Abstract
To identify genetic loci involved in the regulation of organ-specific enzyme activities, a specific histochemical staining protocol was used in combination with quantitative trait locus (QTL) analysis. Using phosphoglucomutase (PGM) as an example, it is shown that enzyme activity can specifically, and with high resolution, be visualized in non-sectioned seedlings of Arabidopsis. The intensities of staining were converted to quantitative data and used as trait for QTL analysis using Landsberg erecta × Cape Verde Islands recombinant inbred lines. Independently, PGM activities were quantified in whole-seedling extracts, and these data were also used for QTL analysis. On the basis of extract data, six significant (P < 0.05) loci affecting PGM activity were found. From the histochemical data, one or more specific QTLs were found for each organ analyzed (cotyledons, shoot apex, hypocotyl, root, root neck, root tip, and root hairs). Loci detected for PGM activity in extracts colocated with loci for histochemical staining. QTLs were found coinciding with positions of (putative) PGM genes but also at other positions, the latter ones supposedly pointing toward regulatory genes. Some of this type of loci were also organ specific. It is concluded that QTL analysis based on histochemical data is feasible and may reveal organ-specific loci involved in the regulation of metabolic pathways.
Carbohydrates constitute the major part of plant biomass, and carbohydrate metabolism is one of the central biochemical pathways in plant cells. Hence understanding carbohydrate metabolism and its regulation is of crucial importance. Over the last few decades, most of the genes encoding the enzymes catalyzing the various steps of carbohydrate metabolic routes, have been unraveled. However, this does not automatically imply that the regulatory mechanisms of these routes have become clear as well. Carbohydrate metabolism as a whole, but also the individual steps of the various pathways, are likely to be controlled by a plethora of genes, encoding the enzymes and regulators at different levels.
Quantitative trait locus (QTL) analysis is a powerful approach to identify genes involved in processes controlled by many genes when genetic variation for these genes is present. This has been shown in Arabidopsis for traits such as flowering time (Koornneef et al., 1998; Ungerer et al., 2002), nitrogen use efficiency (Loudet et al., 2003), and salt tolerance (Quesada et al., 2002). A few papers have shown the potential of this approach in genetic mapping of enzyme activities. Suggestions for possible candidate genes have often been inferred from a similar map position of these QTLs with genes known to be involved in the process under study (Arabidopsis [Mitchell-Olds and Pedersen, 1998]; maize [Prioul et al., 1999]; tomato [Fridman et al., 2002]; rice [Hu et al., 2001]).
Now that the genome of Arabidopsis has been sequenced (Arabidopsis Genome Initiative, 2000), the map position of most genes encoding the enzymes involved in primary metabolism are known, and QTL analysis of enzyme activities would be expected to reveal the already known loci, provided that genetic variation is present in the segregating population under investigation. However, in addition to this, QTL analysis might reveal regulatory loci. Using a QTL approach, Mitchell-Olds and Pedersen (1998) presented evidence for such regulatory loci, affecting the activities of several enzymes involved in primary metabolism in Arabidopsis.
Carbohydrates are primarily synthesized in the leaves (photosynthesis), and then distributed over the various organs of the plants. This may include various cycles of synthesis and breakdown of intermediates, e.g. Suc and starch, both at the sites of synthesis and export (sources) and in the final sinks. Hence, the localization of activities of enzymes involved is important for the understanding of regulation of source-sink interactions.
We took a QTL approach to unravel the regulation of carbohydrate metabolism and source-sink relations one step further by taking into account the localization of enzyme activities. A previously described histochemical method (Sergeeva and Vreugdenhil, 2002), which allowed the detection of the activities of enzymes involved in carbohydrate metabolism in tissue sections, was applied to intact seedlings of Arabidopsis. The obtained staining patterns were (semi-) quantified and used for QTL analysis.
Phosphoglucomutase (PGM) catalyzes the reversible conversion of Glc-6-phosphate to Glc-1-phosphate (Glc1P), an important step determining the further metabolic fate of the carbohydrate: Glc-6-phosphate may enter glycolysis, whereas Glc1P is primarily used for polymer biosynthesis, i.e. starch and cellulose (Periappuram et al., 2000; Fernie et al., 2002).
Using PGM as an example, we show that QTL mapping using data from histochemical analyses results in (a) the identification of loci involved in total enzyme activities in plants, as determined in extracts; (b) QTLs, which affect activities in all organs, and other loci that only affect PGM activity in one or a few organ(s); and (c) colocalization of some loci with structural genes, encoding these enzymes, whereas others do not colocate with structural genes, pointing toward putative regulatory loci, some of which are also organ specific.
RESULTS
PGM Activities in Extracts
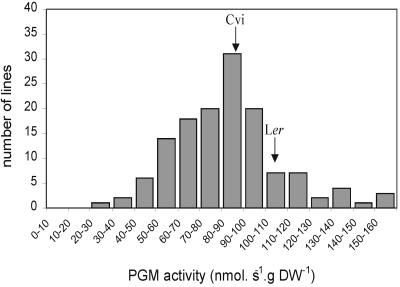
Initially, the activities of PGM in whole-seedling extracts were determined to find out whether the parental lines differed in total PGM activity and whether segregation could be found in the recombinant inbred lines (RILs). Figure 1 shows the frequency distribution of PGM activities in the RILs. Although the parental lines were only marginally different from each other, a wide range of variation was observed among the RILS.
Figure 1.
Frequency distribution of PGM activities in extracts of seedlings of 142 lines of the Cvi × Ler recombinant inbred population. Arrows indicate activities of the parental lines.
The activities in extracts of the RILs were used for QTL analysis. Six significant (P < 0.05) QTLs were found. One additional locus was identified using a lower significance threshold (0.1 < P < 0.05). The effects of the individual loci are summarized in Table I. The major loci at the bottom of chromosome 5 (around 89 centimorgans [cM]) and chromosome 1 (around 115 cM) explained 10.3% and 8.6% of the phenotypic variation, respectively. Total explained variance was 64.2%. The interaction between the loci near markers CD173 and EG113 and between loci near markers AXR-1 and EG75 explained an additional 6.5% of the variation.
Table I.
QTLs for PGM activity in whole-seedling extracts
The chromosome number, genetic location (cM), and nearest marker are given. Significance was calculated based on LOD values (**, P < 0.05; *, P < 0.10). Allelic effects are indicated with + (Ler > Cvi) and - (Ler < Cvi). Total explained variance was 64.2%.
| Chromosome No. Position | Nearest Marker | LOD | Explained Variance | Effect of Ler Allele |
|---|---|---|---|---|
| cM | % | |||
| 1: 7.5 | AXR-1 | 2.82** | 3.6 | — |
| 1: 61.3 | EG113 | 2.57** | 3.4 | — |
| 1: | CD173 | 6.06** | 8.6 | + |
| 115.2 | ||||
| 3: 7.8 | EG75 | 2.47* | 3.1 | + |
| 3: 32.7 | AD92 | 2.60** | 3.3 | — |
| 5: 19.5 | BH107 | 3.71** | 4.8 | + |
| 5: 88.7 | HH445 | 7.49** | 10.3 | — |
The observation that both Landsberg erecta (Ler) and Cape Verde Island (Cvi) have alleles that increase and decrease PGM activity, depending on the locus, is in agreement with the transgression observed in the total population.
Histochemical Staining of PGM Activity
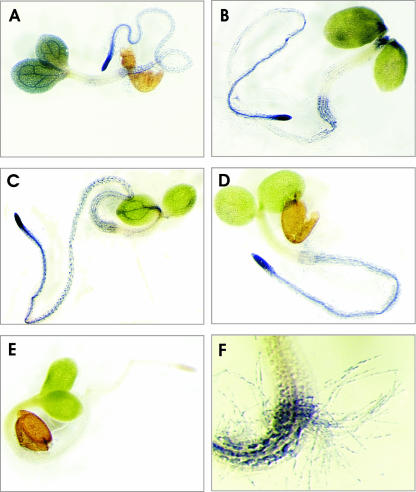
Having demonstrated that PGM activity in seedlings segregates in the Ler × Cvi RIL population, we tested whether PGM activities could be localized using in situ staining, as described previously for tissue sections (Sergeeva and Vreugdenhil, 2002). Whole seedlings were fixed and incubated in the staining mixture without sectioning. Figure 2 shows the results for seedlings of the parental lines. Clearly, precipitation of nitroblue tetrazolium was observed, indicating enzyme activity. This precipitation specifically indicated PGM activity, as judged from the absence of staining in the control, in which Glc1P, being the substrate for the PGM reaction, was omitted (Fig. 2E). Another control, in which NAD was omitted, also showed no staining (data not shown). For Ler, high PGM activity was observed in the cotyledons, especially in the veins. In contrast, PGM activity was nearly absent from the apical meristem and the hypocotyl (Fig. 2A). The root was stained with increasing intensity toward the root tip. Staining was also observed in the root hairs (Fig. 2F).
Figure 2.
Examples of staining for PGM activities in 7-d-old seedlings of Ler (A), Cvi (B), Col (C), PGMp mutant (D), and control (Col; E). In the control, Glc1P was omitted from the incubation. F, A detail of the hypocotyl-root transition, including staining of root hairs (Ler).
Cvi seedlings showed lower staining in the cotyledons, and the specific staining of the veins, as seen in the Ler seedlings, was absent. However, the apex and the lower part of the hypocotyl stained more intense than in Ler seedlings. The staining pattern in the root was similar to that observed in Ler (Fig. 2B).
For comparison, activity staining of PGM is also shown for Columbia (Col) seedlings (Fig. 2C) and the PGMp mutant lacking plastidic PGM activity, in Col background (Fig. 2D; Caspar et al., 1985). The main difference between mutant and wild type was observed in the cotyledons, which hardly stained in the mutant, and was observed much more in the wild type, especially in the veins.
Figure 2F illustrates the high spatial resolution of the method, showing in detail the lower part of the hypocotyl and the upper part of the root, including staining in the root hairs.
Quantification of Staining
By scoring the intensities of staining on a scale, ranging from 0 to 5, indicating no and very intense staining, respectively, we generated quantitative data to be used for QTL analysis. Within a batch of seedlings from the same line, variation was also observed between individual seedlings. Hence, data were based on average patterns derived from 10 to 40 seedlings per line. Activities were quantified separately for cotyledons, shoot apex, hypocotyl, root neck, root, root hairs, and root tip.
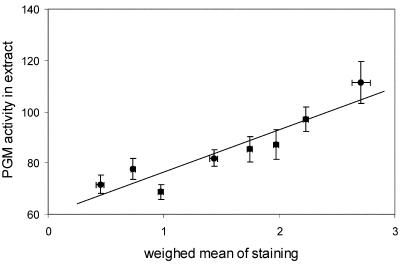
The validity of the scale used for scoring activities was tested by comparing PGM activities as determined in whole-seedling extracts with staining data. To generate staining data for whole seedlings, a weighed sum of data of all organs was calculated for each line, assuming relative proportions per organ to be 40%, 5%, 15%, 25%, 5%, 5%, and 5% for cotyledons, apex, hypocotyl, root, root neck, root tip, and root hairs, respectively. Figure 3 shows that the scale used to score intensities of staining corresponded well with activities as determined in extracts. Slight modifications of the proportional values per organ yielded similar results (data not shown).
Figure 3.
Correlation between PGM activities as quantified in whole-seedling extracts (y axis) and total PGM activity, calculated based on scoring of staining of separate organs (x axis), in the Cvi × Ler recombinant inbred population. PGM activities in extracts are given in nanomoles per second per gram dry weight. For each line, staining figures for the whole seedlings were calculated as a weighed sum of data of each organ, assuming relative proportions per organ to be 40%, 5%, 15%, 25%, 5%, 5%, and 5% for cotyledons, apex, hypocotyl, root, root neck, root tip, and root hairs, respectively. Lines were divided into eight classes based on weighed means of staining and pooled. Averages for pooled lines were calculated and plotted ± se.
QTL Analysis of Activity Staining
Quantitative data, describing activities per organ, were used for further analysis, revealing QTLs for PGM activity in different organs of the Arabidopsis seedling. QTLs affecting the activity of PGM were detected for all organs analyzed. In Table II, the QTLs for the various organs are listed.
Table II.
Staining data QTLs for PGM activity per organ
For further legends, see Table I.
| Organ | Chromosome No. Position | Nearest Marker | LOD | Explained Variance | Total Explained Variance | Effect of Ler Allele |
|---|---|---|---|---|---|---|
| cM | % | % | ||||
| Cotyledon | 1: 115.2 | CD173 | 11.7** | 21.5 | + | |
| 5: 94.9 | GB102 | 2.62** | 4.0 | 38.7 | - | |
| Apex | 1: 29.3 | CH160 | 2.69** | 3.8 | - | |
| 1: 60.3 | GD97 | 3.61** | 5.1 | - | ||
| 1: 115.2 | CD173 | 15.42** | 26.9 | + | ||
| 3: 13.6 | FD111 | 2.66** | 3.7 | - | ||
| 3: 23.1 | GH390 | 3.18** | 4.3 | + | ||
| 3: 65.3 | FD98 | 2.46* | 3.3 | 63.5 | + | |
| Hypocotyl | 1: 24.0 | GD86 | 2.65** | 4.1 | - | |
| 1: 116.2 | CD173 | 12.34** | 22.7 | + | ||
| 5: 3.3 | CH690 | 2.53** | 3.9 | - | ||
| 5: 55.5 | BH96 | 2.41* | 3.7 | 57.5 | + | |
| Root | 1: 115.7 | CD173 | 4.85** | 7.9 | + | |
| 3: 39.1 | GB120 | 2.34* | 3.6 | - | ||
| 5: 26.3 | AD114 | 5.79** | 10.2 | 57.4 | + | |
| Root neck | 1: 24.3 | GD86 | 7.1** | 10.2 | - | |
| 1: 31.7 | CC98 | 3.96** | 5.4 | + | ||
| 1: 54.6 | GB112 | 3.15** | 4.6 | - | ||
| 1: 113.7 | CC318 | 7.87** | 11.6 | + | ||
| 2: 34.9 | BF221 | 3.79** | 5.2 | + | ||
| 3: 25.1 | EC83 | 3.11** | 4.5 | + | ||
| 3: 39.1 | GB210 | 6.7** | 9.6 | - | ||
| 5: 0.5 | FD207 | 3.02** | 4.1 | + | ||
| 5: 28.3 | DF184 | 5.44** | 8.2 | + | ||
| 5: 54.6 | BH96 | 3.1** | 4.5 | 63.3 | + | |
| Root tip | 1: 22.5 | EC66 | 7.12** | 10.0 | - | |
| 1: 29.3 | CH160 | 2.27* | 3.6 | + | ||
| 1: 107.5 | GH157 | 5.19** | 7.5 | + | ||
| 2: 26.7 | FD222 | 4.99** | 6.9 | + | ||
| 2: 69.8 | EC235 | 4.92** | 6.9 | + | ||
| 4: 55.1 | CH70 | 3.05** | 4.4 | - | ||
| 5: 18.2 | BH107 | 6.43** | 9.4 | 64.3 | + | |
| Root hairs | 1: 21.8 | EC66 | 3.72** | 5.3 | - | |
| 1: 115.7 | CD173 | 7.63** | 11.1 | + | ||
| 2: 67.8 | EC235 | 6.21** | 9.2 | + | ||
| 3: 39.1 | GB210 | 5.27** | 7.3 | - | ||
| 3: 83.9 | BH109 | 2.18* | 2.9 | - | ||
| 5: 25.8 | AD114 | 5.3** | 7.8 | 61.1 | + |
At the lower end of chromosome 1, around 110 cM, a clear QTL was present for PGM activity in all organs. The Ler allele increased activity in all organs. The aboveground parts (cotyledons, apex, and hypocotyl) only showed minor additional QTLs in other regions of chromosome 1. In the root (not including the tip, hairs, and root neck), this QTL was the only one observed on chromosome 1. However, for the root tip and root neck region and in root hairs, additional significant QTLs were found around 25 and 50 cM.
Chromosome 2 contained two loci, affecting activities in (parts of) the roots only. On chromosome 3, various loci were found, which were specific for one or only a limited number of organs. Chromosome 4 contained one significant locus, affecting activity in the root tip only.
Chromosome 5 contained various loci affecting PGM activity. At the top of chromosome 5, a locus was found around 25 cM, affecting activities in all parts of the roots, whereas the effects of this locus/loci on the activities in the upper parts of the seedling were not significant. At the lower part of chromosome 5 (around 90 cM), a locus was present that affected PGM activity in cotyledons only. In the middle of this chromosome (around 55 cM), a locus was found that influenced PGM activities in the root neck only.
In all organs, except the root tip, the locus around 110 cM on chromosome 1 explained most of the phenotypic variance. Total explained variance for each organ ranged from 38.7% to 64.3%.
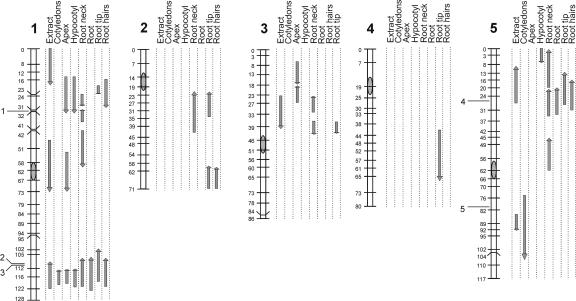
Figure 4 summarizes the observed QTL locations for all chromosomes for the various organs, showing the 2-logarithm-of-odds (LOD) intervals and also including the QTLs observed in whole-seedling extracts and the locations of (putative) PGM genes.
Figure 4.
Ler/Cvi linkage map showing the genetic locations of QTLs affecting PGM activities, determined in whole-seedlings extracts, and in various organs of intact seedlings. The lengths of the arrows indicate the 2-LOD support intervals. The directions of the arrows indicate the Ler allelic phenotype (upward increasing, downward decreasing). The locations of (putative) PGM genes on chromosomes 1 and 5 is indicated: 1, At1g23190; 2, At1g70820; 3, At1g70730; 4, At5g17530; 5, At5g51820.
The loci affecting PGM activities in extracts coincided or partly overlapped with loci found using the histochemical data. However, the opposite was not true, indicating that the latter method was more sensitive and/or specific.
During quantification of the staining patterns, it was found that the staining in the roots and in the hypocotyls was not homogeneous along the lengths of these organs, as can also be seen in Figure 2. Also, the root hairs showed different intensities of staining along the length of the root. Therefore, the intensities of staining of upper and lower parts of hypocotyls, and upper, middle, and lower parts of roots were quantified separately. Similarly, the intensities of staining in the hairs were quantified in three zones along the root. QTL analysis of these data revealed that different loci were involved in controlling PGM activities in various regions of the same organ. As example, QTLs for PGM activities in hairs in different zones is given in Figure 5, for chromosome 1. The predominant locus around 110 cM, which was also observed in the average data (Fig. 4), was only significant in the upper and lower hair zones, but not in the middle one. PGM activity in the lower zone was influenced by a locus at the top of chromosome 1. This locus was not detected for the activity in the other two zones. At other chromosomes also, specific QTLs were found affecting PGM activities in hairs at different positions along the root (data not shown). For roots and hypocotyls also, QTLs were found specifically for various parts of these organs (data not shown). Thus, the histochemical analysis was sensitive enough to reveal differences within organs.
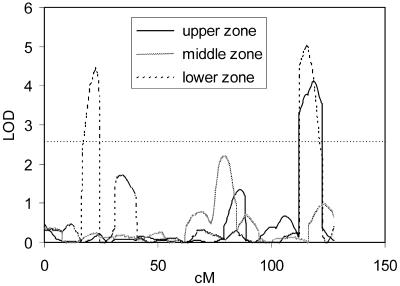
Figure 5.
QTL likelihood maps for the activities of PGM in root hairs at different positions along the root as quantified from histochemical stainings. Only data for chromosome 1 are presented. LODs above 2.6 (horizontal dotted line) are significant at P = 0.05.
Allelic Interactions
Two-way interactions were analyzed for all traits that were significant at P = 0.1, both for extracts and for staining data. From a total of 155 interactions tested, only 10 were significant, i.e. for extracts (two), apex (one), hypocotyl (two), root neck (one), hairs (three), and root tip (one). From these interactions, five included the major locus at the bottom of chromosome 1. For total extracts and for apex and hypocotyl (twice) activities, it was found that when the Ler alleles were present at this locus, thus giving a higher activity, a Cvi allele at the interacting locus synergistically stimulated PGM activity. However, for activities in root hairs, the opposite was found, i.e. a Cvi allele at the interacting locus stimulated PGM activity when a Cvi allele was present at the locus around 110 cM at chromosome 1. Moreover, three cases were observed in which a locus at the top of chromosome 3 interacted with other loci, whereas the main effect of this locus was low, i.e. LODs were between 2.1. and 2.5.
DISCUSSION
Use of Histochemical Data for QTL Analysis
From Figure 2, it can be concluded that detection of PGM activity is feasible in intact (non-sectioned) seedlings of Arabidopsis and allows comparison of relative activities in various organs and tissues.
It was found that staining varies between tissues within the same seedling and also between Arabidopsis accessions (Fig. 2). The latter type of variation was used to perform a QTL analysis, using the segregating RIL population derived from accessions Cvi and Ler (Alonso-Blanco et al., 1998). The validity of the scoring method to quantify the staining patterns was tested in two ways: (a) The first way was by comparing staining data calculated for complete seedlings with PGM activities quantified in whole-seedling extracts (Fig. 3). A positive correlation was observed. From Figure 3, it might be inferred that the relationship deviates from linearity at the lowest activities, indicating lower reliability in this region. (b) The second way was by comparing locations of QTLs, as derived by both methods. It was expected that most, if not all, QTLs found based on extract-data would coincide with QTLs revealed for staining patterns. Figure 4 showed this to be true. Moreover, the directions of the allelic effect of the loci, as detected by the two methods, were the same. Ratings of traits have also been successfully used by others to detect QTLs (Van den Berg et al., 1996).
The histochemical analysis revealed more loci than found by analyzing the extract-data (Fig. 4). This indicates that the staining method is more sensitive, as was also concluded earlier for tissue sections from other species (Sergeeva and Vreugdenhil, 2002). This might be due to activity in a limited number of tissues or cells, which can be seen using histochemical analysis, but is diluted too much when whole-seedling extracts are made.
The reproducibility of the method and the genetic differences are also shown by the relatively high explained phenotypic variance, which was for extract data 64% (Table I). Similar figures (57.5%-64.3%) were observed for all organs as based on the histochemical method, except for the cotyledons (38.7%). The lower value for the cotyledons is likely to be due to the presence of chlorophyll in these organs, obscuring the blue staining and making accurate scoring more difficult. However, a larger number of small effect QTLs can also explain lower R2 values.
Thus, we conclude that histochemical data can successfully be used for QTL analysis and can reveal unique information on organ and tissue specificity that cannot be obtained using extraction protocols.
Organ-Specific QTLs
One locus, identified as based on both extract and staining data, influenced PGM activity in all parts of the plants, i.e. the locus at the lower end of chromosome 1. Moreover, this locus is the predominant one in all organs, except in the root tip, explaining 7.5% to 26.5% of the variation.
The locus at the top of chromosome 1 (25 cM) is observed for most organs (Fig. 4), except in cotyledons and roots. In the latter organs, a QTL was suggested to be present in this region, but just below significance (data not shown). This chromosomal region appears to be complex for this trait, because (a) the 2-LOD interval for the extracts was found just above the 2-LOD intervals for the loci (locus) observed in the separate organs, i.e. 0 to 19 and 14 to 33 cM, respectively; (b) in the root tip, two tightly linked loci were found in this region, with opposite allelic effects (Table II); and (c) the extract data also suggest another locus (22-40 cM), with a positive allelic affect. This locus was just below significance (LOD = 2.16). The complex epistatic interactions of this locus add to the complexity.
Other QTLs were only detected for one or a few organs, e.g. two loci on chromosome 2, being only evident for (parts of) the roots. Furthermore, a locus on chromosome 5 (around 50 cM) only affected activities in the root neck, i.e. the transition from hypocotyl to root. In the other parts of the root, this locus did not appear to affect PGM activity at all.
The detailed analysis of root hairs in various zones along the root (Fig. 5) revealed different QTLs for the various zones. Because root hairs in Arabidopsis are single cells, it shows that the present methods are able to reveal cell-specific QTLs, even discriminating similar cells at different positions.
Comparison between QTLs and Known (Putative) PGM Genes
For Arabidopsis, five (putative) PGM genes have been described (Arabidopsis Genome Initiative, 2000; http://www.arabidopsis.org). Three genes are located on chromosome 1, two of which are very close together (At1g70820 and At1g70730). The other two (putative) PGM genes are on chromosome 5. QTLs affecting PGM activity were found at or near all positions of PGM genes (Fig. 4), although the coincidence is not always perfect, e.g. on chromosome 5, the QTL for PGM activity in extracts seems to be located slightly below the PGM gene at 80 cM. The PGM genes are likely candidate genes for the QTLs found in the corresponding regions. This would indicate that in the RIL population under investigation, variation is present for at least four of five (putative) PGM genes or in their cis-regulatory elements. Because the two PGM genes at the lower end of chromosome 1 are too close together, it cannot be concluded whether only one of the two shows allelic variation between Ler and Cvi, or both.
Gene At5g51820 has been described as a plastidic form of PGM (Caspar et al., 1985; Kofler et al., 2000; Periappuram et al., 2000). We found a QTL coinciding with this gene, and this locus controls variation in the cotyledons only (Fig. 4). Phenotypic analysis of a mutant for this gene revealed that it has severely reduced levels of starch in the leaves, indicating a role for plastidic PGM in starch biosynthesis (Caspar et al., 1985; Corbesier et al., 1998; Kofler et al., 2000; Periappuram et al., 2000). Thus it might be expected that this gene is mainly expressed in organs that are able to synthesize starch. In seedlings, starch has been shown to be present primarily in the cotyledons and also in hypocotyls and in the root tip (Kurata and Yamamoto, 1998). We conclude that the detection of a cotyledon-specific QTL at this position, probably due to allelic variation in the plastidic PGM gene or cis-regulatory elements, further supports the usefulness of the histochemical analysis. Interestingly, a further QTL was suggested (LOD = 2.08; data not shown) at the same chromosomal position for the root tip, in which starch is also present (Kurata and Yamamoto, 1998).
The overall difference in staining patterns comparing the PGMp mutant and the wild type (Col) were small and were only obvious in the cotyledons (Fig. 2). This would imply that the plastidic PGM is mainly active in the cotyledons. This is consistent with the above-described finding that the QTL on chromosome 5, coinciding with the plastidic PGM gene, was evident in cotyledons only. The presence of apparently normal levels of PGM activity in the root tip of the mutant would imply that cytoplasmic rather than plastidic PGM is active in the root tip, or that cytosolic PGM compensates for the plastidic one in this organ, but not, or only partly, in the cotyledons. It has been shown that cytosolic PGM activity can compensate for loss of plastidic PGM activity (Kofler et al., 2000).
Mitchell-Olds and Pedersen (1998) reported only one significant PGM QTL in extracts of the Col × Ler RIL population, i.e. at the lower end of chromosome 5, probably overlapping with the plastidic PGM gene. The reason that we found six QTLs might be the use of seedlings rather than leaves from rosette plants, and/or the fact that allelic differences are present at more loci between Cvi and Ler than between Col and Ler.
Several QTLs did not coincide with structural PGM genes. We suggest that these loci contain regulatory genes affecting PGM activity. For instance, on chromosome 2, two loci were found that significantly affected activities in root tip and root hairs (around 65 cM) or root neck and root tip (around 30 cM). The latter locus was suggested in extracts as well, although at low significance (LOD = 2.07). The putative regulating loci might be trans-acting factors, influencing gene expression, or might act via other mechanisms at the level of transcription, translation, or posttranslational control of enzyme activity. The 2-LOD intervals are too large to justify speculation about the genes underpinning these regulatory effects, and further fine-mapping would be required to identify these genes.
Allelic Interactions
The analyses of two-way interactions revealed that the loci around 115 cM on chromosome 1 and at the top of chromosome 3 (around 8 cM) were involved in most of the significant allelic interactions. Interestingly, the former one coincides with a structural PGM gene, suggesting that the interaction alleles might be trans-acting modifiers of PGM expression or regulate PGM at the protein level, e.g. in an allosteric way. The locus at the top of chromosome 3 does not overlap with a structural PGM gene and might point to an important regulator.
It has been reported that the cytosolic PGM activity might (partly) compensate for loss of chloroplastic PGM (Kofler et al., 2000). This might have shown up as allelic interaction between this locus and another PGM-linked locus. However, no interaction between the locus at the lower end of chromosome 5, overlapping with the chloroplast PGM gene, and one of the other loci was observed (data not shown).
CONCLUSIONS
In conclusion, histochemical data can be used successfully for QTL analysis, revealing different types of loci, i.e. (a) coinciding with structural genes, suggesting allelic variation for the enzyme under investigation; and (b) other loci, presumably involved in regulating enzyme activity. Moreover, organ-specific loci can be detected, allowing conclusions to be drawn on organ-specific expression of both structural and regulatory genes. The observation that tissue-specific natural variation for enzyme activities is present indicates that for large-scale analysis of genetic material in plant breeding and functional genomics, the pooling of tissues to obtain a single value for a certain genotype implies the risk that relevant differences in tissue variation remain undetected.
We are currently analyzing activities of other enzymes involved in primary metabolism to find possible loci regulating carbohydrate metabolic pathways.
MATERIALS AND METHODS
Plant Material
The RIL population derived from crosses between the laboratory strain Ler and the accession Cvi was used. These RILs have previously been characterized for amplified fragment-length polymorphism and cleaved-amplified polymorphic sequence markers (Alonso-Blanco et al., 1998).
Seeds were sown in 6-cm petri dishes with a single layer of moist filter paper and placed at 4°C in darkness for 7 d to break dormancy. They were then transferred to 20°C in the light with a 16-h/8-h day/night cycle and grown for 7 d. Samples for analysis were taken 4 to 6 h after the start of the light period and immediately frozen in liquid nitrogen (for enzyme extraction) or fixed (for histochemical staining). Samples for the two types of assays were taken from independent experiments. Due to low germination rates or poor seedling vigor, some RILs were discarded from the analyses, resulting in 142 RILs for PGM assays in extracts and 133 RILs for the histochemical staining.
PGM Assay
To determine overall PGM activity, samples of about 75 mg fresh weight were taken. The samples were frozen in liquid nitrogen, lyophilized, weighed, and homogenized. Enzyme extraction was done at 0°C to 4°C in 0.6 mL of 50 mm HEPES (pH 7.4), 5 mm MgCl2, 1 mm EGTA, 1 mm EDTA, 10% (v/v) glycerol, 0.1% (w/v) bovine serum albumin (BSA), and 5 mm dithiothreitol (DTT), with addition of 10 mg of insoluble polyvinylpyrrolidone. The homogenate was centrifuged for 5 min at 15,000g. The supernatant was frozen in liquid nitrogen and stored at -80°C until further analysis.
PGM activity was determined spectrophotometrically. In a microtiter plate, 210 μL of 50 mm HEPES (pH 7.4), 1 mm EDTA, 1 mm EGTA, 5 mm MgCl2, 5 mm DTT, 0.1% (w/v) BSA, 1 unit mL-1 Glc-6-phosphate dehydrogenase, 1 mm NAD, and 0.02 mm Glc-1,6-bisphosphate were added to 20 μL of enzyme extract. After mixing and waiting for 3 min, substrate (20 μL of 50 mm Glc1P) was added, and absorption at 340 nm was measured during 15 min with a microtiterplate reader (Versamax, Molecular Devices, Sunnyvale, CA) at 25°C. Also, blank incubations were done for each extract in which the substrate was omitted.
Localization of Activities
Seedlings (10-40 per line) were fixed in 2% (w/v) paraformaldehyde with 2% (w/v) polyvinylpyrrolidone 40 and 0.001 m DTT, pH 7.0 at 4°C for 1 h and then rinsed overnight in water at 4°C and refreshed at least five times to remove soluble carbohydrates.
Staining for PGM activity was as described by Sergeeva and Vreugdenhil (2002). PGM activity was visualized by incubating the seedlings in 0.5 mL of incubation medium in Eppendorf vials in a water bath at 30°C for 1 h. The reaction medium consisted of 42 mm HEPES-NaOH buffer (pH 7.4), 4.2 mm MgCl2, 0.84 mm EDTA, 0.84 mm EGTA, 0.084% (w/v) BSA, 1.4 mm NAD, 1 unit of Glc-6-phosphate dehydrogenase, 0.03% (w/v) nitroblue tetrazolium, and 4.35 mm Glc1P. It was checked that substrates and enzymes were added in excess. In control reactions, Glc1P or NAD was omitted. After the incubation period, the enzyme reaction was terminated by rinsing the seedlings in distilled water.
The seedlings were studied for staining with a binocular (Leica, Wetzlar, Germany); the intensities of staining patterns were (semi-) quantified on an arbitrary scale, ranging from 0 to 5, indicating no and very intense staining, respectively.
QTL Analysis
To map QTLs using the RIL population, a set of 99 markers covering most of the Arabidopsis genetic map was selected from the previously published RIL Ler/Cvi map (Alonso-Blanco et al., 1998). These markers spanned 482 cM, with an average distance between consecutive markers of 5 cM and the largest genetic distance being 12 cM.
The computer program MapQTL v4.0 (Plant Research International, Wageningen-University and Research Centre, Wageningen, The Netherlands) was used to identify and locate QTLs linked to the molecular markers using both interval mapping and multiple-QTL model mapping methods as described in its reference manual (http://www.plant.wageningen-ur.nl/products/mapping/mapqtl/). The estimated additive effect and the percentage of variance explained by each QTL as well as the total variance explained by all of the QTLs affecting a trait were obtained with MapQTL in the final multiple-QTL model mapping model. For this, different cofactor markers were tested around the putative QTL positions (van Ooijen and Maliepaard, 1996), selecting as final cofactors the closest marker to each QTL, i.e. those maximizing the LOD score. A LOD score threshold of 2.6 was applied to declare the presence of a QTL, which corresponds to a general genome-wide significance of P = 0.05 for normally distributed data, as was determined by extensive simulation experiments (van Ooijen, 1999). We verified this threshold for interval mapping by applying the permutation test to each data set (1,000 repetitions) and found P = 0.05 LOD thresholds between 2.5 and 2.6 for all traits. Two-LOD support intervals were established as ≈ 95% confidence intervals (van Ooijen, 1992).
PGM Genes
Five genes annotated as (putative) PGM genes by the International Arabidopsis Genome Project (http://www.arabidopsis.org) were considered to be strong candidates for QTLs affecting PGM activity, i.e. At1g23190, At1g70730, At1g70820, At5g17530, and At5g51820. The position of these genes were placed on the genetic AFLP map by positioning AFLP markers directly on the genome with in silico restriction fragment analysis (Peters et al., 2001; Fig. 4). For the full set of markers, we refer to the Web site http://www.dpw.wau.nl/natural/.
Acknowledgments
We gratefully acknowledge the help of Dr. Leonie Bentsink in the QTL mapping.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027615.
This work was supported in part by the European Union (grant no. QLRT-2000 - 01097). L.I.S. was supported by the North Atlantic Treaty Organization-Linkage (grant no. CLG 978850). J.J.B.K. was supported by Netherlands Organization for Scientific Research Program Genomics (grant no. 050-10-029).
References
- Arabidopsis Genome Initiative (2000) Analysis of the gene sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259-271 [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville CR (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase. Plant Physiol 79: 11-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G (1998) The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206: 131-137 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Swiedrych A, Tauberger E, Lytovchenko A, Trethewey RN, Willmitzer L (2002) Potato plants exhibiting combined antisense repression of cytosolic and plastidial isoforms of phosphoglucomutase surprisingly approximate wild type with respect to the rate of starch synthesis. Plant Physiol Biochem 40: 921-927 [Google Scholar]
- Fridman E, Liu YS, Carmel-Goren L, Gur A, Shoresh M, Pleban T, Eshed Y, Zamir D (2002) Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol Genet Genomics 266: 821-826 [DOI] [PubMed] [Google Scholar]
- Hu B, Wu P, Liao CY, Zhang WP, Ni JJ (2001) QTLs and epistasis underlying activity of acid phosphatase under phosphorus sufficient and deficient condition in rice (Oryza sativa L.). Plant Soil 230: 99-105 [Google Scholar]
- Kofler H, Hausler RE, Schulz B, Groner F, Flugge UI, Weber A (2000) Molecular characterisation of a new mutant allele of the plastid phosphoglucomutase in Arabidopsis, and complementation of the mutant with the wild-type cDNA. Mol Gen Genet 263: 978-986 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49: 345-370 [DOI] [PubMed] [Google Scholar]
- Kurata T, Yamamoto KT (1998) Petit1, a conditional growth mutant of Arabidopsis defective in sucrose-dependent elongation growth. Plant Physiol 118: 793-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Pedersen D (1998) The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics 149: 739-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periappuram C, Steinhauer L, Barton DL, Taylor DC, Chatson B, Zou JT (2000) The plastidic phosphoglucomutase from Arabidopsis: a reversible enzyme reaction with an important role in metabolic control. Plant Physiol 122: 1193-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, Gerats T (2001) A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol 127: 1579-1589 [PMC free article] [PubMed] [Google Scholar]
- Prioul JL, Pelleschi S, Sene M, Thevenot C, Causse M, de Vienne D, Leonardi A (1999) From QTLs for enzyme activity to candidate genes in maize. J Exp Bot 50: 1281-1288 [Google Scholar]
- Quesada V, Garcia-Martinez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130: 951-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Vreugdenhil D (2002) In situ staining of activities of enzymes involved in carbohydrate metabolism in plant tissues. J Exp Bot 53: 361-370 [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Modliszewski JL, Mackay TFC, Purugganan MD (2002) Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160: 1133-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg JH, Ewing EE, Plaisted RL, McMurry S, Bonierbale MW (1996) QTL analysis of potato tuberization. Theor Appl Genet 93: 307-316 [DOI] [PubMed] [Google Scholar]
- van Ooijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84: 803-811 [DOI] [PubMed] [Google Scholar]
- van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613-624 [DOI] [PubMed] [Google Scholar]
- van Ooijen JW, Maliepaard C (1996) MapQTL Version 4.0: Software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen, The Netherlands