.41 | 1.79 | 0.59 | 0.68 | 0.69 | 0.84 | 0.27 | 0.40 |
| 1000 | 0.86 | 1.08 | 0.39 | 0.44 | 0.43 | 0.52 | 0.16 | 0.24 | |
| 3000 | 0.43 | 0.54 | 0.2 | 0.23 | 0.22 | 0.26 | 0.09 | 0.13 | |
| Cover* | 500 | 91.39 | 94.15 | 94.46 | 95.28 | 94.46 | 94.97 | 91.49 | 95.28 |
| 1000 | 92.76 | 94.5 | 93.70 | 94.17 | 92.56 | 93.36 | 92.82 | 93.76 | |
| 3000 | 93.93 | 94.34 | 94.09 | 94.67 | 93.52 | 94.18 | 93.52 | 94.59 | |
Coverage of 95% bootstrap percentile confidence interval
Results for the semiparametric estimators of TG and STG are provided in Table 2. Values of W corresponding to its 10th and 50th percentile in the population are considered. Again the estimators of TG and STG appear to be approximately unbiased for both models at sample size 500 or larger. Satisfactory coverage of their percentile bootstrap confidence interval can be achieved at sample size 500 or larger for the one-marker model but requires a larger sample size (1,000 or larger) for the two-marker model. For both models, the large standard errors for estimation of STG at sample size 500 reflect the impact of having a small estimated disease prevalence difference in the denominator of the STG in this rare disease setting; power to detect any surrogate value is in general larger for TG compared to STG, although the difference diminishes with a sample size as large as 3,000.
Table 2.
Performance of the semiparametric estimator for estimating TG and STG.
| One-marker model | Two-marker model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| W=1.72 | W=3.00 | W=1.72 | W=3.00 | ||||||
| n | TG 0.053 |
STG 0.206 |
TG 0.032 |
STG 0.388 |
TG 0.08 |
STG 0.313 |
TG 0.043 |
STG 0.517 |
|
| Bias | 500 | 0.02 | 0.08 | 0.002 | 0.05 | 0.04 | 0.15 | 0.01 | 0.13 |
| 1000 | 0.01 | 0.03 | −8e-4 | 0.01 | 0.02 | 0.08 | 0.01 | 0.07 | |
| 3000 | 0.002 | 0.01 | 1.2e-5 | 0.005 | 0.01 | 0.03 | 0.001 | 0.02 | |
| SE | 500 | 0.04 | 14.95 | 0.02 | 4.66 | 0.04 | 1.81 | 0.02 | 22.38 |
| 1000 | 0.03 | 0.21 | 0.02 | 0.29 | 0.03 | 0.13 | 0.02 | 0.75 | |
| 3000 | 0.02 | 0.08 | 0.01 | 0.11 | 0.02 | 0.06 | 0.01 | 0.07 | |
| Cover* | 500 | 92.37 | 95.74 | 95.14 | 97.52 | 81.23 | 87.59 | 92.51 | 93.23 |
| 1000 | 95.86 | 96.19 | 94.39 | 96.79 | 88.4 | 87.59 | 93.7 | 94.3 | |
| 3000 | 95.11 | 94.92 | 93.48 | 94.72 | 91.33 | 90.03 | 93.74 | 93.35 | |
| Power† | 500 | 29.73 | 9.12 | 30.92 | 11.99 | 30.43 | 11.08 | 27.02 | 13.35 |
| 1000 | 47.13 | 22.79 | 52.94 | 32.29 | 46.13 | 26.5 | 48.55 | 32.68 | |
| 3000 | 95.11 | 89.77 | 98.04 | 92.11 | 95.37 | 92.89 | 93.61 | 92.76 | |
Coverage of 95% bootstrap percentile confidence interval
Power to detect nonzero TG (STG) based on wald test using bootstrap standard error
Note P(Y = 1|Z = 0, W) and P(Y = 1|Z = 1, W) are 0.31 and 0.16 at W = 1.72 and 0.075 and 0.032 at W = 3.0.
Table 3 shows results for evaluating incremental value of marker 2 by assessing the increase in TG and STG achieved by adding marker 2 to the one-marker model. Confidence intervals based on the percentiles of bootstrap samples provide satisfactory coverage of the TG or STG difference. Based on both TG and STG, power to detect increased surrogacy with the additional marker 2 ranges from 10% to 50% for sample sizes varying from 500 to 3,000.
Table 3.
Performance of the semiparametric estimators to detect a higher TG and STG with a two-marker model compared with a one-marker model.
| W=1.7 | W=3.0 | ||||
|---|---|---|---|---|---|
| n | TG diff 0.027 |
STG diff 0.107 |
TG diff 0.011 |
STG diff 0.129 |
|
| Bias | 500 | 0.01 | 0.03 | 0.003 | 0.04 |
| 1000 | 0.01 | 0.03 | 0.002 | 0.03 | |
| 3000 | 0.002 | 0.01 | 3e-4 | 0.004 | |
| SE | 500 | 0.04 | 15.7 | 0.02 | 23.1 |
| 1000 | 0.03 | 0.14 | 0.01 | 0.82 | |
| 3000 | 0.02 | 0.06 | 0.01 | 0.09 | |
| Cover* | 500 | 97.72 | 99.28 | 98.76 | 97.1 |
| 1000 | 97.24 | 97.98 | 96.3 | 96.97 | |
| 3000 | 96.09 | 96.61 | 93.87 | 93.87 | |
| Power† | 500 | 14.29 | 8.59 | 11.39 | 7.97 |
| 1000 | 24.08 | 19.3 | 22.13 | 19.3 | |
| 3000 | 57.5 | 50.26 | 52.35 | 49.48 | |
Coverage of 95% bootstrap percentile confidence interval
Power to detect increase in TG (STG) — probability that 95% bootstrap percentile confidence interval does NOT cover zero
Note P(Y = 1|Z = 0, W) and P(Y = 1|Z = 1, W) are 0.31 and 0.16 at W = 1.72 and 0.075 and 0.032 at W = 3.0.
Finally, to evaluate the sensitivity of our estimators to violation of the CB condition, we modify the two-marker setting such that 5% of placebo subjects have a low-level but positive S value and analyze the data assuming CB. Results for estimation of TG and STG are presented in Tables 4 and 5 in Web Supplementary Appendix F. Comparing those to Tables 2 and 3 in the main text, we found that this small violation of CB has minimal impact on bias and coverage of the summary measure estimators for both the one-marker and two-marker models as well as for the increment due to the addition of marker 2. Under the small violation of CB, power to detect surrogate value (Web Supplementary Table 4) or increase in surrogate value (Web Supplementary Table 5) is again larger for TG compared to STG. In general the power is decreased when CB is violated, but only by about 5%.
4. Illustration
We illustrate our methods using data from the ‘Step’ HIV vaccine efficacy trial, where 3000 HIV seronegative participants were 1:1 randomized to receive the MRKAd5 HIV-1 Gag/Pol/Nef vaccine or placebo, pre-stratified by sex, baseline adenovirus type 5 (Ad5) neutralization titer, and study site. The primary objective of this study was to evaluate the effect of the candidate HIV vaccine (Z) on HIV infection diagnosis (Y) within 3 years of randomization (Buchbinder et al., 2008). Only one woman became HIV infected. Among 1836 men participants, the observed HIV infection rate was about twice as high in vaccine recipients than placebo recipients. It is of particular interest to assess if the vaccine effect on the magnitude of HIV-specific T cell response (S) to the Gag, Pol, and Nef HIV proteins measured τ = 8 weeks after randomization can predict the apparent elevated infection risk in the vaccine arm. The study cohort consists of 906 vaccine and 915 placebo recipients uninfected at week 8 visit, of which a fraction ρ̂1 = 0.045 and ρ̂0 = 0.028 became HIV infected. The magnitude of immune responses in placebo recipients was similar to background response levels, suggesting the appropriateness of Case CB. In the vaccine arm, immune responses to Gag, Pol and Nef were available in 35 (85.4%) infected and 203 (23.5%) uninfected men.
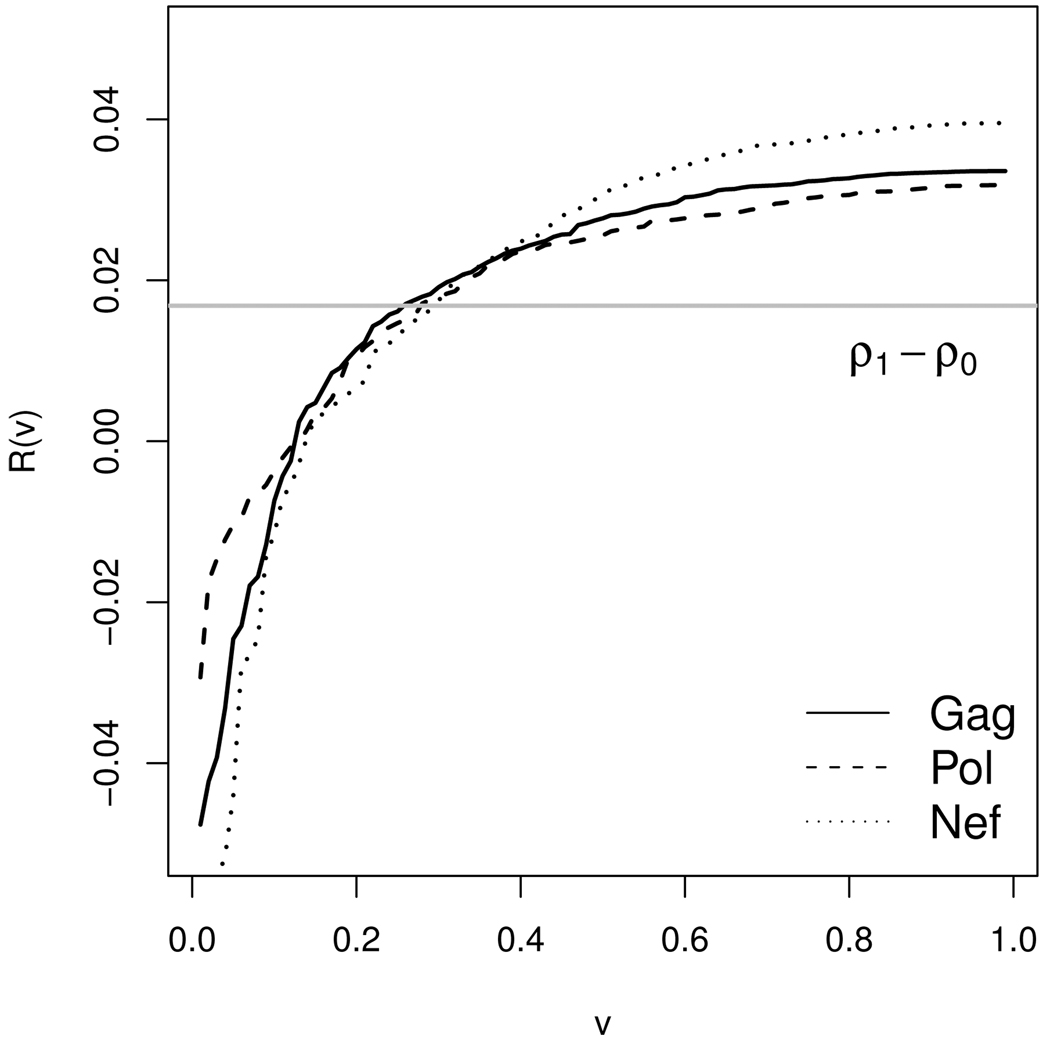
We assessed principal surrogacy for each of the three protein-specific biomarkers separately. For each marker, a probit risk model was assumed conditional on treatment (Z) and log-transformed biomarker given vaccine (S(1)): risk(Z){S(1)} = Φ{β0 + β1Z + β2S(1) + β3ZS(1)}. A semiparametric location model with mean of S(1) modeled linearly over log-transformed baseline Ad5 titer (W) was used to predict the unobserved S(1) value. Figure 1 displays the estimated predictiveness curves for Gag, Pol, and Nef, defined as the quantile plot of risk(1){S(1)} − risk(0){S(1)} to acknowledge the harmful vaccine effect. It appears that the variability in risk difference for Nef is slightly larger compared to that for Gag and Pol. Table 4 shows estimates and 95% CIs for TG, STG, PAE, and AS for each marker. STG (and TG) are slightly larger for Nef compared to Gag and Pol, although none of the pairwise comparisons are significant. The standardized TG estimates are 0.36, 0.29, and 0.50 respectively for Gag, Pol, and Nef, corresponding to optimal sensitivity plus specificity being 1.36, 1.29, and 1.50, suggesting modest surrogate value. When PAE and AS are considered, they are not significantly different between any two markers either. Note that in this example, PAE is smaller than 0.5 and AS is negative. This is due to the negative interaction coefficient between S(1) and Z (consequently a larger S(1) is associated with a smaller difference between risk(1) and risk(0)). This analysis suggests that each marker has little surrogate value at best, albeit the analysis has limited power/precision given the small sample size.
Figure 1.
Estimated quantile curve of risk(1) − risk(0) of T cell response magnitudes to Gag, Pol, and Nef for predicting the vaccine effect on HIV infection (Step trial).
Table 4.
Estimates of summary measures of principal surrogate value for T cell response magnitudes to Gag, Pol, and Nef, for predicting the vaccine effect on HIV infection (Step trial).
| Estimand | Estimate (95% CI) | P-value for pairwise comparison | ||||
|---|---|---|---|---|---|---|
| Gag | Pol | Nef | Gag-Pol | Gag-Nef | Pol-Nef | |
| TG | 0.014 (0.001,0.054) | 0.011 (0.001, 0.049) | 0.019 (0.001, 0.055) | 0.680 | 0.415 | 0.337 |
| STG | 0.36 (0.01, 5.33) | 0.29 (0.008, 4.36) | 0.50 (0.009, 5.54) | 0.983 | 0.980 | 0.954 |
| PAE* | 0.16 (0.06, 0.72) | 0.16 (0.05, 0.76) | 0.14 (0.07, 0.73) | 0.946 | 0.925 | 0.887 |
| AS* | −1.33 (−5.80, 0.30) | −1.15 (−4.76, 0.28) | −1.49 (−5.69, 0.28) | 0.832 | 0.820 | 0.720 |
PAE = |EAE|/(|EAE| + |EDE|), where EAE ≡ E[CEPrisk{S(1)}|S(1) > 0] = β1 + β3E{S(1)} and EDE ≡ E[CEPrisk{S(1)}|S(1) = 0] = β1
AS ≡ |CEP(U)| − |EDE| = |β1 + β3U| − |β1|, where U is the maximum of S(1) (in the data).
Note that in Gilbert and Hudgens (2008), the general form of CEPrisk is defined as h [risk(0){S(1)}, risk(1){S(1)}] for some contrasting function h. Here in the simulation studies we follow their choice of the contrast function: h(x, y) = Φ−1(x) − Φ−1(y). This yields a simple linear contrast: h [risk(0){S(1)}, risk(1){S(1)}] = Φ−1 [risk(0){S(1)}] − Φ−1 [risk(1){S(1)}] = β1 + β3S(1).
5. Discussion
In this article, we study the problem of surrogate marker evaluation based on the same set of assumptions A1–A4 made by GH.
The major focus of GH is the one-marker setting. While the estimation methods proposed in their paper can be well-extended to accommodate more than one marker, the summary measures of surrogate value are essentially defined based on the single marker value itself (i.e., PAE and AS proposed in GH). With the number of markers in the model increasing, it becomes more and more difficult to quantify an individual marker’s contribution. Instead, characterizing the joint effect of multiple markers together using a metric that can be compared between risk models becomes essential. This is what we are trying to achieve here using a new graphical tool and its summary measure. We proposed a graphical tool for characterizing the distribution of risk difference between randomized treatment arms as a function of marker values, and used this tool to put different risk models on the same scale for comparison with respect to their principal surrogate value. In particular, we proposed a clinically meaningful summary measure (standardized total gain) derived from the risk difference distribution as a basis for inference. This summary measure is appealing given that it characterizes the capacity of the model for classifying subjects into treatment effective and ineffective categories. It has a limitation of being well defined only based on the arithmetic difference between risk(0) and risk(1). Depending on the scientific question, the treatment effect may be represented by a different type of contrast (e.g., the risk ratio (GH)), in which case alternative summary measures may be preferred.
The graphical tool can be applied to multiple markers because of its focus on the distribution of risk difference instead of the distribution of the marker, and has application to guide vaccine/treatment development. For example, identifying the region of marker values with large risk differences may provide a lead for refinement of the vaccine or treatment. In practice, the predictiveness curve and its summary measure can be used to compare markers or to evaluate incremental value of a new marker. Then, for a chosen model, we can further explore how the risk difference depends on individual marker values.
Including more markers increases model complexity and poses a challenge to estimation. Here we are interested in continuous markers and continuous or discrete baseline covariates. The existing nonparametric method discretizing continuous variables has unsatisfactory performance when the marker’s performance is evaluated conditional on covariates. The fact that the fully parametric method relies on the assumption about the joint distribution of the baseline covariates and the markers is also unappealing. Here we developed a semiparametric approach for estimation. An easy-to-implement EM algorithm is employed to maximize the estimated likelihood. The method works either in a standard randomized trial or when a close-out placebo vaccination (CPV) component is added to help identify and estimate risk(0). In addition to developing the standardized total gain and using the close-out design, this work extends GH by providing a method for evaluating and comparing surrogate value of multiple biomarkers, and for providing a more robust method for estimation that naturally handles continuous biomarker and continuous or discrete covariates. The method accommodates two-phase sampling designs, commonly used in clinical trials. On the other hand, the semiparametric estimator based on EM algorithm does take more computation time compared to the parametric method in GH. While GH explicitly allows the continuous marker to be subject to left-censoring, our new work does not address this issue. This is a topic of current research.
Under the baseline predictor strategy utilized in GH, with multiple biomarkers we need to be able to predict fairly well each of the biomarkers. The more biomarkers the greater the challenge in accomplishing this. The CPV strategy is particularly attractive as the number of biomarkers increases, because its effectiveness to predict the biomarkers does not decline with the number of biomarkers. By extending from the one to at least two biomarkers setting, we also face all of the challenges faced in model selection for ordinary regression modeling, such as collinearity. In practice, we can consider different approaches to handle collinearity such as selecting biomarkers measuring different biological functions, or reduce the dimension of markers using techniques such as principal components analysis.
In practice it is important to check the validity of the parametric structural models for risk(1) and risks(0) specified by A4. In a standard trial design, while it is straightforward to test goodness-of-fit of models for risk(1), models for risk(0) cannot be tested. Fortunately the CPV design provides a way, based on the equation
| (5) |
from which it is apparent that P{S(1)|Y (0) = 0, W} is identified from the CPV sample, P{Y (0) = 1|W} is identified from placebo subjects, and P{S(1)|W} is identified from active treatment subjects. Therefore a goodness-of-fit test can be constructed based on the difference between risk(0) obtained under A4 and that obtained based on (5).
The meaningful interpretation of the standardized total gain (STG) as a measure of classification accuracy relies on an extra assumption (A5, monotonicity), even though our method for its estimation does not require this assumption. Thus again we recommend the STG for settings where monotonicity is plausible, for example placebo-controlled trials where there is a significant overall beneficial treatment effect. We leave it to other work to explore relaxation of this assumption, which would be important for trials of two active treatments.
We developed our method for the scenario of Constant Biomarkers (CB). However, for placebo-controlled trials the method can also be applied to the general case that S(0) varies. For example, in an influenza vaccine trial, with biomarker(s) immune response(s) to influenza targets, S(0) will vary due to prior flu-illnesses. With interest in the risk conditional on both S(1) and S(0), we can enhance the study design by measuring the anti-influenza immune response(s) at baseline for subjects assigned to active treatment, which can substitute for S(0). Then vaccine arm subjects have data on both potential biomarkers (S(1), S(0)), which allows direct application of our semiparametric method. The semiparametric location-scale model may be employed to estimate the distribution of S(1) conditional on W and S(0).
Finally, with various summary measures of surrogate value developed in the literature, an important objective is to evaluate the comparative performance of the summary measures in terms of discrimination, predictiveness, etc. It does not appear possible to address these questions directly based on a single trial, as what is needed is meta-analysis of multiple trials, or at least meta-analysis of sub-sets of one very large trial. Meta-analysis would allow assessing, across study units, the correlation of treatment effects on the biomarker(s) with treatment effects on the clinical endpoint (for example such methods are developed and discussed in Daniels and Hughes (1997); Molenberghs et al. (2002, 2008)). If a summary measure is a good predictor of the level of clinical treatment efficacy, then trials with high surrogate value (according to the measure) will have a tight correlation, and trials with low surrogate value according to the measure will have low correlation. This kind of assessment could be formalized into a metric for comparing the predictiveness of different summary measures.
Supplementary Material
Acknowledgments
This work was supported by NIH grant 2R37AI054165-08.
Footnotes
Supplementary Materials
Web Appendices referenced in Sections 2.2–2.4 and 3[p7] are available under the Paper Information link at the Biometrics website http://www.biometrics.tibs.org.
References
- Breslow NE, Wellner JA. Weighted likelihood for semiparametric models and two-phase stratified samples, with application to cox regression. Scandinavian Journal of statistics. 2008;34(1):86–102. doi: 10.1111/j.1467-9469.2007.00574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzykowski T, Molenberghs G, Buyse M. The Evaluation of Surrogate Endpoints. London: Springer; 2005. [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JRA, Corey L, Robertson MN the Step Study Protocol Team. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bura E, Gastwirth JL. The binary regression quantile plot: assessing the importance of predictors in binary regression visually. Biometrical Journal. 2001;43(1):5–21. [Google Scholar]
- Buyse M, Molenberghs G, Burzykowski T, Renard D, Geys H. The validation on surrogate endpoints in meta-analyses of randomized experiments. Biostatistics. 2000;1:49–67. doi: 10.1093/biostatistics/1.1.49. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Hughes MD. Meta-analysis for the evaluation of potential surrogate markers. Statistics in Medicine. 1997;16:1965–1982. doi: 10.1002/(sici)1097-0258(19970915)16:17<1965::aid-sim630>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Fleming TR, DeMets DL. Surrogate endpoints in clinical trials: Are we being misled? Annals of Internal Medicine. 1996;125:605–613. doi: 10.7326/0003-4819-125-7-199610010-00011. [DOI] [PubMed] [Google Scholar]
- Follmann DA. Augmented designs to assess immune response in vaccine trials. Biometrics. 2006;62:1161–1169. doi: 10.1111/j.1541-0420.2006.00569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis CE, Rubin DB. Principal stratification in causal inference. Biometrics. 2002;58:21–29. doi: 10.1111/j.0006-341x.2002.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman L, Graubard B, Schatzkin A. Statistical validation of intermediate endpoints for chronic disease. Statistics in Medicine. 1992;11:167–178. doi: 10.1002/sim.4780110204. [DOI] [PubMed] [Google Scholar]
- Gallop R, Small D, Lin J, Elliott M, Joffe M, Ten Have T. Mediation analysis with principal stratification. Statistics in Medicine. 2009;28:1108–1130. doi: 10.1002/sim.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PB, Hudgens MG. Evaluating candidate principal surrogate end-points. Biometrics. 2008;64(4):1146–1154. doi: 10.1111/j.1541-0420.2008.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heagerty PJ, Pepe MS. Semiparametric estimation of regression quantiles with application to standardizing weight for height and age in US children. Applied Statistics. 1999;48(4):533–551. [Google Scholar]
- Huang Y, Pepe MS. A parametric ROC model based approach for evaluating the predictiveness of continuous markers in case-control studies. Biometrics. 2009;65:1133–1144. doi: 10.1111/j.1541-0420.2009.01201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pepe MS, Feng Z. Evaluating the predictiveness of a continuous marker. Biometrics. 2007;63(4):1181–1188. doi: 10.1111/j.1541-0420.2007.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffe MM, Greene T. Related causal frameworks for surrogate outcomes. Biometrics. 2009;65:530–538. doi: 10.1111/j.1541-0420.2008.01106.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Taylor JMG, Elliott MR. A bayesian approach to surrogacy assessment using principal stratification in clinical trials. Biometrics. 2010;66:523–531. doi: 10.1111/j.1541-0420.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Fleming TR, Gruttola VD. Estimating the proportion of treatment effect explained by a surrogate marker. Statistics in Medicine. 1997;16(13):1515–1527. doi: 10.1002/(sici)1097-0258(19970715)16:13<1515::aid-sim572>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Molenberghs G, Buyse M, Geys H, Renard D, Burzykowski T, Alonso A. Statistical challenges in the evaluation of surrogate endpoints in randomized trials. Controlled Clinical Trials. 2002;23(6):607–625. doi: 10.1016/s0197-2456(02)00236-2. [DOI] [PubMed] [Google Scholar]
- Molenberghs G, Burzykowski T, Alonso A, Assam P, Tilahun A, Buyse M. The meta-analytic framework for the evaluation of surrogate endpoints in clinical trials. Journal of Statistical Planning and Inference. 2008;138(2):432–449. [Google Scholar]
- Pepe MS, Feng Z, Huang Y, Longton GM, Prentice R, Thompson IM, Zheng Y. Integrating the predictiveness of a marker with its performance as a classifier. American Journal of Epidemiology. 2008;167(3):362–368. doi: 10.1093/aje/kwm305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Fleming TR. A nonparametric method for dealing with mismeasured covariate data. JASA. 1991;86(413):108–113. [Google Scholar]
- Prentice RL. Surrogate endpoints in clinical trials: Definition and operational criteria. Statistics in Medicine. 1989;8:431–440. doi: 10.1002/sim.4780080407. [DOI] [PubMed] [Google Scholar]
- Robins JM, Greenland S. Identifiability and exchangeability of direct and indirect effects. Epidemiology. 1992;3:143–155. doi: 10.1097/00001648-199203000-00013. [DOI] [PubMed] [Google Scholar]
- Taylor J, Wang Y, Thibaut R. Counterfactual links to the proportion of treatment effect explained by a surrogate marker. Biometrics. 2005;61:1102–1111. doi: 10.1111/j.1541-0420.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- Weir CJ, Walley RJ. Statistical evaluation of biomarkers as surrogate endpoints: a literature review. Statistics in Medicine. 2006;25:183–203. doi: 10.1002/sim.2319. [DOI] [PubMed] [Google Scholar]
- Wolfson J, Gilbert PB. Statistical identifiability and the surrogate endpoint problem, with application to vaccine trials. Biometrics. 2010;66(4):1153–1161. doi: 10.1111/j.1541-0420.2009.01380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youden Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.