Abstract
Invertase activity is thought to play a regulatory role during early kernel development by converting sucrose originating from source leaves into hexoses to support cell division in the endosperm and embryo. Invertases are regulated at the posttranslational level by small protein inhibitors, INVINHs. We found that in maize (Zea mays), an invertase inhibitor homolog (ZM-INVINH1) is expressed early in kernel development, between 4 and 7 d after pollination. Invertase activity is reduced in vitro in the presence of recombinant ZM-INVINH1, and inhibition is attenuated by pre-incubation with sucrose. The presence of a putative signal peptide, fractionation experiments, and ZM-INVINH1::green fluorescent protein fusion experiments indicate that the protein is exported to the apoplast. Moreover, association of ZM-INVINH1 with the glycoprotein fraction by concanavalin A chromatogaphy suggests that ZM-INVINH1 interacts with an apoplastic invertase during early kernel development. ZM-INVINH1 was localized to the embryo surrounding region by in situ analysis, suggesting that this region forms a boundary, compartmentalizing apoplast invertase activity to allow different embryo and endosperm developmental rates.
Kernel development in maize (Zea mays) proceeds through a series of tightly regulated, overlapping stages. After double fertilization, during the prestorage phase, two distinct cell types are established: the triploid endosperm and the diploid embryo. Despite clearly different cell fates, the embryo and endosperm both rely upon photosynthate from source leaves transported through the maternal pedicel region of the developing kernel, ending at the terminal phloem cells. The presence of Suc-hydrolyzing enzymes, which produce hexose sugars from Suc, have been identified as critical for the establishment of the prestorage phase of seed development, and Suc hydrolysis is an important component of realizable plant yield (Cheng and Chourney, 1999; Weschke et al., 2003). The “invertase control hypothesis,” largely based on work from dicots (Wobus and Weber, 1999), is supported in maize by the presence of a cell wall invertase, INCW2, localized to the basal endosperm transfer layer (Talercio et al., 1999). Mutations in this gene result in miniature kernels (the mn1 mutation) and have a severely reduced endosperm (Cheng et al., 1996; Vilhar et al., 2002). Recently, the association of a IVR2, a soluble invertase expressed during early kernel development, with seed yield under conditions of limiting photosynthesis suggests that soluble invertases also play a significant role in providing hexose sugars to support cell division during the prestorage phase (Andersen et al., 2002), as has been previously suggested (Zinselmeier et al., 1999).
Invertases exhibit complex regulation at the transcriptional and posttranscriptional levels in response to developmental, environmental, and carbohydrate signals (Sturm, 1999). In addition, small (<20-kD) inhibitor proteins (INVINH) have been associated with invertase preparations in a number of dicot plant species (Pressey, 1994; Weil et al., 1994; Krausgrill et al., 1996, 1998; Greiner et al., 1998, 2000) since their first biochemical characterization (Pressey, 1966; Jaynes and Nelson, 1971). Recently, the first gene encoding an INVINH was characterized from tobacco (Nicotiana tabacum) and was shown to have inhibitory activity by assaying the effect of recombinant protein on invertase activity in vitro (Greiner et al., 1998). Transgenic studies, ectopically expressing tobacco NtINVINH1, demonstrated that overexpression reduces invertase activity in vivo and reduces the cold-induced sweetening of potato (Solanum tuberosum) tuber (Greiner et al., 1999). In addition, the activity of apoplastic versions of INVINH can be attenuated by the addition of low concentrations of Suc (Sander et al., 1996), suggesting the existence of a three-way regulatory switch involving invertase, INVINH, and Suc that serves to regulate sink strength and carbohydrate partitioning in plants (Krausgrill et al., 1998).
Here, we present the characterization of an invertase inhibitor from maize and demonstrate inhibitory activity by reducing invertase activity in vitro with recombinant ZM-INVINH1. Characterization of ZM-INVINH1 reveals that it is an early kernel-specific, embryo surrounding region (ESR)-localized, and apoplast-targeted protein. The role of ZM-INVINH1 may be to compartmentalize invertase activity within the early kernel to allow the endosperm and embryo to follow different rates of cell division and distinct developmental programs.
RESULTS
Identification of ZM-INVINH1
To identify invertase inhibitor homologs from maize, we screened DuPont/Pioneer databases with previously characterized dicot INVINH protein sequences. Three distinct sequences had a similar size and shared sequence homology with dicot INVINH proteins. One contig had an expressed sequence tag (EST) representation that suggested it was kernel specific, with two EST sequences originating from coencytic embryo sac libraries 4 d after pollination (DAP). Full-length sequence analysis revealed an open reading frame of 528 nucleotides, with a deduced amino acid sequence of 176 residues (Fig. 1). The other two INVINH-related genes in maize were restricted to either tassel libraries or libraries constructed from maize cell culture (GenBank accession nos. AX214357 and AX214336, respectively). On the basis of protein size and homology, these sequences represent the closest maize homologs to dicot INVINH proteins present in the public and our proprietary databases. To more clearly define the role of invertase inhibitor proteins in seed development, we concentrated on the EST with a library distribution limited to early kernel development and designate it as ZM-INVINH1.
Figure 1.
Nucleotide and deduced amino acid sequence of the ZM-INVINH1 gene, including 587 bp upstream of the ATG. The deduced amino acid sequence is presented above the DNA sequence. Putative TATA sites are underlined, and the site of the longest cDNA clone is indicated by a double underline.
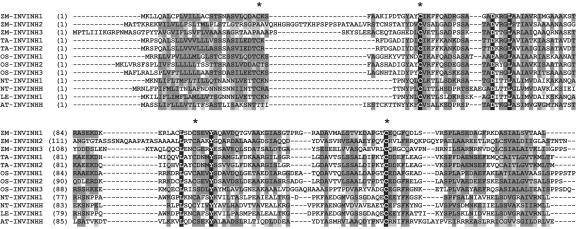
ZM-INVINH1-deduced protein sequence has an apparent signal peptide sequence of 22 amino acid residues resulting in a mature protein of 17.7 kD, a size that is in keeping with other INVINH-related proteins (Pressey, 1994; Weil et al., 1994). Analysis of the full-length protein sequence by PSORT (http://psort.nibb.ac.jp/) suggests that the protein is targeted to the apoplast. Comparison of ZM-INVINH1 protein sequence with other INVINH-related gene products from dicots shows the conservation of four Cys residues as well as other amino acids spaced throughout the protein (Fig. 2).
Figure 2.
Comparison of ZM-INVINH1 with other invertase inhibitor-like proteins. Alignment of the deduced amino acid sequence of INVINH-like proteins from maize (ZM-INVINH1, ZM-INVINH2, and ZM-INVINH3), wheat (TA-INVINH1 and TA-INVINH2), rice (OS-INVINH1, OS-INVINH2, and OS-INVINH3), as well as previously published sequences from tobacco (NT-INVINH1 and NT-INVINHH), tomato (Lycopersicon esculentum; LE-INVINH1), and Arabidopsis (AT-INVINHH). Conserved Cys residues are indicated by asterisks.
Comparison of ZM-INVINH1 with the closest homologs in grass species reveals a higher degree of similarity, approaching 50% across the entire protein region. In the most recent public rice (Oryza sativa) sequence, three INVINH-related sequences exist, and we have identified an additional sequence (CAC69344). Two of the rice INVINH1-like sequences are in close proximity on chromosome four, separated by 1,437 bp, suggesting duplication (AL606658). The fourth rice sequence is present on chromosome 2 (AP004069). Two wheat (Triticum aestivum) INVINH-related sequences have also been identified in libraries from floral tissue 3 and 7 DAP (Fig. 2).
Effect of Recombinant ZM-INVINH1 on Maize Invertase Activity
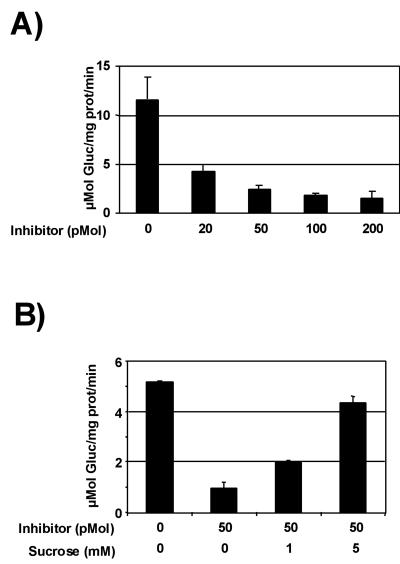
To determine whether ZM-INVINH1 functions as an invertase inhibitor in vitro, we produced ZM-INVINH1 recombinant protein and assayed its ability to reduce invertase activity in a crude insoluble enzyme preparation from 10-DAP kernels. Figure 3 demonstrates that increasing the concentration of inhibitor in invertase reactions results in a quantitative decrease in invertase activity. The presence of 20 pmol of inhibitor protein reduces invertase activity by approximately 3-fold. A characteristic feature of invertase inhibitor proteins is that their inhibitory activity is reduced by the presence of Suc in the reaction. In Figure 3B, we demonstrate that inclusion of Suc concentrations as low as 1 mm has a significant attenuating effect on the reduction of invertase activity in vitro. In a separate experiment, we compared the ability of recombinant ZM-INVINH1 to reduce the activity of soluble and insoluble invertase activity (Table I). In this experiment, 20 pmol of recombinant ZM-INVINH1 reduced kernel soluble invertase activity by 5.3-fold and insoluble invertase by approximately 7-fold.
Figure 3.
Recombinant ZM-INVINH1 inhibits invertase activity in vitro. ZM-INVINH1 produced in Escherichia coli was pre-incubated with an enzyme extraction from maize kernels harvested at 10 DAP. A, Invertase activity measured after pre-incubation with 20 to 200 pmol of recombinant ZM-INVINH1. B, Effect of Suc on ZM-INVINH1 inhibition of invertase activity. Pre-incubation of 10-DAP kernel enzyme extract with 50 pmol of ZM-INVINH1 and 1 or 5 mm Suc. The data are presented as a mean ± sd of three reactions.
Table I.
Effect of recombinant invertase inhibitor on soluble and insoluble invertase activity from maize (n = 3). Twenty picomoles of recombinant invertase inhibitor was pre-incubated with soluble and insoluble preparations of kernel protein.
| Protein Preparation
|
Invertase Activity
|
||
|---|---|---|---|
| —Inh | +Inh | -Fold Reduction | |
| μmol | reducing sugar mg-1 | protein min-1 | |
| Maize soluble | 0.21 | 0.04 | 5.3 |
| Maize insoluble | 8.7 | 1.23 | 7.1 |
Localization of ZM-INVINH1
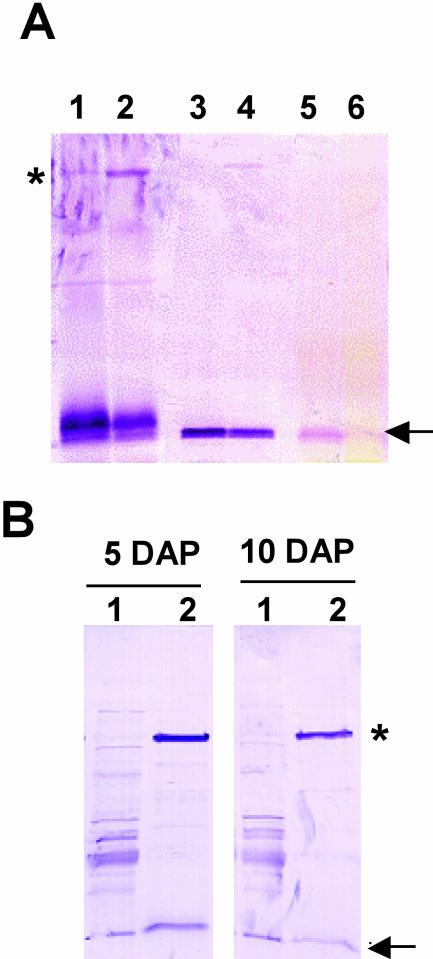
An apparent signal peptide suggests that ZM-INVINH1 is partitioned to the apoplast, where it targets apoplastic invertase. Apoplastic invertase is ionically bound to the cell wall and can be extracted with high-salt buffers, such as 1 m NaCl. We therefore used high-salt extraction, combined with western blotting to confirm the partitioning of ZM-INVINH1 in the cell wall fraction. Figure 4A shows that ZM-INVINH1 is present in significant proportion in the high-salt fraction and present in lower levels in the particulate fraction after the salt wash. Another diagnostic feature of invertases is N-glycosylation (Sturm, 1999), and association of non-glycosylated INVINH proteins with invertase has been indirectly shown by affinity purification with conconavalin A (ConA) chromatography (Weil et al., 1994; Greiner et al., 2000). Figure 4B demonstrates that ConA chromatography partitions ZM-INVINH1 into the glycosylated protein fraction, providing further evidence for a physical interaction between ZM-INVINH1 and invertase enzymes in maize. A high molecular mass protein, with an apparent size of approximately 75 kD, is present in western blots with ConA preparations, and a faint band of similar size appears in western-blot analysis of total protein preparations (Fig. 4A, lanes 1 and 2).
Figure 4.
Fractionation of ZM-INVINH1. A, Proteins isolated from 5-DAP (odd lanes) and 10-DAP (even lanes) kernels were fractionated into soluble (lanes 1 and 2), high-salt wash (lanes 3 and 4), and insoluble fractions (lanes 5 and 6) and subjected to western-blot analysis with antibody raised against ZM-INVINH1. Fifty micrograms of total soluble protein and 20 μg of high-salt and insoluble protein were separated by SDS-PAGE, and western-blot analysis was performed with ZM-INVINH1 antibody. B, ZM-INVINH1 associates with the glycoprotein fraction in maize kernels. Glycoproteins were isolated from 5- and 10-DAP kernels by ConA chromatography, and ZM-INVINH1 was detected by western-blot analysis. Lanes 1 and 2 correspond to ConA chromatography flow through and eluate, respectively. A large arrow represents ZM-INVINH1, and an asterisk indicates a high Mr cross-reacting protein.
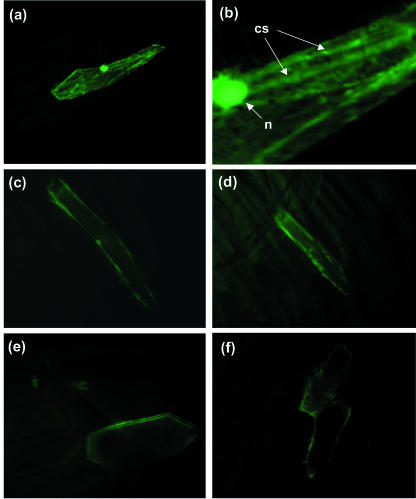
To further characterize the intercellular location of ZM-INVINH1, we used green fluorescent protein (GFP) targeting by bombardment of a ZM-INVINH1::GFP fusion into onion epidermal cells. GFP alone, under the control of a constitutive promoter, fluoresces throughout the cell, including the nucleus (Fig. 5, A and B). A higher magnification clearly shows cytoplasmic streams (Fig. 5B), indicating that the protein is partitioned to the cytoplasm as has been previously discussed (Scott et al., 1999). GFP fused to the C terminus of ZM-INVINH1 has a distinct pattern of fluorescence: No signal is detectable in the nucleus or vacuole, no cytoplasmic streams are evident, and ZM-INVINH1-targeted GFP fluorescence is restricted to the outer edges of the onion cells (Fig. 5, C-F). Fluorescent signal was shown to spread to adjacent cells as shown in Figure 5F, which is a diagnostic feature of small, nonanchored apoplastic proteins. In addition, a strong fluorescence signal required incubation of the bombarded cells with neutral pH, further suggesting that the fusion is targeted to the apoplast, where a low pH quenches the fluorescence signal (Scott et al., 1999).
Figure 5.
ZM-INVINH1 is targeted to the apoplast. Particle bombardment into onion epidermal cells was performed with GFP alone (A and B) or fused to ZM-INVINH1 at the C terminus (E and F) under the control of the maize ubiquitin promoter. Cytoplasmic streams and the nucleus evident in GFP bombardments are indicated by cs and n, respectively.
Expression and Localization of ZM-INVINH1
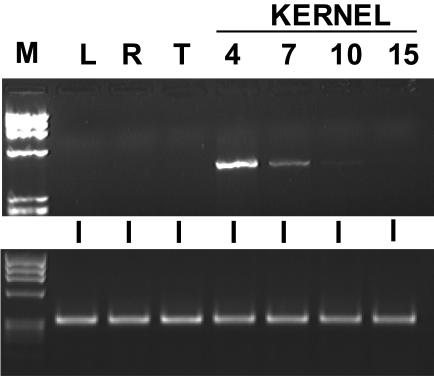
EST distribution for ZM-INVINH1 was restricted to 4-DAP libraries and suggests a low level of expression because only two ESTs are present. Consistent with this, expression was not reliably detected by northern-blot analysis. To verify the restricted spatial and temporal pattern of expression, we performed reverse transcriptase (RT)-PCR using RNA isolated from leaf, root, and tassel tissues, as well as from a range of early kernel development time points (Fig. 6). No message was detectable by RT-PCR in any tissue other than early kernel development, and ZM-INVINH1 transcript accumulation decreased dramatically after 4 DAP to almost undetectable levels by 10 DAP.
Figure 6.
Expression analysis of ZM-INVINH1 by RT-PCR. Top, Total RNA isolated from mature leaf (L), root (R), and tassel (T), as well as from whole kernels harvested 4, 7, 10, and 15 DAP was subjected to RT-PCR to amplify the ZM-INVINH1 transcript. A Mr marker (M) is included. Bottom, A control reaction using primers designed to amplify maize tubulin was also performed.
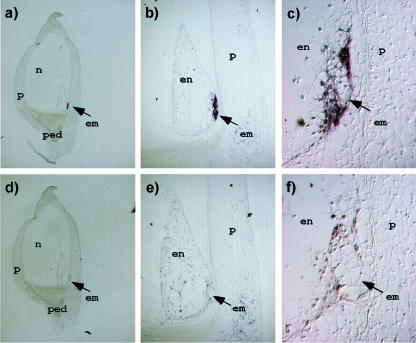
To further characterize the role of ZM-INVINH1 in the invertase-mediated control of early kernel development, we used in situ analysis to determine the spatial pattern of ZM-INVINH1 expression. In situ analysis was performed on 5-DAP kernels because this is close to the peak of expression as determined by RT-PCR (Fig. 6). Figure 7 demonstrates that expression of ZM-INVINH1 is restricted to a very defined region surrounding the embryo. This region, called the ESR, has been previously characterized as one of the four cell types of the developing endosperm (Olsen, 2001). Previously, a group of small proteins with a similar pattern of expression in maize have been characterized (ESR1-3; Opsahl-Ferstad et al., 1997; Benello et al., 2000), suggesting the ESR is a distinct compartment important for endosperm and embryo development (Bommert and Werr, 2001).
Figure 7.
In situ analysis of ZM-INVINH1 in 5-DAP kernels. Embedded and longitudinal kernel sections were hybridized with an antisense (A-C) or sense transcript (D-F) generated from ZM-INVINH1. ped, pedicel; n, nucellus; p, pericarp; en, endosperm; em, embryo.
DISCUSSION
Invertase inhibitor proteins were first characterized biochemically as copurifying proteins in invertase enzyme preparations (Pressey, 1966; Jaynes and Nelson, 1971) and dicot genes encoding INVINH proteins with demonstrated activity in vitro (Greiner et al., 1998) and in vivo (Greiner et al., 1999) were recently characterized. Analysis of sequences from maize and other monocots indicates that there are at least two to three INVINH-related proteins in these species as well. Furthermore, phylogenetic tree analysis indicates that individual sequences share a greater degree of homology across species than exists for INVINH-related sequences within either species alone. Taken together, sequence analyses suggest that there are distinct classes of INVINH-related sequences within each species, potentially reflecting distinct invertase inhibitory activities or function. Identification is further complicated by structurally related proteins that have been demonstrated to have distinct inhibitory activity (Camardella et al., 2000). Recombinant INVINH protein expression and in vitro demonstration of inhibitory activity may be the most definitive means to distinguish INVINH proteins from those with similar structure but distinct function.
ZM-INVINH1 produced as a recombinant protein inhibits maize invertase in a dose-dependent manner, establishing it as an invertase inhibitor. Similar to other INVINH proteins with demonstrated inhibitor activity, inhibition is attenuated by the presence of low Suc concentrations. In addition, ZM-INVINH1 associates with the glycoprotein fraction despite having no obvious glycosylation sites, as has been demonstrated for dicot INVINH proteins (Sander et al., 1996; Krausgrill et al., 1998; Greiner et al., 2000). Taken together, biochemical characterization indicates that ZM-INVINH1 is the first invertase inhibitor gene identified from a monocot species.
ZM-INVINH1 is an early kernel, ESR-localized, apoplastic protein. The question therefore arises: Which invertase is ZM-INVINH1 inhibiting? At least three possible invertase targets for ZM-INVINH1 can be envisioned. First, ZM-INVINH1 could be inhibiting INCW2 present at low levels in the ESR region. INCW2 has been localized to the basal endosperm transfer layer by immunolocalization (Cheng et al., 1996) and in situ analysis (Talercio et al., 1999). However, it is possible that INCW2 may be present in the ESR at levels undetectable by either of these methods. A second possibility is that vacuolar-targeted IVR2, present in the ESR, is mis-localized to the cell wall. IVR2 has been localized to the pedicel region by in situ analysis (Andersen et al., 2002), and we have demonstrated a strong in situ hybridization signal for IVR2 transcript in the ESR at 5 DAP (Y. Wang and N.J. Bate, unpublished data). A third possibility is that ZM-INVINH1 functions to inhibit an as yet uncharacterized apoplastic invertase (INCW). In addition to INCW2, there are at least three cell wall invertases (INCW1, INCW3, and INCW4) that all have expression detectable in early kernels (Talercio et al., 1999; Comparot et al., 2003), albeit at very low levels relative to INCW2 and IVR2. Another related possibility is that ZM-INVINH1 functions as insurance against the activity of any apoplastic invertase activity in the cells surrounding the embryo, whether derived from INCW activity or mis-directed IVR activity. The ability of ZM-INVINH1 to inhibit both soluble and insoluble invertase activities suggests that this may be the case.
The importance of Suc/hexose ratio and the role of invertases in development has been well documented. Transported Suc acts as a carbon source for both the developing embryo and endosperm, yet these two cell types have distinct developmental programs and rates of cell division. By 4 DAP, the embryo has between eight and 32 nuclei, whereas the endosperm develops much more rapidly, having approximately 250 nuclei at the same stage (Randolph, 1936). Furthermore, the basal endosperm cells begin to differentiate at approximately 6 DAP (Kiesselbach, 1949), whereas embryo cells differentiate later. ZM-INVINH1 may therefore serve as a barrier to protect the slower developing embryo from hexoses supplied from an apoplastic invertase early post-pollination, allowing the proper developmental rate of the embryo. The transient presence of ZM-INVINH1 suggests that by 7 to 10 DAP, the inhibitory barrier to invertase is no longer necessary.
The ESR is a region of cells within the endosperm, and three highly related proteins have been localized to the ESR by in situ localization and promoter::GUS fusion experiments (Opsahl-Ferstad et al., 1997; Benello et al., 2000). ESR1-3 are small proteins with some structural homology to the developmental ligand CLV3 from Arabidopsis (Brand and Simon, 2000), suggesting the possibility that ESR proteins play a developmental role in maize. ESR-localized ZM-INVINH1, which is about 2-fold larger than ESR1-3, share several features, most notably their ESR localization and apoplast targeting. Interestingly, a mammalian protein, uterglobin (also known as Clara2 and blastokinin), is a small protein that plays a signaling role and also serves as an enzyme inhibitor (Mukherjee et al., 1999). An intriguing possibility is that ZM-INVINH1 has a dual-function: as an invertase inhibitor and also performing a signaling role as has been suggested for the ESR1-3 proteins. Despite similar temporal and spatial expression, there is little similarity between ESR upstream regulatory regions and the 587 bp of ZM-INVINH1 upstream sequence presented here (Fig. 1). Repeated sequences conserved between ESR1-3 promoters are absent, for example. Alignment of ESR1-3 and ZM-INVINH1 promoter regions identifies two small conserved sequences: AGCATA and T(A/T) AAAAT, present at -170 and -100, relative to the translational start site of ZM-INVINH1. More detailed analysis of the ZM-INVINH1 and ESR1-3 promoters will determine whether these sequences are important for ESR-localized expression.
Identification of ZM-INVINH1 represents the first monocot invertase inhibitor gene to be characterized. In addition, the work presented here provides a bridge between invertase activity, known to be important for the control of carbohydrate partitioning, and the regulation of events during early kernel development by showing expression of ZM-INVINH1 in the ESR, an important and intriguing cluster of cells surrounding the embryo. ZM-INVINH1 therefore also provides a useful marker for early events in the establishment of the embryo and endosperm.
MATERIALS AND METHODS
Gene Isolation and Characterization
To characterize invertase inhibitor homologs from maize (Zea mays), we screened Pioneer/DuPont EST databases using a consensus sequence generated from alignment of previously characterized invertase homologs from dicot species. Full-length sequence for three maize invertase homologs were generated, and effort was concentrated on one homolog, designated ZM-INVINH1, present only in EST libraries from 4-DAP kernels. Rice (Oryza sativa) and wheat (Triticum aestivum) INVINH1-like cDNAs were characterized by screening Pioneer/DuPont EST databases using the ZM-INVINH1 sequence.
Expression by RT-PCR
Mature leaves from 12-leaf (V12) plants, whole roots from 18-leaf (V18) plants, and preshed tassel or kernels 4, 7, 10, or 15 DAP were excised from field-grown public inbred (B73) plants. Total RNA was isolated using the Plant RNeasy kit from Qiagen (Valencia, CA). Total RNA (1 μg) was treated with amplification grade DNase (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions, and RT reactions were performed using the Thermoscript kit according to manufacturer's instructions (Invitrogen) using an oligo(dT) primer. Primers specific to ZM-INVINH1 were generated for RT-PCR: CCAGTCAAGGGGACCATGAA and ATGCATTGGAACCCCTGCTCACAGGTGC. Primers designed to maize tubulin were used as a control for loading and amplification. PCR conditions were optimized, and amplification was cycled 25 times.
Recombinant Protein Expression and Invertase Activity
Recombinant protein was produced using the pET expression system from Novagen (Madison, WI) by cloning the HindIII site immediately downstream of the ATG start, in frame with the poly-His tag of pET28. NotI from the ZM-INVINH1 vector was used at the 3′ end. Recombinant protein was produced in BL21 (Lys) cells according to standard protocols, and affinity chromatography was performed using a cesium column and FPLC. Protein purity was estimated at >75%, and protein concentration determined using Coomassie PLUS (Pierce, Rockford, IL) using bovine serum albumin as a standard. Recombinant protein was dialyzed against 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride. For long-term storage, recombinant protein preparations were combined with an equal volume of 50% (w/v) glycerol, aliquoted into working volumes, and stored at -80°C.
Inhibition of invertase activity was measured using protein preparations from 10-DAP kernels from a public inbred (B73), isolated as described by Cheng et al., 1996. Excised kernels, ground in liquid nitrogen and stored at -80°C, were mixed with cold isolation buffer (10 mm Tris, pH 7.5, and 1 mm phenylmethylsulfonyl fluoride) in a 1:4 (w/v) ratio. The slurry was centrifuged in an Eppendorf centrifuge at full speed for 10 min, the supernatant was removed (soluble fraction), and the pellet was washed five times with cold isolation buffer by resuspension and centrifugation. Washed pellet fractions were incubated for 30 min on ice with isolation buffer plus 1 m NaCl. After centrifugation, the supernatant was dialyzed overnight against isolation buffer using micro-slide-a-lyzer dialysis units from Pierce. Protein was quantified by Bradford reagent (Bio-Rad Laboratories, Hercules, CA). Invertase activity was measured by adapting a protocol of the Nelson's assay (Jaynes and Nelson, 1971) for 96-well plates. Fifty microliters of soluble or insoluble enzyme preparation was incubated with 2% (w/v) Suc at 37°C for 30 min before transfer to Somagyi's reagent and heating to 100°C for 10 min. After addition of Nelson's reagent to the mixture, reducing sugars were measured by A520 in a spectrophotometer (SpectraMax 190, Molecular Devices, Sunnyvale, CA). ZM-INVINH1 and Suc treatments were performed by pre-incubation of the reaction mixture with known concentrations of recombinant protein or Suc for 10 min before addition of 2% (w/v) Suc.
In Situ Analysis
Greenhouse-grown inbred maize (public inbred B73) was grown in a greenhouse in Johnston, Iowa. Kernels were taken from the middle of ears harvested at 5 DAP. Longitudinal median sections containing embryos were obtained and immediately fixed in 4% (w/v) paraformaldehyde (in phosphate-buffered saline [PBS; 100 mm sodium phosphate and 100 mm NaCl, pH 7.5]), which was prepared in diethyl pyrocarbonate-treated deionized, distilled water. Fixation, sectioning, and in situ conditions were as published previously (Woo et al., 2001). Digoxigenenin (DIG)-labeled sense and antisense RNA probes were synthesized in vitro by transcription of linearized pBluescript II KS+ plasmids with T3 or T7 RNA polymerase (Roche Diagnostics). For ZM-INVINH1, a HindIII/SmaI fragment was used to reduce nonspecific binding from the poly(A) sequence present in the cDNA. After hybridization, slides were washed three times in 50% (v/v) formamide, 2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 0.2× SSC for a total of 120 min. Between these stringent washes, the slides were treated with 10 μg mL-1 RNase A for 30 min at 37°C. DIG-labeled probes were detected with anti-DIG antibody conjugated with alkaline phosphatase (AP) and an AP color reaction (Roche Diagnostics), which produces a red precipitate.
GFP Localization
To visualize subcellular localization of ZM-INVINH1, we used GFP imaging as has been previously described (Scott et al., 1999). A control plasmid (GFP) and the ZM-INVINH1 fusion plasmid (the entire coding sequence fused in-frame to GFP), both under the control of the maize ubiquitin promoter, were bombarded into onion epidermal cells with 1-μm Tungsten particles using a Biolistic 100 (Bio-Rad Laboratories; placed onto Murashige and Skoog plates, with 10 mg L-1 l-ascorbic acid) at 650 pounds per square inch. Bombarded cells were incubated for 48 h before incubation with 20 mm PIPES buffer pH 7.0 (Scott et al., 1999) for 3 h. GFP fluorescence was visualized by epifluorescence microscopy using a microscope (SMZ1500, Nikon, Tokyo).
Protein Isolation and Analysis
For fractionation studies, the lower one-third of 5- and 10-DAP kernels was ground in liquid nitrogen and stored at -80°C. Total proteins were extracted by grinding frozen tissue with 4 volumes (v/w) of extraction buffer (50 mm Tris, pH 8.0, 100 mm KCl, 5% [w/v] glycerol, 10 mm dithiothreitol, 2% [w/v] polyvinylpolypyrrolidone, 1 mm EDTA, and 4 mm PefaBloc [Roche Diagnostics]) before centrifugation at full speed in an Eppendorf microfuge at 4°C. Total soluble proteins were removed, and the pellet was washed by adding 10 volumes of extraction buffer, mixing, and centrifuging as above for a total of five times. The insoluble pellet was incubated for 1 h at 4°C in three volumes of 1 m NaCl and 50 mm Tris, pH 7.5, before centrifugation. Pellets from this fraction represent the insoluble fraction. Sodium chloride wash fractions were dialyzed overnight at 4°C as for invertase enzyme assays. Fifty micrograms of total soluble protein and 20 μg of high-salt and insoluble fractions were separated by SDS-PAGE for western analysis.
For affinity purification of the glycoprotein fraction, proteins were subjected to ConA chromatography. Whole-kernel fresh frozen tissue was incubated with extraction buffer (50 mm citric acid, pH 4.6, 500 mm NaCl, 5 mm dithiothreitol, and 1 mm Pefabloc [Roche Diagnostics]), filtered through four layers of cheesecloth, and centrifuged for 10 min at 10,000 rpm in a JA17 rotor (Beckman Coulter, Fullerton, CA). The supernatant was removed, and NH4SO4 added to 90% (w/v) and stirred on ice for 90 min. After centrifugation as above, the pellets were resuspended in 2 mL of column buffer (50 mm sodium acetate, pH 6.3, 500 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, 1 mm MnCl2, and 1 mm Pefabloc). The resuspended proteins were dialyzed overnight against two changes of column buffer using Slide-A-lyzer dialysis membranes (3000-D molecular weight cut-off, Pierce). Hi-Trap ConA columns (Pharmacia, Uppsala) were equilibrated with 10 mL of column buffer using a syringe. Samples were added to the column at a rate of one drop every 2 to 3 s. An aliquot was removed from the column flow-through for analysis before washing immobilized proteins with five column volumes of column buffer. Proteins were eluted with 3 mL of 15% (w/v) methyl-α-glucopyranoside and concentrated in a centricon-10 microconcentrator (Ambion, Austin, TX). Thirty micrograms of protein from flow through and affinity-purified fractions was used for western-blot analysis.
Antibodies were raised against recombinant ZM-INVINH1 in New Zealand White rabbits at HTI Bioproducts (Ramona, CA), raised on a maize-free diet. Affinity-purified His tag fusions of ZM-INVINH1 were denatured in 5 m urea for 5 min at 100°C and used as antigens. Antibody titer was determined using purified antigen, and crude serum was used as a 1:2,000 (v/v) dilution in western-blot experiments.
For western-blot analysis, proteins were separated by SDS-PAGE, blotted onto PVDF membranes (Bio-Rad Laboratories), and blocked according to standard protocols issued by the manufacturer, using either 5% (w/v) nonfat milk or Superblocker (Pierce). Primary antibody was diluted 1:2,000 in PBS with 0.1% (v/v) Tween 20 for several hours to overnight. Blots were washed several times in PBS with 0.1% (v/v) Tween 20 before incubating with a secondary antibody (goat anti-rabbit, AP conjugated, Sigma-Aldrich, St. Louis) for 1 h. Blots were subsequently washed before visualizing immunoreactive signal by AP color development.
Distribution of Materials
Novel materials described in this publication may be available for noncommercial research purposes upon acceptance and signing of a material transfer agreement. In some cases, such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors, or controllers of all or parts of the material. Obtaining any permissions will be the sole responsibility of the requestor.
Acknowledgments
We are grateful to Gabrielle Tordensen and Tom Davis and the protein production facility at Pioneer for working with us to produce functional recombinant ZM-INVINH. Odd-Arne Olsen and Cunxi Wang provided helpful and insightful discussion. Virginia Crane and Jeanne Sandahl helped with bombardment, and Bill Gordon-Kamm helped with GFP localization. In addition, we are grateful to Jeff Mullen, Chris Zinselmeier, and other members of the Yield Stability Group for helpful suggestions and productive collaboration.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027466.
References
- Andersen MN, Asch F, Wu Y, Jensen CR, Næsted H, Mogensen VO, Koch KE (2002) Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiol 130: 591-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benello J-F, Opsahl-Ferstad H-G, Perez P, Dumas C, Rogowsky PM (2000) Esr genes show different levels of expression in the same region of maize endosperm. Gene 246: 219-227 [DOI] [PubMed] [Google Scholar]
- Bommert P, Werr W (2001) Gene expression patterns in the maize caryopsis: clues to decisions in embryo and endosperm development. Gene 271: 131-142 [DOI] [PubMed] [Google Scholar]
- Brand U, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617-619 [DOI] [PubMed] [Google Scholar]
- Camardella L, Carrotore V, Ciardiello MA, Servillo L, Balestrieri C, Giovane A (2000) Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur J Biochem 267: 4561-4565 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Chourney P (1999) Genetic evidence that invertase-mediated release of hexoses is critical for appropriate carbon partitioning and normal seed development in maize. Theor Appl Genet 98: 485-495 [Google Scholar]
- Cheng W-H, Taliercio EW, Chourey PS (1996) The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8: 971-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot S, Lingiah G, Martin T (2003) Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J Exp Bot 54: 595-604 [DOI] [PubMed] [Google Scholar]
- Greiner S, Koster U, Lauer K, Rosenkranz H, Vogel R, Rausch T (2000) Plant invertase inhibitors: expression in cell culture and during plant development. Aust J Plant Physiol 27: 807-814 [Google Scholar]
- Greiner S, Krausgrill S, Rausch T (1998) Cloning of a tobacco apoplasmic invertase inhibitor: proof of function of the recombinant protein and expression analysis during plant development. Plant Physiol 116: 733-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Rausch T, Sonnewald U, Herbers K (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold-induced sweetening of potato tubers. Nat Biotechnol 17: 708-711 [DOI] [PubMed] [Google Scholar]
- Jaynes TA, Nelson OE (1971) An invertase inactivator in maize endosperm and factors affecting inactivation. Plant Physiol 47: 629-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesselbach TA (1949) The Structure and Reproduction of Corn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Krausgrill S, Greiner S, Koster U, Vogel R, Rausch T (1998) In transformed tobacco cells the apoplasmic invertase inhibitor operates as a regulatory switch of cell wall invertase. Plant J 13: 275-280 [Google Scholar]
- Krausgrill S, Sander A, Greiner S, Weil M, Rausch T (1996) Regulation of cell wall invertase by a proteinaceous inhibitor. J Exp Bot 47: 1193-1198 [DOI] [PubMed] [Google Scholar]
- Mukherjee AB, Kundu GC, Mantile-Selvaggi G, Yuan CJ, Mandal AK, Chattopadhyay S, Zheng F, Pattabiraman N, Zhang Z (1999) Uteroglobin: a novel cytokine? Cell Mol Life Sci 55: 771-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O-A (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52: 233-267 [DOI] [PubMed] [Google Scholar]
- Opsahl-Ferstad H-G, Le Deunff E, Dumas C, Rogowsky PM (1997) ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J 12: 235-246 [DOI] [PubMed] [Google Scholar]
- Pressey R (1966) Separation and properties of potato invertase and invertase inhibitor. Arch Biochem Biophys 113: 667-674 [DOI] [PubMed] [Google Scholar]
- Pressey R (1994) Invertase inhibitor in tomato fruit. Phytochemistry 36: 543-546 [Google Scholar]
- Randolph LF (1936) Developmental morphology of the caryopsis in maize. J Agric Res 53: 881-916 [Google Scholar]
- Sander A, Krausgrill S, Greiner S, Weil M, Rausch T (1996) Sucrose protects cell wall invertase but not vacuolar invertase against proteinaceous inhibitors. FEBS Lett 385: 171-175 [DOI] [PubMed] [Google Scholar]
- Scott A, Wyatt P-L, Tsou D, Robertson D, Stromgren A (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125-1132 [DOI] [PubMed] [Google Scholar]
- Sturm A (1999) Invertases: primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121: 1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talercio E, Kim J-Y, Mahe A, Shaker S, Choi J, Cheng W-H, Prioul J-L, Chourey PS (1999) Isolation, characterization and expression analyses of two cell wall invertase genes in maize. J Plant Physiol 155: 197-204 [Google Scholar]
- Vilhar B, Kladnik A, Blejec A, Chourey PS, Dermastia M (2002) Cytometrical evidence that the loss of seed weight in the miniature1 seed mutant of maize is associated with reduced mitotic activity in the developing endosperm. Plant Physiol 129: 23-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M, Krausgrill S, Schuster A, Rausch T (1994) A 17-kDa Nicotiana tabacum cell-wall peptide acts as an in-vitro inhibitor of the cell-wall isoform of acid invertase. Planta 193: 438-445 [DOI] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U (2003) The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. Plant J 33: 395-411 [DOI] [PubMed] [Google Scholar]
- Wobus U, Weber H (1999) Sugars as signal molecules in plant seed development. Biol Chem 380: 937-944 [DOI] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297-2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier C, Jeong B-R, Boyer JS (1999) Starch and the control of kernel number in maize at low water potential. Plant Physiol 121: 25-35 [DOI] [PMC free article] [PubMed] [Google Scholar]