Abstract
N2′-Pyrene-functionalized 2′-amino-α-L-LNAs (Locked Nucleic Acids) display extraordinary affinity toward complementary DNA targets due to favorable preorganization of the pyrene moieties for hybridization-induced intercalation. Unfortunately, the synthesis of these monomers is challenging (~20 steps, <3% overall yield), which has precluded full characterization of DNA-targeting applications based on these materials. Access to more readily accessible functional mimics would be highly desirable. Here we describe short synthetic routes toward a series of O2′-intercalator-functionalized uridine and N2′-intercalator-functionalized 2′-N-methyl-2′-aminouridine monomers and demonstrate – via thermal denaturation, UV-visible absorption and fluorescence spectroscopy experiments – that several of them mimic the DNA-hybridization properties of N2′-pyrene-functionalized 2′-amino-α-L-LNAs. For example, oligodeoxyribonucleotides (ONs) modified with 2′-O-(coronen-1-yl)methyluridine monomer Z, 2′-O-(pyren-1-yl)methyluridine monomer Y or 2′-N-(pyren-1-ylmethyl)-2′-N-methylaminouridine monomer Q, display prominent increases in thermal affinity toward complementary DNA relative to reference strands (average ΔTm/mod up to +12 °C), pronounced DNA-selectivity, and higher target specificity than 2′-amino-β-L-LNA benchmark probes. In contrast, ONs modified with 2′-O-(2-napthyl)uridine monomer W, 2′-O-(pyren-1-yl)uridine monomer X or 2′-N-(pyren-1-ylcarbonyl)-2′-N-methylaminouridine monomer S display very low affinity toward DNA targets. This demonstrates that even conservative alterations in linker chemistry, linker length and surface area of the appended intercalators have marked impact on DNA-hybridization characteristics. Straightforward access to high-affinity building blocks such as Q/Y/Z is likely to accelerate their use in DNA-targeting applications within nucleic acid based diagnostics, therapeutics, and material science.
INTRODUCTION
Development of chemically modified oligonucleotides continues to attract remarkable attention due to their wide-ranging applications within fundamental research, diagnostics, therapeutics and materials science.1 Representative applications include modulation of gene expression at the DNA2 or RNA level,3 detection of DNA/RNA targets including those with single nucleotide polymorphisms (SNPs),4 and the use of nucleic acids as supramolecular scaffolds for organization of chromophores.5
As part of our ongoing interest in functionalized variants6 of affinity- and specificity-enhancing LNA (Locked Nucleic Acid)7 and α-L-LNA,7c,8 we recently developed oligodeoxyribonucleotides (ONs) modified with N2′-pyrene-functionalized 2′-amino-α-L-LNA monomers.9 The ability of these monomers to precisely and site-specifically position intercalators in DNA duplex cores results in exceptional affinity toward complementary DNA targets and has enabled us to: a) form dense pyrene arrays within duplex cores,10 b) develop probes for fluorescent discrimination of SNPs in DNA targets under non-stringent conditions,11 and c) target mixed-sequence regions in double-stranded DNA via the so-called ‘Invader LNA’ approach.12 The synthesis of the N2′-intercalator-functionalized 2′-amino-α-L-LNA thymine phosphoramidites is unfortunately very challenging. For example, the corresponding phosphoramidite of 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA thymine monomer L is obtained in ~3% yield over ~20 steps from diacetone-α-D-glucose (Fig. 1).9b,13 Access to synthetically more readily accessible mimics of N2′-intercalator-functionalized 2′-amino-α-L-LNA monomers would be highly desirable to evaluate the full scope of the aforementioned DNA-targeting applications.
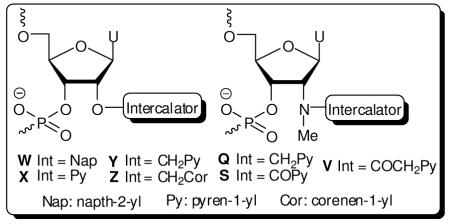
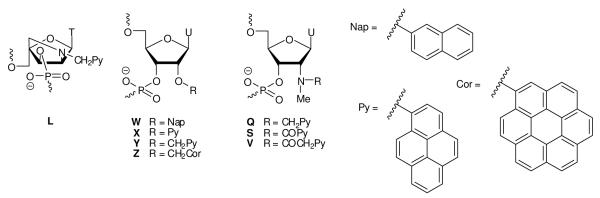
Figure 1.
Chemical structures of monomers studied herein: 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA thymine monomer L, O2′-intercalator-functionalized uridine monomers W-Z and N2′-intercalator-functionalized 2′-N-methyl-2′-aminouridine monomers Q/S/V.
We identified O2′-intercalator-functionalized RNA and N2′-intercalator-functionalized 2′-N-methyl-2′-amino-DNA monomers (Fig. 1) as potential structural and functional mimics of N2′-intercalator-functionalized 2′-amino-α-L-LNA. We based this hypothesis on studies showing that a) ONs modified with 2′-O-(pyren-1-yl)methyluridine monomer Y or 2′-N-methyl-2′-N-(pyren-1-ylmethyl)-2′-amino-DNA monomer Q display high thermal affinity toward complementary DNA,14,15 and that b) the pyrene moieties intercalate between the nucleobase of the monomer and the 3′-flanking nucleoside.16 Motivated by this, we set out to a) develop synthetic routes toward a series of O2′-intercalator-functionalized uridine and N2′-intercalator-functionalized 2′-N-methyl-2′-aminouridine phosphoramidites and b) compare hybridization properties of the correspondingly modified ONs with those of 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA ‘benchmark’ probes (Fig. 1).
RESULTS AND DISCUSSION
Synthesis of O2′-Intercalator-functionalized Uridine Phosphoramidites
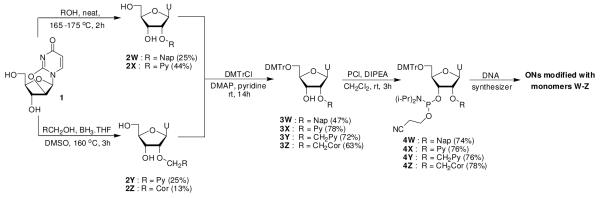
Four phosphoramidites 4W-4Z were selected as synthetic targets in this series as this allowed us to: a) test whether known monomer Y17 indeed is a mimic of ‘benchmark’ monomer L, and b) systematically evaluate the influence of linker length (monomer X vs Y) and aromatic surface area (monomer W vs X; monomer Y vs Z) on the hybridization properties of modified ONs (Scheme 1).
Scheme 1.
Synthesis of O2′-intercalator-functionalized uridine phosphoramidites 4W-4Z.
Sekine and coworkers recently described a microwave-based approach in which 2,2′-anhydrouridine 1 is treated with neat phenols to afford O2′-arylated uridines.18 We adapted this approach for conventional heating sources and used 2-naphtol and 1-pyrenol19 to obtain 2W18 and 2X in 25% and 44% yield, respectively (Scheme 1). Treatment of 120 with pyren-1-ylmethanol or coronen-1-ylmethanol under similar conditions failed to afford desired products 2Y and 2Z in acceptable yields, requiring development of an alternative approach.
Yamana and coworkers previously installed arylmethyl groups at the O2′-position of 3′,5′-ditrityluridine via Williamson etherification.17,21 We found the initial O3′,O5′-tritylation of uridine22 and the protecting group manipulations following the O2′-functionalization step to be cumbersome. Instead, the reaction between 1 and tris(pyrene-1-ylmethyl) or tris(coronene-1-ylmethyl) borate - generated in situ upon addition of pyren-1-ylmethanol23 or coronen-1-ylmethanol24 to borane - afforded 2Y and 2Z in modest but robust yields (Scheme 1).25 The two-step route to nucleoside 2Y from uridine proceeds in similar yield (~20%) as the original three-step approach,17 but is more convenient. To our knowledge, this is the first time that the trialkyl borate mediated opening26 of O2,O2′-anhydrouridine 1 has been used to install large arylmethylethers at the C2′-position of nucleosides. Subsequent O5′-dimethoxytritylation afforded nucleosides 3W-3Z in 47-78% yield, which upon treatment with 2-cyanoethyl-N,N-diisopropylchlorophosporamidite (PCl-reagent) and Hünig’s base provided target phosphoramidites 4W-4Z in 74-78% % yield (Scheme 1).
Synthesis of N2′-Pyrene-functionalized 2′-N-Methyl-2′-Aminodeoxyuridine Phosphoramidites
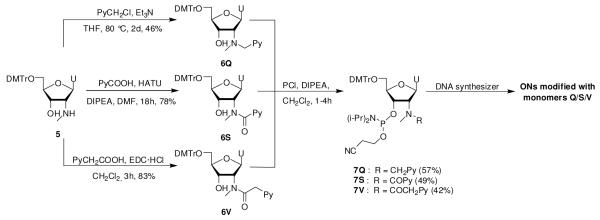
In this series, three phosphoramidites 7Q/7S/7V were selected as synthetic targets to a) evaluate whether known monomer Q15 is a mimic of benchmark monomer L, and b) to study the influence of linker chemistry (monomer Q vs S) and length (monomer S vs V) on hybridization properties of correspondingly modified ONs (Scheme 2).
Scheme 2.
Synthesis of N2′-intercalator-functionalized uridine phosphoramidites 7Q/7S/7V.
Wengel and coworkers previously reported a route to 2′-N-methyl-2′-N-(pyren-1-ylmethyl)-2′-aminouridine phosphoramidite 7Q from 5′-O-dimethoxytrityl-2′-aminouridine, which entails N2′-functionalization via reductive amination, O5′-detritylation, N2′-methylation under acidic conditions, O5′-tritylation and O3′-phophitylation (~7% overall yield over five steps).15 In order to decrease the number of protection/deprotection steps and to introduce N2′-moieties at a later stage, we instead decided to utilize nucleoside 5 as a starting material, which is easily obtained from 5′-O-dimethoxytrityl-2,2′-anhydrouridine in ~73% yield over three steps.27 Direct N2′-alkylation of 5 using pyren-1-ylmethyl chloride afforded the desired product 6Q in 46% yield.25 Interestingly, reductive amination of 5 using 1-pyrenecarbaldehyde and sodium triacetoxyborohydride28 or sodium cyanoborohydride failed to afford 6Q in acceptable yields due to prominent formation of the corresponding cyclic N2′,O3′-hemiaminal ether (see supporting information, Scheme S1). While formation of this byproduct was avoided by prior protection of the O3′-position of 5 as a TBDMS-ether, the increased steric bulk resulted in low yields during the subsequent reductive amination (results not shown).
HATU-mediated coupling between nucleoside 5 and 1-pyrenecarboxylic acid afforded N2′-acylated nucleoside 6S in 78% yield, while EDC-mediated coupling between nucleoside 5 and 1-pyreneacetic acid furnished 6V in 83% yield.25 Subsequent O3′-phosphitylation of 6Q/6S/6V using similar conditions as for the synthesis of 4W-4Z afforded phosphoramidites 7Q/7S/7V only in moderate yields (42-57%), presumably due to the increased steric bulk at the N2′-position.
Synthesis of Modified ONs and Setup of Thermal Denaturation Studies
Intercalator-functionalized phosphoramidites 4W/4X/4Y/4Z/7Q/7S/7V were incorporated into ONs via machine-assisted solid-phase DNA synthesis using the following hand-coupling conditions (activator; coupling time; approximate coupling yield): monomer W/X/Y (4,5-dicyanoimidazole; 15min; ~98%/~98%/~98%); monomer Z (4,5-dicyanoimidazole; 35min; ~90%); monomers Q/S/V (5-(bis-3,5-trifluoromethylphenyl)-1H-tetrazole [Activator 42]; 15min; ~95%/~89%/~99%). Suitable activators were identified through initial screening of common activators (results not discussed). The selected mixed-sequence 9-mers have been previously used to study ONs modified with 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA thymine monomer L.9b ONs containing a single incorporation in the 5′-GBG ATA TGC context are denoted V1, W1, Y1, Z1, Q1, S1, V1 and L1, respectively. Similar conventions apply for ONs in the B2-B7 series (Table 1). In addition, the following descriptive nomenclature is used: O2′-Nap (W-series), O2′-Py (X-series), O2′-PyMe (Y-series), O2′-CorMe (Z-series), N2′-PyMe (Q-series), N2′-PyCO (S-series), N2′-PyAc (V-series) and N2′-PyMe-α-L-LNA (L-series). Reference DNA and RNA strands are denoted D1/D2 and R1/R2, respectively.
Table 1.
Tm-values of duplexes between B1-B7 and complementary DNA targetsa
|
Tm [ΔTm/mod] (°C) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ON | Duplex | B = | T | W | X | Y | Z | Q | S | V | L9b |
|
B1 D2 |
5′-GBG ATA TGC 3′-CAC TAT ACG |
29.5 | 21.5 [−8.0] |
26.5 [−3.0] |
34.5 [+5.0] |
34.0 [+4.5] |
34.5 [+5.0] |
23.5 [−6.0] |
29.0 [−0.5] |
36.5 [+7.0] |
|
|
| |||||||||||
|
B2 D2 |
5′-GTG ABA TGC 3′-CAC TAT ACG |
29.5 | 24.5 [−5.0] |
33.5 [+4.0] |
42.0 [+12.5] |
49.5 [+20.0] |
43.5 [+14.0] |
32.5 [+3.0] |
35.5 [+6.0] |
43.5 [+14.0] |
|
|
| |||||||||||
|
B3 D2 |
5′-GTG ATA BGC 3′-CAC TAT ACG |
29.5 | 24.5 [−5.0] |
26.0 [−3.5] |
37.5 [+8.0] |
40.5 [+11.0] |
ND | ND | ND | 40.0 [+10.5] |
|
|
| |||||||||||
|
D1 B4 |
5′-GTG ATA TGC 3′-CAC BAT ACG |
29.5 | 16.5 [−13.0] |
26.0 [−3.5] |
33.0 [+3.5] |
36.0 [+6.5] |
31.0 [+1.5] |
23.5 [−6.0] |
30.5 [+1.0] |
36.0 [+6.5] |
|
|
| |||||||||||
|
D1 B5 |
5′-GTG ATA TGC 3′-CAC TAB ACG |
29.5 | 24.5 [−5.0] |
30.5 [+1.0] |
41.0 [+11.5] |
45.5 [+16.0] |
42.5 [+13.0] |
33.5 [+4.0] |
36.0 [+6.5] |
45.0 [+15.5] |
|
|
| |||||||||||
|
D1 B6 |
5′-GTG ATA TGC 3′-CAC BAB ACG |
29.5 | ND | 25.5 [−2.0] |
43.5 [+7.0] |
47.0 [+8.8] |
43.5 [+7.0] |
ND | 37.0 [+3.8] |
ND | |
|
| |||||||||||
|
B7 D2 |
5′-GBG ABA BGC 3′-CAC TAT ACG |
29.5 | ND | 25.5 [−1.3] |
51.0 [+7.2] |
ND | ND | ND | ND | ND | |
ΔTm = change in Tm’s relative to unmodified reference duplex; Tm’s determined as the maximum of the first derivative of melting curves (A260 vs T) recorded in medium salt buffer ([Na+] = 110mM, [Cl−] = 100mM, pH 7.0 (NaH2PO4/Na2HPO4)), using 1.0 μM of each strand. Tm’s are averages of at least two measurements within 1.0 °C; A = adenin-9-yl DNA monomer, C = cytosin-1-yl DNA monomer, G = guanin-9-yl DNA monomer, T = thymin-1-yl DNA monomer. ND = not determined. For structures of monomers Q-Z see Figure 1.
The thermal affinity of B1-B7 toward complementary DNA or RNA targets was evaluated via UV thermal denaturation experiments using medium salt buffers that mimic physiological ionic strengths ([Na+] = 110 mM, Tables 1 and 2). As expected, all denaturation curves display sigmoidal monophasic transitions (Figs. S1 and S2). Changes in thermal denaturation temperatures (Tm-values) of modified duplexes are discussed relative to Tm-values of unmodified reference duplexes, unless otherwise mentioned. The effect on duplex thermostability upon exchanging thymine moieties (reference ONs) with uracil moieties (modified ONs) is recognized (ΔTm/modification [ΔTm/mod] ~ −0.5 °C)29 but otherwise disregarded.
Table 2.
Tm-values of duplexes between B1-B7 and complementary RNA targetsa
|
Tm [ΔTm/mod] (°C) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ON | Duplex | B = | T | W | X | Y | Z | Q | S | V | L9b |
|
B1 R2 |
5′-GBG ATA TGC 3′-CAC UAU ACG |
26.5 | 21.0 [−5.5] |
22.5 [−4.0] |
24.5 [−2.0] |
21.5 [−5.0] |
22.0 [−4.5] |
14.0 [−12.5] |
20.0 [−6.5] |
27.0 [+0.5] |
|
|
| |||||||||||
|
B2 R2 |
5′-GTG ABA TGC 3′-CAC UAU ACG |
26.5 | 16.0 [−10.5] |
22.5 [−4.0] |
30.5 [+4.0] |
37.0 [+10.5] |
29.0 [+2.5] |
18.0 [−8.5] |
28.0 [+1.5] |
31.5 [+5.0] |
|
|
| |||||||||||
|
B3 R2 |
5′-GTG ATA BGC 3′-CAC UAU ACG |
26.5 | 18.0 [−8.5] |
14.5 [−12.0] |
26.5 [±0.0] |
30.5 [+4.0] |
ND | ND | ND | 28.0 [+1.5] |
|
|
| |||||||||||
|
R1 B4 |
5′-GUG AUA UGC 3′-CAC BAT ACG |
24.5 | 16.5 [−8.0] |
16.5 [−8.0] |
20.0 [−4.5] |
22.5 [−2.0] |
17.5 [−7.0] |
9.0 [−15.5] |
16.0 [−8.5] |
23.5 [−1.0] |
|
|
| |||||||||||
|
R1 B5 |
5′-GUG AUA UGC 3′-CAC TAB ACG |
24.5 | 19.5 [−5.0] |
21.5 [−3.0] |
27.0 [+2.5] |
30.5 [+6.0] |
27.0 [+2.5] |
16.0 [−8.5] |
26.5 [+2.0] |
32.0 [+7.5] |
|
|
| |||||||||||
|
R1 B6 |
5′-GUG AUA UGC 3′-CAC BAB ACG |
24.5 | ND | 14.5 [−5.0] |
24.0 [−0.3] |
11.0 [−6.8] |
20.0 [−2.3] |
ND | 19.0 [−2.8] |
ND | |
|
| |||||||||||
|
B7 D2 |
5′-GBG ABA BGC 3′-CAC UAU ACG |
26.5 | ND | 15.5 [−3.7] |
27.5 [+0.3] |
ND | ND | ND | ND | ND | |
For conditions of thermal denaturation experiments, see Table 1.
Thermal Affinity towards Complementary DNA/RNA – O2′-Intercalator-functionalized Uridine Monomers W-Z
The effects on thermal DNA affinity upon incorporation of O2′-functionalized uridine monomers W-Z into ONs vary dramatically (ΔTm/mod = −13.0 °C to +20.0 °C, Table 1). The observed trends in DNA affinity of singly modified ONs (Z ≥ L > Y >> X >> V) suggest that: a) 2′-O-(pyren-1-yl)methyluridine monomer Y is a mimic of 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA benchmark monomer L with respect to DNA-hybridization properties and binding mode albeit slightly lower duplex thermostabilization is observed (ΔTm/mod = +3.5 °C to +12.5 °C vs +6.5 °C to +15.5 °C, for Y1-Y5 and L1-L5, respectively, Table 1); b) increasing the intercalator surface area leads to additional increases in DNA duplex thermostability (O2′-PyMe monomer Y → O2′-CorMe monomer Z, ΔTm/mod = +4.5 to +20.0 °C, Table 1); c) shortening the linker between the furanose and pyrene moiety by one CH2-group results in markedly lower DNA-affinity (O2′-PyMe monomer Y → O2′-Py monomer X, ΔTm/mod = −3.5 to +4.0 °C, Table 1); and d) concomitant reduction in aromatic surface area results in additionally decreased DNA duplex thermostability (O2′-Py monomer X → O2′-Nap monomer W, Tm/mod = −13.0 to −5.0 °C, Table 1). The results underline that intercalation is an important binding mode for monomers Y and Z and corroborate earlier studies demonstrating that pyrene and coronene moieties engage in efficient π-π stacking with flanking nucleobases.30,31 The shorter linker of monomers W and X, in contrast, seems to reduce the likelihood of an intercalative binding mode. The magnitude of DNA duplex thermostabilization is highly sequence-dependent, in particular for ONs modified with monomer Z. Particularly pronounced stabilization is observed when Z is centrally incorporated and flanked by purines (e.g., compare Tm/mod-values for Z2 and Z4). This most likely reflects strong π-π stacking efficiency with large 3′-flanking purines.9b,14
Singly modified ONs B1-B5 display similar relative trends in thermal affinity toward complementary RNA (Z ~ L > Y >> X > W) as toward DNA except that the resulting duplexes are considerably less stable (Tm/mod = −12.0 °C to +10.5 °C, Table 2). In fact, none of the monomers consistently results in increased thermal affinity relative to reference strand D1. Substantial DNA-selectivity, defined as ΔΔTm (DNA-RNA) = Tm (vs DNA) - ΔTm (vs RNA) > 0 °C, is therefore observed (Table 3). The DNA-selectivity of O2′-CH2Py-modified Y1-Y5 and O2′-CH2Cor-modified Z1-Z5 is especially pronounced and comparable to that of the corresponding N2′-PyMe-α-L-LNA L1-L5 (Table 3). The extensive DNA-selectivity of doubly O2′-CH2Cor-modified Z6 is particularly noteworthy and hints at interesting DNA-targeting applications using multilabeled ONs (ΔΔTm (DNA-RNA) = 31.0 °C, Table 3). DNA-selective hybridization is typically observed for ONs modified with intercalating moieties,9b,14,32,33 since intercalation favors the less compressed helix geometry of DNA:DNA duplexes. In contrast, the majority of O2′-Py-modified X1-X7 and all O2′-Nap-modified W1-W5, in particular, display much lower DNA selectivity, indicating that intercalative binding modes are less prominent with these monomers than with Y and Z monomers (Table 3).
Table 3.
DNA-selectivity of B1-B7.a
| ΔΔTm (DNA-RNA) [°C] |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ON | Duplex | B = | W | X | Y | Z | Q | S | V | L9b |
|
B1 D2 |
5′-GBG ATA TGC 3′-CAC TAT ACG |
−2.5 | +1.0 | +7.0 | +9.5 | +9.5 | +6.5 | +6.0 | +6.5 | |
|
| ||||||||||
|
B2 D2 |
5′-GTG ABA TGC 3′-CAC TAT ACG |
+5.5 | +8.0 | +8.5 | +9.5 | +11.5 | +11.5 | +4.5 | +9.0 | |
|
| ||||||||||
|
B3 D2 |
5′-GTG ATA BGC 3′-CAC TAT ACG |
+3.5 | +8.5 | +8.0 | +7.0 | ND | ND | ND | +9.0 | |
|
| ||||||||||
|
D1 B4 |
5′-GTG ATA TGC 3′-CAC BAT ACG |
+5.0 | +4.5 | +8.0 | +8.5 | +8.5 | +9.5 | +9.5 | +7.5 | |
|
| ||||||||||
|
D1 B5 |
5′-GTG ATA TGC 3′-CAC TAB ACG |
0 | +4.0 | +9.0 | +10.0 | +10.5 | +12.5 | +4.5 | +8.0 | |
|
| ||||||||||
|
D1 B6 |
5′-GTG ATA TGC 3′-CAC BAB ACG |
ND | +6.0 | +14.5 | +31.0 | +18.5 | ND | +13.0 | ND | |
|
| ||||||||||
|
B7 D2 |
5′-GBG ABA BGC 3′-CAC TAT ACG |
ND | +7.0 | +20.5 | ND | ND | ND | ND | ND | |
DNA selectivity defined as ΔΔTm (DNA-RNA) = ΔTm (vs DNA) - ΔTm (vs RNA).
Thermal Affinity towards Complementary DNA/RNA – N2′-Pyrene-functionalized 2′-N-Methyl-2′-Aminodeoxyuridine Monomers Q/S/V
The highly variable effects on thermal affinity toward DNA targets upon incorporation of N2′-pyrene-functionalized 2′-N-methyl-2′-aminodeoxyuridine monomers Q/S/V into ONs are intriguing considering the very conservative differences in linker chemistry and linker length (ΔTm/mod = −6.0 °C to +14.0 °C, Table 1). The observed trends in DNA duplex thermostabilization of singly modified ONs (L>Q>>V>S) suggest that: a) 2′-N-(pyren-1-ylmethyl)-2′-N-methylaminouridine monomer Q indeed is a mimic of N2′-PyMe-α-L-LNA monomer L with respect to DNA-hybridization properties and binding mode (Tm/mod = +1.5 to +14.0 °C, Table 1); b) monomers where the pyrene moiety is attached via N2′-alkyl linkers are preferred over those with N2′-alkanoyl linkers (compare Tm/mod for S1-S5 = −6.0 to +4.0 °C with data for Q1-Q5, Table 1); and c) extending the N2′-alkanoyl linker between the furanose and pyrene moiety as in ONs modified with 2′-N-(pyren-1-ylmethycarbonyl)-2′-aminouridine monomer V, partially reverses the detrimental effects of N2′-acylation on DNA duplex thermostability (N2′-PyCO monomer S → N2′-PyAc monomer V, ΔTm/mod = −0.5 to +6.5 °C, Table 1). ONs with central incorporations of N2′-acylated 2′-N-aminouridine monomers (i.e., without the 2′-N-methyl group of monomers Q/S/V) are known to display low affinity toward DNA targets.34,35 This suggests that the low thermal affinity observed for ONs in the S- and V-series is a consequence of the 2′-N-amido substituent rather than the 2′-N-methyl group. We speculate that the rigid 2′-N-alkanoyl linker positions the intercalator in an unsuitable position for affinity-enhancing intercalation and/or that increased solvation of the linker stabilizes the single-stranded state rendering hybridization less energetically favorable.
The hybridization characteristics of ONs modified with monomers Q/S/V share several points of similarity with the corresponding ONs modified with monomers W-Z: a) thermostabilization of DNA duplexes is highly sequence-dependent; b) incorporation of two monomers as next-nearest neighbors leads to additive increases in DNA duplex thermostability (i.e., compare ΔTm/mod-values for Q4-Q6, Table 1); c) duplexes with complementary RNA are dramatically destabilized (ΔTm/mod = −15.5 °C to +2.5 °C, Table 2); and d) extensive DNA selectivity is observed which is most pronounced for N2′-PyMe-modified Q1-Q5 and N2′-PyCO-modified S1-S5 (ΔΔTm (DNA-RNA) up to +12.5 °C, Table 3). The latter is in agreement with the suggested intercalative pyrene binding mode for monomer Q,15 while the pronounced DNA-selectivity of S1-S5 is more surprising given the low DNA duplex thermostability. This indicates that intercalation of the pyrene moiety remains an important binding mode for monomer S but that it interferes with duplex formation. The less pronounced DNA selectivity of V1-V6 suggests that intercalative pyrene binding modes are less dominant for monomer V.
To sum up, the thermal denaturation data suggest that O2′-PyMe monomer Y, O2′-CorMe monomer Z and N2′-PyMe monomer Q are mimics of N2′-PyMe-α-L-LNA monomer L and attractive candidates for DNA targeting applications.
Mismatch Discrimination
The Watson-Crick specificity of singly modified ONs (B2-series) was evaluated using DNA (Table 4) or RNA targets (Table S4) with mismatched nucleotides opposite of the incorporation site. The B2-series ONs generally display reduced DNA target specificity relative to reference strand D1 which is in agreement with observations on other intercalator-functionalized ONs.9b,36,37 For example, O2′-PyMe-modified Y2 and O2′-CorMe-modified Z2 discriminate dT-mismatches very poorly relative to D1 (compare Tm-values for D1/Y2/Z2 against targets with a dT-mismatch, Table 4) while dC- and dG-mismatches almost are as efficiently discriminated as with D1. N2′-PyMe-α-L-LNA L2 has a similar specificity profile but discriminates the dT-mismatch even more poorly. Interestingly, N2′-PyMe-modified Q2 displays substantially better discrimination of dC- and dT-mismatches than Y2, Z2 and L2 rendering it as the most specific of the high-affinity DNA-targeting modifications. Even minor changes in linker chemistry and length have marked influence on target specificity. Thus, N2′-PyCO-modified S2 displays similar DNA target specificity as Y2/Z2/L2, while N2′-PyAc-modified V2 discriminates DNA mismatches more poorly. Similarly, O2′-Nap-modified W2 exhibits very poor target specificity, while O2′-Py-modified X2 displays much higher target specificity than W2 or O2′-PyMe-modified Y2 (Table 4). Structural models (e.g., NMR solution structures) will be necessary to fully understand the molecular underpinnings of these irregular effects.
Table 4.
Discrimination of Mismatched DNA Targets by B2-series and Reference Strands.a
| DNA: 3′-CAC TBT ACG |
||||||
|---|---|---|---|---|---|---|
|
Tm [°C] |
ΔTm[°C] |
|||||
| ON | Sequence | B = | A | C | G | T |
| D1 | 5′-GTG ATA TGC | 29.5 | −16.5 | −9.5 | −17.0 | |
| W2 | 5′-GTG AWA TGC | 24.5 | −11.0 | +2.0 | −3.5 | |
| X2 | 5′-GTG AXA TGC | 33.5 | <−23.5 | −7.0 | −13.0 | |
| Y2 | 5′-GTG AYA TGC | 42.0 | −13.0 | −5.0 | −6.5 | |
| Z2 | 5′-GTG AZA TGC | 49.0 | −13.5 | −6.0 | −7.0 | |
| Q2 | 5′-GTG AQA TGC | 43.5 | −22.0 | −3.5 | −12.0 | |
| S2 | 5′-GTG ASA TGC | 32.5 | −11.5 | −9.0 | −8.5 | |
| V2 | 5′-GTG AVA TGC | 35.5 | −11.5 | +1.5 | −3.5 | |
| L2 9b | 5′-GTG ALA TGC | 43.5 | −12.5 | −5.5 | −3.5 | |
For conditions of thermal denaturation experiments, see Table 1. Tm-values of fully matched duplexes are shown in bold. ΔTm = change in Tm relative to fully matched DNA:DNA duplex.
The specificity of ONs with two modifications positioned as next-nearest neighbors (B6-series) was evaluated against DNA targets with a single central mismatched nucleotide opposite of the central 2′-deoxyadenosine residue (Table 5). Interestingly, this probe design generally results in improved mismatch discrimination relative to reference strand D2 with specificity decreasing in the following manner: Y6>X6>Z6~Q6>V6; A- and G-mismatches are particularly efficiently discriminated. Subsequent experiments will reveal if these ‘intercalator-capped fidelity cassettes’ can be used as general motifs to develop ONs with improved target specificity.
Table 5.
Discrimination of mismatched DNA targets by B6-series and reference strands.a
| DNA : 5′-GTG ABA ACG | ||||||
|---|---|---|---|---|---|---|
|
Tm [°C] |
ΔTm [°C] |
|||||
| ON | Sequence | B = | T | A | C | G |
| D2 | 3′-CAC TAT ACG | 29.5 | −17.0 | −15.5 | −9.0 | |
| X6 | 3′-CAC XAX ACG | 25.5 | <−15.5 | −14.0 | −14.5 | |
| Y6 | 3′-CAC YAY ACG | 43.5 | −24.0 | −17.0 | −14.0 | |
| Z6 | 3′-CAC ZAZ ACG | 47.0 | −19.5 | −13.0 | −11.0 | |
| Q6 | 3′-CAC QAQ ACG | 43.5 | −21.5 | −10.5 | −13.5 | |
| V6 | 3′-CAC VAV ACG | 37.0 | −14.5 | −13.5 | −11.0 | |
For conditions of thermal denaturation experiments, see Table 1. Tm-values of fully matched duplexes are shown in bold. ΔTm = change in Tm relative to fully matched DNA:DNA duplex.
Optical spectroscopy
Absorption and steady state fluorescence emission spectra of ONs modified with monomers Q/S/V/X/Y in the presence or absence of complementary DNA/RNA targets were recorded to gain additional insight into the binding modes of the pyrene moieties. Intercalation of pyrene moieties is known to induce bathochromic shifts of pyrene absorption peaks.38 In agreement with this, ONs modified with monomers Q/S/V/Y display significant bathochromic shifts upon hybridization with DNA targets (average Δλmax = 2.8-5.0 nm, respectively, Table 6; see also Fig. S4, and Tables S5 and S6). Hybridization with complementary RNA leads to smaller bathochromic shifts as intercalation is less favorable in the increasingly compressed A-type duplexes (average Δλmax = 2.0-4.5 nm, Table 6). O2′-Py-modified X1-X5 display distinctly smaller hybridization-induced bathochromic shifts than the other pyrene-functionalized ONs (average Δλmax ~0.6 nm, Table 6), which substantiates the trends from thermal denaturation studies suggesting that intercalation of the pyrene moiety is a less important binding mode for monomer X.
Table 6.
Average bathochromic shifts of pyrene absorption bands (λem ~350 nm) upon hybridization of pyrene-functionalized ONs with complementary DNA/RNA targets.a
| Average Δλmax (nm) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B= | X |
Y |
Q |
S |
V |
L9b |
||||||
| +DNA |
+RNA |
+DNA |
+RNA |
+DNA |
+RNA |
+DNA |
+RNA |
+DNA |
+RNA |
+DNA |
+RNA |
|
| 0.6±1.8 | 0.6±1.3 | 3.4±1.1 | 2.6±0.9 | 5.0±0.8 | 2.3±1.7 | 2.8±1.0 | 2.0±1.8 | 4.5±0.6 | 4.5±1.3 | 2.5±1.0 | 1.0±0.8 | |
Values are averages of measurements ‘performed for B1-B5 at 5 °C (Q/S/V), 7 °C (X/Y) or room temperature (L)9b. Buffer conditions are as for thermal denaturation experiments. “±” denotes standard deviation. For full data set, see Table S5 and S6.
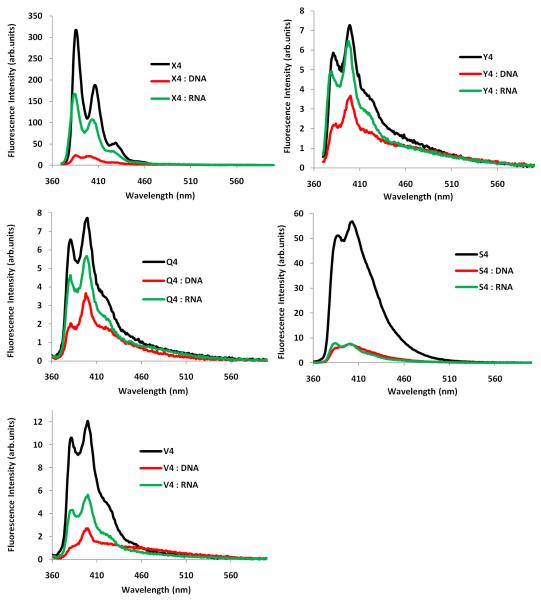
The steady-state fluorescence emission spectra (λex = 350 nm; λem = 360-600 nm; T = 5 °C) display the two expected vibronic bands I and III at λem = 382 ± 3 nm and 402 ± 3 nm, respectively, as well as a small shoulder at ~420 nm (Fig. 2). Hybridization of ONs modified with monomers Q/S/V/X/Y with complementary targets, and DNA in particular, results in reduced fluorescence emission intensity (see Fig. 2 for B4-series; see Figs. S5-S9 for all series). This trend corroborates pyrene intercalation as nucleobase moieties are known to quench pyrene fluorescence via photoinduced electron transfer (PET) with guanine and cytosine moieties being the strongest quenchers.39 In agreement with this, duplexes involving the Y/Q/V-series (intercalation is important binding mode) display far lower emission intensity than those involving the X-series (intercalation is less important binding mode). Sequence-specific deviations from general trends are observed in some cases (e.g., Q2/S1/S4/S5, see Figs. S7-S8), which likely reflects a trend-defying duplex geometry or positioning of the pyrene moiety. For additional discussion of fluorescence emission data, please see the Supporting Information.
Figure 2.
Fluorescence emission spectra of B4-series in presence or absence of complementary DNA/RNA (λex = 350 nm; T = 5 °C; [ON] = 1.0 μM). Note that different Y-axes are used.
On balance, the data from the thermal denaturation and optical spectroscopy studies (DNA-selectivity; mismatch discrimination; UV-vis; fluorescence) suggest that intercalation of the attached hydrocarbon is a possible binding mode for all studied monomers. Intercalation is least important for monomers W and X, while being a dominant binding mode for monomers Y, Z and Q.
CONCLUSION
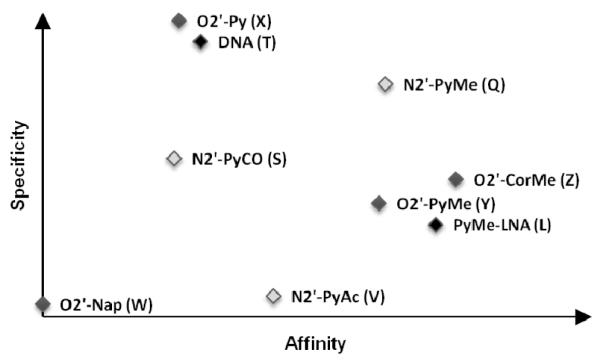
We have developed short routes toward a series of O2′-intercalator-functionalized uridine and N2′-intercalator-functionalized 2′-N-methyl-2′-aminouridine phosphoramidites. Thermal denaturation studies of correspondingly modified ONs reveal that even very conservative changes in linker chemistry, linker length or surface area of the intercalator, translate into markedly different DNA-hybridization characteristics (Fig. 3). We propose the following guidelines for design of high-affinity DNA-targeting monomers based on uridine or 2′-N-methyl-2′-aminouridine scaffolds: a) direct attachment of intercalators to the O2′-position of uridine should be avoided (e.g., monomer W and X), b) attachment of intercalators to the N2′-position of 2-N-methyl-2′-aminouridine via alkanoyl linkers should be avoided (e.g., monomer S and V), and c) attachment of large intercalators via a methylene linker is desirable (e.g., monomers Q/Y/Z). Thus, ONs modified with 2′-O-(pyren-1-yl)methyluridine monomer Y, 2′-O-(coronen-1-yl)methyluridine monomer Z, or 2′-N-(pyren-1-ylmethyl)-2′-N-methylaminouridine monomer Q display similar thermal affinity toward DNA complements (average ΔTm/mod between +8 and +12 °C) and higher target specificity than ONs modified with 2′-N-(pyren-1-yl)methyl-2′-amino-α-L-LNA benchmark monomer L (Fig. 3). The highly DNA-selective hybridization of these ONs, and of the doubly O2′-CH2Cor-modified Z6 in particular (ΔΔTm (DNA-RNA) = 31.0 °C), hints at interesting applications within DNA diagnostics. Straightforward access to these monomers will likely spur their use within nucleic acid based diagnostics, therapeutics, and material science.10-12,40-42 Studies along these lines are ongoing and will be reported shortly.
Figure 3.
Qualitative representation of DNA-hybridization characteristics (affinity vs specificity) of O2′-intercalator-functionalized uridine (light grey) and N2′-intercalator-functionalized 2′-N-methyl-2′-aminouridine (dark grey) monomers studied herein relative to reference monomers L and T (black).
EXPERIMENTAL SECTION
General Experimental Section
Unless otherwise noted, reagents and solvents were commercially available, of analytical grade and used without further purification. Petroleum ether of the distillation range 60-80°C was used. Solvents were dried over activated molecular sieves: acetonitrile and THF (3Å); CH2Cl2, 1,2-dichloroethane, N,N′-diisopropylethylamine and anhydrous DMSO (4Å). Water content of “anhydrous” solvents was verified on Karl-Fisher apparatus. Reactions were conducted under argon whenever anhydrous solvents were used. Reactions were monitored by TLC using silica gel coated plates with a fluorescence indicator (SiO2-60, F-254) which were visualized a) under UV light and/or b) by dipping in 5% conc. H2SO4 in absolute ethanol (v/v) followed by heating. Silica gel column chromatography was performed with Silica gel 60 (particle size 0.040–0.063 mm) using moderate pressure (pressure ball). Evaporation of solvents was carried out under reduced pressure at temperatures below 45 °C. After column chromatography, appropriate fractions were pooled, evaporated and dried at high vacuum for at least 12h to give the obtained products in high purity (>95%) as ascertained by 1D NMR techniques. Chemical shifts of 1H NMR (500 MHz), 13C NMR (125.6 MHz), and/or 31P NMR (121.5 MHz) are reported relative to deuterated solvent or other internal standards (80% phosphoric acid for 31P NMR). Exchangeable (ex) protons were detected by disappearance of signals upon D2O addition. Assignments of NMR spectra are based on 2D spectra (HSQC, COSY) and DEPT-spectra. Quaternary carbons are not assigned in 13C NMR but verified from HSQC and DEPT spectra (absence of signals). MALDI-HRMS spectra of compounds were recorded on a Q-TOF mass spectrometer using 2,5-dihydroxybenzoic acid (DHB) as a matrix and polyethylene glycol (PEG 600) as an internal calibration standard.
General protocol for coupling between 1 and phenols (ArOH) to prepare 2W/2X (description for ~1.33 mmol scale)
The appropriate phenol and 2,2′-anhydrouridine 117 were placed in a sealed pressure tube (specific quantities of substrates and reagents given below) and heated (165 °C for 2W; 175 °C for 2X) until analytical TLC indicated full conversion (~2h). The resulting crude was purified by silica gel column chromatography (2-4% MeOH in CH2Cl2, v/v) to afford nucleoside 2W/2X (yields specified below).
2′-O-(Napth-2-yl)uridine (2W)
A mixture of 2,2′-anhydrouridine 1 (1.00 g, 4.42 mmol) and 2-napthol (2.40 g, 22.1 mmol) were reacted and purified as described above to afford nucleoside 2W (0.41 g, 25%) as a light brown solid. Rf: 0.4 (10% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 393.1039 ([M+Na]+, C19H18N2O6·Na+, Calc. 393.1057); 1H NMR (DMSO-d6) δ 11.34 (s, 1H, ex, NH), 8.03 (1H, d, J = 8.1 Hz, H6), 7.82-7.85 (ap d, 2H, Nap), 7.73-7.75 (1H, d, J = 8.3 Hz, Nap), 7.44-7.48 (ap t, 1H, Nap), 7.41 (d, 1H, J = 2.5 Hz, Nap), 7.34-7.37 (ap t, 1H, Nap), 7.23-7.25 (dd, 1H, J = 9.1 Hz, J = 2.5 Hz, Nap), 6.14 (d, 1H, J = 4.9 Hz, H1′), 5.68 (d, 1H, J = 8.1 Hz, H5), 5.46 (d, 1H, ex, J = 6.4 Hz, 3′-OH), 5.25 (t, 1H, ex, J = 5.2 Hz, 5′-OH), 5.02 (ap t, 1H, H2′), 4.43-4.46 (m, 1H, H3′), 4.02-4.05 (m, 1H, H4′), 3.74-3.78 (m, 1H, H5′), 3.65-3.69 (m, 1H, H5′); 13C NMR (DMSO-d6) δ 162.9, 155.4, 150.5, 140.4 (C6), 133.9, 129.2 (Nap), 128.7, 127.4 (Nap), 126.6 (Nap), 126.4 (Nap), 123.8 (Nap), 118.9 (Nap), 108.9 (Nap), 102.0 (C5), 86.4 (C1′), 85.2 (C4′), 79.3 (C2′), 68.2 (C3′), 60.4 (C5′). While 2W is obtained in lower yield than via a recently published microwave-assisted approach (~50% yield), the current approach does not require access to microwave reactors. The 1H NMR data for 2W are in agreement with the literature report.18
2′-O-(Pyren-1-yl)uridine (2X)
A mixture of 2,2′-anhydrouridine 1 (0.30 g, 1.33 mmol) and 1-pyrenol19 (0.86 g, 3.97 mmol) were reacted and purified as described above to afford nucleoside 2X (0.26 g, 44 %) as a pale yellow solid. Rf: 0.4 (10% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 467.1217 ([M+Na]+, C25H20N2O6·Na+, Calc. 467.1214); 1H NMR (DMSO-d6) δ 11.36 (s, 1H, ex, NH), 8.45 (d, 1H, J = 9.3 Hz, Py), 8.20-8.24 (m, 3H, Py), 8.14 (d, 1H, J = 9.3 Hz, Py), 7.99-8.09 (m, 4H, H6, Py), 7.86 (1H, d, J = 8.5 Hz, Py), 6.35 (d, 1H, J = 4.6 Hz, H1′), 5.68 (d, 1H, J = 8.2 Hz, H5), 5.63 (br s, 1H, ex, 3′-OH), 5.30 (br s, 1H, ex, 5′-OH), 5.22-5.25 (ap t, 1H, H2′), 4.55-4.56 (m, 1H, H3′), 4.19-4.21 (m, 1H, H4′), 3.81-3.84 (ap d, 1H, H5′), 3.72-3.75 (ap d, 1H, H5′); 13C NMR (DMSO-d6) δ 162.9, 151.7, 150.5, 140.3 (C6), 131.0, 130.9, 127.1 (Py), 126.4 (Py), 125.6 (Py), 125.2, 125.0 (Py), 124.8 (Py), 124.5 (Py), 124.3 (Py), 123.9, 121.1 (Py), 120.1, 111.9 (Py), 102.0 (C5), 86.6 (C1′), 85.4 (C4′), 80.9 (C2′), 68.5 (C3′), 60.4 (C5′).
General protocol for coupling between 1 and arylmethyl alcohol (ArCH2OH) for the preparation of 2Y/2Z (description for ~44.2 mmol scale)
The appropriate aromatic alcohol (ArCH2OH), NaHCO3 and 1.0M BH3 in THF were placed in a pressure tube, suspended in anhydrous DMSO and stirred under an argon atmosphere at rt until effervescence ceased (~10 min). At this point, 2,2′-anhydrouridine 120 was added (specific quantities of substrates and reagents given below), the pressure tube was purged with argon and sealed, and the reaction was heated at ~160 °C until analytical TLC indicated full conversion (~3h). At this point, the reaction mixture was poured into water (200 mL), stirred for 30 min and diluted with EtOAc (500 mL). The organic phase was washed with water (4 × 200 mL), evaporated to dryness, and the resulting crude purified by silica gel column chromatography (2-4%, MeOH in CH2Cl2, v/v) to afford a residue, which was precipitated from cold acetone to obtain nucleoside 2 (yields specified below).
2′-O-(Pyren-1-yl-methyl)uridine (2Y)
2,2′-Anhydrouridine 1 (10.00 g, 44.2 mmol), pyren-1-ylmethanol23 (20.5 g, 88.4 mmol), NaHCO3 (0.73 g. 8.80 mmol), 1.0 M BH3 in THF (24.5 mL, 22.0 mmol) and anhydrous DMSO (40 mL) were mixed, reacted, worked up, and purified as described above to afford nucleoside 2Y (5.04 g, 25 %) as a white solid. Rf: 0.4 (10 % MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 458.1480 ([M]+, C26H22N2O6+, Calc. 458.1472); 1H NMR (DMSO-d6): δ 11.29 (s, 1H, ex, NH), 8.37-8.39 (d, 1H, J = 9.3 Hz, Py), 8.29-8.31 (m, 2H, Py), 8.24-8.26 (d, 1H, J = 7.7 Hz, Py), 8.17-8.19 (m, 3H, Py), 8.06-8.12 (m, 2H, Py), 7.82 (d, 1H, J = 8.4 Hz, H6), 6.04 (d, 1H, J = 5.1 Hz, H1′), 5.43-5.50 (m, 2H, H5, CH2Py), 5.37 (d, 1H, ex, J = 5.7 Hz, 3′-OH), 5.28-5.30 (d, 1H, J = 12.0 Hz, CH2Py), 5.11 (t, 1H, ex, J = 4.9 Hz, 5′-OH), 4.26-4.31 (m, 1H, H3′), 4.18-4.21 (m, 1H, H2′), 3.96-3.97 (m, 1H, H4′), 3.64-3.68 (m, 1H, H5′), 3.59-3.62 (m, 1H, H5′); 13C NMR (DMSO-d6) δ 162.9, 150.6, 140.1 (C6), 131.4, 130.7, 130.2, 128.7, 127.4 (Py), 127.3 (Py), 127.0 (Py), 126.2 (Py), 125.3 (Py), 124.5 (Py), 124.0 (Py), 123.8, 123.5 (Py), 101.7 (C5), 86.2 (C1′), 85.4 (C4′), 80.9 (C2′), 69.8 (CH2Py), 68.5 (C3′), 60.6 (C5′). 1H NMR data for nucleoside 2Y are in agreement with data where nucleoside 2Y was obtained via a different route.17 Full experimental details on the preparation and characterization of 2Y have, to our knowledge, not been previously published.
2′-O-(Coronen-1-yl-methyl)uridine (2Z)
2,2′-Anhydrouridine 1 (1.40g, 6.19 mmol), coronen-1-ylmethanol24 (4.08 g, 12.4 mmol), NaHCO3 (0.104 g. 1.24 mmol), 1.0 M BH3 in THF (3.5 mL, 3.1 mmol) and anhydrous DMSO (40 mL) were mixed, reacted, worked up and purified as described above with a minor modification. The precipitate that formed upon pouring the reaction mixture into water was collected by filtration, washed with water (3 × 100 mL) and purified by column chromatography to afford nucleoside 2Z (450 mg, 13 %) as a pale yellow solid. Rf: 0.4 (10 % MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 579.1537 ([M+Na]+, C34H24N2O6·Na+, Calc. 579.1532); 1H NMR (DMSO-d6) δ 11.30 (br d, ex, 1H, J = 1.9 Hz, NH), 9.13-9.15 (d, 1H, J = 8.7 Hz, Cor), 8.93-9.03 (m, 10H, Cor), 7.82 (d, 1H, J = 8.0 Hz, H6), 6.18 (d, 1H, J = 4.9 Hz, H1′), 5.85-5.88 (d, 1H, J = 12.1 Hz, CH2Cor), 5.67-5.69 (d, 1H, J = 12.1 Hz, CH2Cor), 5.52 (d, 1H, ex, J = 5.5 Hz, 3′-OH), 5.38 (dd, 1H, J = 8.0 Hz, 1.9 Hz, H5), 5.11 (ap t, 1H, ex, 5′-OH), 4.37-4.43 (m, 2H, H2′, H3′), 4.03-4.06 (m, 1H, H4′), 3.63-3.71 (m, 2H, H5′); 13C NMR (DMSO-d6) δ 162.9, 150.6, 140.1 (C6), 132.1, 128.2, 128.1, 127.9, 127.4, 126.5, 126.23 (Cor), 126.20 (Cor), 126.1 (Cor), 126.0 (Cor), 122.5 (Cor), 121.8, 121.5, 121.4, 121.23, 121.17, 101.7 (C5), 86.3 (C1′), 85.5 (C4′), 81.1 (C2′), 70.7 (CH2Cor), 68.6 (C3′), 60.6 (C5′).
General DMTr-protection protocol for the preparation of 3W-3Z (description for ~2.2 mmol scale)
The appropriate nucleoside 3 (specific quantities given below) was co-evaporated twice with anhydrous pyridine (15 mL) and redissolved in anhydrous pyridine. To this was added 4,4′-dimethoxytritylchloride (DMTrCl) and N,N-dimethyl-4-aminopyridine (DMAP), and the reaction mixture was stirred at rt until TLC indicated complete conversion (~14h). The reaction mixture was diluted with CH2Cl2 (70 mL) and the organic phase sequentially washed with water (2 × 70 mL) and sat. aq. NaHCO3 (2 × 100 mL). The organic phase was evaporated to near dryness and the resulting crude co-evaporated with absolute EtOH and toluene (2:1, v/v, 3 × 6 mL) and purified by silica gel column chromatography (0-5%, MeOH in CH2Cl2, v/v) to afford nucleoside 3 (yields specified below).
2′-O-(Napth-2-yl)-5′-O-(4,4′-dimethoxytrityl)uridine (3W)
Nucleoside 2W (150 mg, 0.40 mmol), DMTrCl (240 mg, 0.60 mmol) and DMAP (~6 mg) in anhydrous pyridine (7 mL) were mixed, reacted, worked up and purified as described above to afford nucleoside 3W (120 mg, 47%) as a pale yellow foam. Rf: 0.6 (5%, MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 695.2379 ([M+Na]+, C40H36N2O8·Na+, Calc. 695.2364); 1H NMR (DMSO-d6) δ 11.40 (d, 1H, ex, J = 2.1 Hz, NH), 7.83-7.86 (m, 3H, H6, Nap), 7.68 (d, 1H, J = 8.2 Hz, Nap), 7.24-7.45 (m, 13H, DMTr, Nap), 6.90-6.92 (d, 4H, J = 7.1 Hz, DMTr), 6.06 (d, 1H, J = 3.2 Hz, H1′), 5.51 (d, 1H, ex, J = 7.1 Hz, 3′-OH), 5.38-5.40 (m, 1H, H5), 5.12-5.14 (m, 1H, H2′), 4.51-4.55 (m, 1H, H3′), 4.14-4.17 (m, 1H, H4′), 3.75 (s, 6H, 2 × CH3O), 3.32-3.41 (m, 2H, H5′); 13C NMR (DMSO-d6) 162.9, 158.1, 155 162.9, 158.1, 155.5, 150.3, 144.6, 140.6 (C6), 135.4, 135.2, 133.9, 129.8 (Ar), 129.1 (Nap), 128.8, 127.9 (Ar), 127.7 (Ar), 127.5 (Nap), 126.8 (Ar), 126.6 (Nap), 126.4 (Ar), 123.8 (Ar), 119.0 (Ar), 113.2 (Ar), 109.0 (Ar), 101.8 (C5), 87.6 (C1′), 85.9, 82.6 (C4′), 79.1 (C2′), 68.5 (C3′), 62.7 (C5′), 55.0 (CH3O). The described protocol is similar to an independently developed and recently published protocol; 13C NMR data recorded in CDCl3 are in agreement with this report.43 NMR spectra recorded in DMSO-d6 have, to our knowledge, not been provided for this compound.
2′-O-(Pyren-1-yl)-5′-O-(4,4′-dimethoxytrityl)uridine (3X)
Nucleoside 2X (230 mg, 0.52 mmol), DMTrCl (0.30 g, 0.78 mmol) and DMAP (~9 mg) in anhydrous pyridine (8 mL) were mixed, reacted, worked up and purified as described above to afford nucleoside 3X (0.30 g, 78%) as a light yellow foam. Rf: 0.6 (5%, MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 769.2504 ([M+Na]+, C46H38N2O8·Na+, Calc. 769.2520); 1H NMR (DMSO-d6): δ 11.32 (s, 1H, ex, NH), 8.49 (d, 1H, J = 9.2 Hz, Py), 8.20-8.24 (m, 3H, Py), 8.14 (d, 1H, J = 9.2 Hz, Py), 8.08-8.10 (d, 1H, J = 9.1 Hz, Py), 7.99-8.04 (m, 2H, Py), 7.86-7.89 (m, 2H, H6, Py), 7.43-7.45 (m, 2H, DMTr), 7.24-7.36 (m, 7H, DMTr), 6.90-6.92 (m, 4H, DMTr), 6.25 (d, 1H, J = 3.2 Hz, H1′), 5.69 (d, 1H, ex, J = 6.8 Hz, 3′-OH), 5.38 (d, 1H, J = 8.2 Hz, H5), 5.35-5.37 (m, 1H, H2′), 4.61-4.65 (m, 1H, H3′), 4.34-4.37 (m, 1H, H4′), 3.74 (s, 6H, 2 × CH3O), 3.46-3.50 (m, 1H, H5′), 3.38-3.41 (m, 1H, H5′); 13C NMR (DMSO-d6) δ 162.9, 158.1, 151.7, 150.3, 144.6, 140.5 (C6), 135.4, 135.1, 131.1, 131.0, 129.78 (DMTr), 129.76 (DMTr), 127.9 (DMTr), 127.1 (Py), 126.8 (DMTr), 126.4 (Py), 125.6 (Py), 125.3, 125.1 (Py), 124.9, 124.5 (Py), 124.3 (Py), 124.0, 121.2 (Py), 120.1, 113.2 (DMTr), 112.3 (Py), 101.7 (C5), 87.8 (C1′), 85.9, 82.7 (C4′), 80.7 (C2′), 68.7 (C3′), 62.7 (C5′), 55.0 (CH3O).
2′-O-(Pyren-1-yl-methyl)-5′-O-(4,4′-dimethoxytrityl)uridine (3Y)
Nucleoside 2Y (1.02 g, 2.20 mmol), DMTrCl (1.29 g, 3.30 mmol) and DMAP (~18 mg) in anhydrous pyridine (20 mL) were mixed, reacted, worked up and purified as described above to afford 3Y (1.20 g, 72 %) as pale yellow foam. Rf: 0.6 (5%, MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 783.2698 ([M+Na]+, C47H40N2O8·8Na+, Calc. 783.2677); 1H NMR (DMSO-d6) δ 11.36 (d, 1H, ex, J = 1.9 Hz, NH), 8.41-8.43 (d, 1H, J = 9.3 Hz, Py), 8.30-8.32 (m, 2H, Py), 8.14-8.25 (m, 5H, Py), 8.07-8.10 (t, 1H, J = 7.7 Hz, Py), 7.63 (d, 1H, J = 8.1 Hz, H6), 7.17-7.34 (m, 9H, DMTr), 6.82-6.86 (m, 4H, DMTr), 6.02 (d, 1H, J = 3.9 Hz, H1′), 5.48-5.50 (d, 1H, J = 12.1 Hz, CH2Py), 5.45 (d, 1H, ex, J = 6.3 Hz, 3′-OH), 5.36-5.38 (d, 1H, J = 12.1 Hz, CH2Py), 5.13 (dd, 1H, J = 8.1 Hz, 1.9 Hz, H5), 4.34-4.38 (m, 1H, H3′), 4.21-4.24 (m, 1H, H2′), 4.05-4.09 (m, 1H, H4′), 3.71 (s, 3H, CH3O), 3.69 (s, 3H, CH3O), 3.20-3.24 (m, 2H, H5′); C NMR (DMSO-d6) δ 162.8, 158.08, 158.06, 150.4, 144.5, 140.0 (C6), 135.3, 135.0, 131.3, 130.7, 130.2, 129.71 (DMTr), 129.66 (DMTr), 128.7, 127.8 (DMTr), 127.6 (DMTr), 127.4 (Py), 127.3 (Py), 127.0 (Py), 126.7 (DMTr), 126.2 (Py), 125.3 (Py), 124.5 (Py), 124.0, 123.8, 123.4 (Py), 113.20 (DMTr), 113.17 (DMTr), 101.4 (C5), 87.1 (C1′), 85.9, 83.1 (C4′), 80.6 (C2′), 69.9 (CH2Py), 68.7 (C3′), 62.8 (C5′), 55.0 (CH3O). The protocol and 1H NMR data recorded in CDCl3 for 3Y are in agreement with those from a related protocol.17 Full experimental details on the preparation and characterization of 3Y have not been previously reported.
2′-O-(Coronen-1-yl-methyl)-5′-O-(4,4′-dimethoxytrityl)uridine (3Z)
Nucleoside 2Z (250 mg, 0.45 mmol), DMTrCl (262 mg, 0.67 mmol) and DMAP (~15 mg) in anhydrous pyridine (6 mL) were mixed, reacted, worked up and purified as described above to afford nucleoside 3Z (245 mg 63%) as a yellow foam. Rf: 0.6 (5%, MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 881.2824 ([M+Na]+, C55H42N2O8·Na+, Calc. 881.2839); 1H NMR (DMSO-d6) δ 11.40 (br d, 1H, ex, J = 1.9 Hz, NH), 9.14-9.16 (d, 1H, J = 8.8 Hz, Cor), 8.96-9.02 (m, 9H, Cor), 8.88-8.90 (d, 1H, J = 8.5 Hz, Cor), 7.63 (d, 1H, J = 8.1 Hz, H6), 7.29-7.31 (d, 2H, J = 7.4 Hz, DMTr), 7.11-7.22 (m, 7H, DMTr), 6.74-6.79 (m, 4H, DMTr), 6.18 (d, 1H, J = 4.1 Hz, H1′), 5.87-5.90 (d, 1H, J = 12.6 Hz, CH2Cor), 5.75-5.78 (d, 1H, J = 12.6 Hz, CH2Cor), 5.59 (d, 1H, ex, J = 6.3 Hz, 3′-OH), 5.06 (dd, 1H, J = 8.1 Hz, 1.9 Hz, H5), 4.46-4.50 (m, 1H, H3′), 4.40-4.43 (m, 1H, H2′), 4.15-4.18 (m, 1H, H4′), 3.63 (s, 3H, CH3O), 3.58 (s, 3H, CH3O), 3.32-3.34 (m, 1H, H5′), 3.25-3.27 (m, 1H, H5′); 13C NMR (DMSO-d6) δ 162.8, 158.02, 157.97, 150.4, 144.4, 140.0 (C6), 135.3, 135.0, 132.2, 129.7 (DMTr), 129.6 (DMTr), 128.3, 128.2, 128.0, 127.7 (DMTr), 127.59 (DMTr), 127.56, 126.61 (DMTr), 126.59, 126.41 (Cor), 126.36 (Cor), 126.3 (Cor), 126.23 (Cor), 126.21, 126.19, 126.1 (Cor), 122.6 (Cor), 121.9, 121.6, 121.5, 121.4, 121.35, 121.28, 113.13 (DMTr), 113.09 (DMTr), 101.4 (C5), 87.1 (C1′), 85.9, 83.2 (C4′), 80.7 (C2′), 70.7 (CH2Cor), 68.8 (C3′), 62.8 (C5′), 54.9 (CH3O), 54.8 (CH3O).
General phosphitylation protocol for the preparation of 4W-4Z (description for ~1 mmol scale)
The appropriate nucleoside 3 (specific quantities of substrates and reagents given below) was co-evaporated with anhydrous 1,2-dicholoroethane (4 mL) and redissolved in anhydrous CH2Cl2. To this was added N,N-diisopropylethylamine (DIPEA) and 2-cyanoethyl-N,N-diisopropylchlorophosporamidite (PCl-reagent) and the reaction mixture was stirred at rt until TLC indicated complete conversion (~3h), whereupon abs. EtOH (2 mL) and CH2Cl2 (20 mL) were sequentially added to the solution. The organic phase was washed with sat. aq. NaHCO3 (10 mL), evaporated to near dryness, and the resulting residue purified by silica gel column chromatography (40-70% EtOAc in petroleum ether, v/v) to afford the corresponding phosphoramidite 4 (yields specified below).
2′-O-(Napth-2-yl)-3′-O-(N,N-diisopropylamino-2-cyanoethoxyphosphinyl)-5′-O-(4,4′-dimethoxytrityl)uridine (4W)
Nucleoside 3W (100 mg, 0.15 mmol), DIPEA (106 μL, 0.59 mmol) and PCl-reagent (66 μL, 0.23 mmol) in anhydrous CH2Cl2 (1.5 mL) were mixed, reacted, worked up and purified as described above to afford phosphoramidite 4W (95 mg, 74 %) as a white foam. Rf: 0.8 (5% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 895.3462 ([M+Na]+, C49H53N4O9P·Na+, Calc. 895.3448); 31P NMR (CDCl3) δ 151.0, 150.9. The reaction yield compares favorably with an independently developed protocol utilizing 2-cyanoethyl N,N,N’,N’-tetraisopropylphosphorodiamidite (PN2-reagent) and 1H-tetrazole as phosphitylation reagent and activator, respectively (~61% yield).40 The 31P NMR data are in agreement with literature data.43
3′-O-(N,N-Diisopropylamino-2-cyanoethoxyphosphinyl)-2′-O-(pyren-1-yl)-5′-O-(4,4′-dimethoxytrityl)uridine (4X)
Nucleoside 3X (0.28 g, 0.38 mmol), DIPEA (268 μL, 1.50 mmol) and PCl-reagent (167 μL, 0.75 mmol) in anhydrous CH2Cl2 (2.5 mL) were mixed, reacted, worked up and purified as described above to afford phosphoramidite 4X (0.27 g, 76 %) as a white foam. Rf: 0.8 (5% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 969.3608 ([M+Na]+, C55H55N4O9P·Na+, Calc. 969.3604); 31P NMR (CDCl3) δ 149.8, 149.4.
3′-O-(N,N-Diisopropylamino-2-cyanoethoxyphosphinyl)-2′-O-(pyren-1-yl-methyl)-5′-O-(4,4′-dimethoxytrityl)uridine (4Y)
Nucleoside 3Y (0.58 g, 0.76 mmol), DIPEA (0.53 mL, 3.05 mmol) and PCl-reagent (340 μL, 1.53 mmol) in anhydrous CH2Cl2 (5 mL) were mixed, reacted, worked up and purified as described above to afford phosphoramidite 4Y (0.56 g, 76 %) as a white foam. Rf: 0.8 (5% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 983.3767 ([M+Na]+, C56H57N4O9P·Na+, Calc. 983.3761); 31P NMR (CDCl3) δ 150.3, 150.2. The observed reaction yield and 31P NMR data are comparable to the reported protocol that utilizes PN2-reagent and 1H-tetrazole as phosphitylation reagent and activator, respectively.14 HRMS data have not been previously reported for this compound.
2′-O-(Coronen-1-yl-methyl)-3′-O-(N,N-diisopropylamino-2-cyanoethoxyphosphinyl)-5′-O-(4,4′-dimethoxytrityl)uridine (4Z)
Nucleoside 3Z (240 mg, 0.28 mmol), DIPEA (200 μL, 1.11 mmol) and PCl-reagent (125 μL, 0.56 mmol) in anhydrous CH2Cl2 (6 mL) were mixed, reacted, worked up and purified as described above to afford phosphoramidite 4Z (230 mg, 78 %) as a light yellow foam. Rf: 0.8 (5% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 1081.3864 ([M+Na]+, C64H59N4O9P·Na+, Calc. 1081.3917); 31P NMR (CDCl3) δ 150.2.
2′-Amino-2′-deoxy-2′-N-methyl-2′-N-(pyren-1-yl-methyl)-5′-O-(4,4′-dimethoxytrityl)uridine (6Q)
Known nucleoside 527 (200 mg, 0.36 mmol) was co-evaporated with anhydrous 1,2-dichloroethane (2 × 4 mL) and redissolved in anhydrous THF (5 mL). Pyrene-1-ylmethylchloride (205 mg, 0.37 mmol) and triethylamine (0.52 mL, 3.73 mmol) were added and the reaction mixture was heated at reflux for two days, whereupon the solvent was evaporated off. The crude residue was taken up in a mixture CHCl3 and sat. aq. NaHCO3 (50 mL, 3:2, v/v) and the layers were separated. The aqueous phase was extracted with CHCl3 (2 × 20 mL) and the combined organic phase was evaporated to dryness. The resulting residue was purified by silica gel column chromatography (0-1.25 % MeOH/CH2Cl2, v/v) to afford nucleoside 6Q as a yellow foam (129 mg, 46 %). Rf: 0.5 (60%, EtOAc in petroleum ether, v/v); MALDI-HRMS m/z 774.3156 ([M+H]+ C48H43N3O7·H+ 1, Calc 774.3174); 1H NMR (DMSO-d6) δ 11.41 (d, 1H, ex, J = 1.7 Hz, NH), 8.50 (d, 1H, J = 9.1 Hz, Py), 8.01-8.29 (m, 8H, Py), 7.63 (d, 1H, J = 8.2 Hz, H6), 7.20-7.38 (m, 9H, DMTr), 6.85-6.89 (m, 4H, DMTr), 6.43 (d, 1H, J = 8.2 Hz, H1′), 5.56 (d, 1H, ex, J = 5.2 Hz, 3′-OH), 5.43 (dd, 1H, J = 8.3 Hz, 1.7 Hz, H5), 4.41-4.50 (m, 3H, CH2Py, H3′), 4.06-4.08 (m, 1H, H4′), 3.71 (s, 3H, CH3O), 3.70 (s, 3H, CH3O), 3.44-3.48 (dd, 1H, J = 8.3 Hz, 8.1 Hz, H2′), 3.28-3.31 (m, 1H, H5′, overlap with H2O), 3.16-3.20 (dd, 1H, J = 10.6 Hz, 3.6 Hz, H5′), 2.33 (s, 3H, CH3N); 13C NMR (DMSO-d6) δ 162.7, 158.08, 158.07, 150.6, 144.5, 140.2 (C6), 135.4, 135.1, 132.7, 130.7, 130.3, 130.2, 129.71 (DMTr), 129.67 (DMTr), 129.2, 128.0 (Py), 127.8 (DMTr), 127.6 (DMTr), 127.3 (Py), 126.9 (Py), 126.8 (Py), 126.7 (DMTr), 126.1 (Py), 125.01, 124.98 (Py), 124.4 (Py), 124.1, 124.0 (Py), 123.9, 113.20 (DMTr), 113.17 (DMTr), 102.1 (C5), 85.9, 85.1 (C4′), 83.4 (C1′), 71.3 (C3′), 67.8 (C2′), 64.1 (C5′), 57.4 (CH2Py), 55.0 (CH3O), 38.6 (CH3N). 13C NMR data recorded in CDCl3 are in agreement with literature reports where 6Q was obtained via a different synthetic route.15 Full experimental details on the preparation and characterization of 6Q have not been previously published.
2′-Amino-2′-deoxy-2′-N-methyl-2′-N-(pyren-1-yl-carbonyl)-5′-O-(4,4′-dimethoxytrityl)uridine (6S)
Nucleoside 527 (150 mg, 0.27 mmol) was co-evaporated with anhydrous 1,2-dichloroethane (2 × 5 mL), dissolved in anhydrous DMF (4.5 mL) and added to a pre-stirred (1h at rt) solution of 1-pyrenecarboxylic acid (100 mg, 0.40 mmol), O-(7-azabenzotriazole-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HATU, 125 mg, 0.32 mmol) and DIPEA (0.12 mL, 0.70 mmol) in anhydrous DMF (4.5 mL). The reaction mixture was stirred for 17 h, whereupon it was diluted with EtOAc (50 mL) and sequentially washed with sat. aq. NaHCO3 (20 mL) and H2O (2 × 20 mL). The organic phase was evaporated to dryness and the resulting residue purified by silica gel column chromatography (0-2% MeOH in CH2Cl2, v/v) to afford nucleoside 6S as a white foam (164 mg, 78 %). Rf = 0.4 (5% MeOH in CH2Cl2, v/v). MALDI-HRMS m/z 788.2985 ([M+H]+, C48H41N3O8·H+, calc. 788.2966); 1H NMR (CDCl3) δ 8.65 (br s, 1H, NH, ex), 7.97-8.28 (m, 9H, Py), 7.85 (d, 1H, J = 8.3 Hz, H6), 7.20-7.44 (m, 9H, DMTr), 6.80-6.90 (m, 4H, DMTr), 6.78 (d, 1H, J = 6.0 Hz, H1′), 5.42-5.48 (m, 1H, H5), 4.80-4.90 (m, 2H, H2′, H3′), 4.28-4.43 (m, 1H, H4′), 3.77 (s, 6H, CH3O), 3.48-3.63 (m, 2H, H5′), 2.98 (s, 3H, NCH3), traces of a second rotamer are observed; 13 C NMR (CDCl3) δ 174.4, 162.9, 159.0, 150.5, 144.5, 140.0 (C6), 135.6, 135.5, 132.3, 131.4, 131.0, 130.44 (DMTr), 130.42 (DMTr), 129.4 (Py), 128.7 (Py), 128.5 (DMTr), 128.3 (DMTr), 127.4, 126.7 (Py), 126.1 (Py), 126.0 (Py), 125.0 (Py), 124.8, 124.7, 124.3 (Py), 113.6 (DMTr), 113.3 (DMTr), 103.2 (C5), 87.3, 86.6 (C4′), 85.3 (C1′), 71.8 (C3′/C2′), 65.8 (C2′/C3′), 63.0 (C5′), 55.5 (CH3O), 38.5 (NCH3). A trace impurity of grease was observed at 29.9 ppm.
2′-Amino-2′-deoxy-2′-N-methyl-2′-N-(pyren-1-yl-methylcarbonyl)-5′-O-(4,4′-dimethoxytrityl)uridine (6V)
Known nucleoside 527 (158 mg, 0.28 mmol) was co-evaporated with 1,2-dichloroethane (2 × 5 mL) and subsequently dissolved in anhydrous CH2Cl2 (8 mL). To this was added 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide hydrochloride (EDC·HCl, 73 mg, 0.38 mmol) and pyrene-1-ylacetic acid (108 mg, 0.41 mmol). The reaction mixture was stirred under argon at rt for 3h whereupon the reaction mixture was diluted with CH2Cl2 (30 mL) and sequentially washed with sat. aq. NaHCO3 (20 mL) and H2O (3 × 15 mL). The organic phase was evaporated to dryness and the resulting residue was purified by silica gel column chromatography (0-3 % i-PrOH/CH2Cl2, v/v) to afford a rotameric mixture (2:5 by 1HNMR) of nucleoside 6V (187 mg, 83 %) as a brown foam. Rf: 0.5 (5%, i-PrOH in CH2Cl2, v/v); MALDI-HRMS m/z 801.3078 ([M]+, C49H43N3O8, calc. 801.3045); 1H NMR (DMSO-d6) δ 11.52 (d, 1H, J = 1.7 Hz, NH(A)), 11.46 (d, 0.4H, J = 1.7 Hz, NH(B)), 8.05-8.31 (m, 11.2H, Py(A) + Py(B)), 7.89 (d, 1H, J = 7.8 Hz, Py(A)), 7.82 (d, 0.4H, J = 7.8 Hz, Py(B)), 7.67-7.70 (m, 1.4H, H6(A) + H6(B)), 7.13-7.43 (m, 12.6H, DMT(A+B)), 6.78-6.87 (m, 5.6H, DMT(A+B)), 6.42 (d, 1H, J = 8.0 Hz, H1′(A)), 6.31 (d, 0.4H, J = 5.5 Hz, H1′(B)), 5.92 (d, 0.4H, ex, J = 4.9 Hz, 3′-OH(B)), 5.76 (d, 1H, ex, J = 4.9 Hz, 3′-OH(A)), 5.38 (dd, 1H, J = 8.2 Hz, 1.7 Hz, H5(A)), 5.33 (dd, 0.4H, J = 8.2 Hz, 1.7 Hz, H5(B)), 5.11-5.17 (m, 1H, H2′(A)), 4.80-4.86 (m, 0.4H, H2′(B)), 4.58-4.63 (d, 1H, J = 16.5 Hz, CH2Py(A)), 4.51-4.56 (d, 0.4H, J = 16.5 Hz, CH2Py(B)), 4.37-4.49 (m, 2.4H, 1 x CH2Py(A), H3′(A), H3′(B)), 4.30-4.37 (d, 0.4H, J = 16.5 Hz, CH2Py(B)), 4.10-4.16 (m, 1.4H, H4′(A) + H4′(B)), 3.68 (s, 2.4H, CH3O(B)), 3.64 (s, 3H, CH3O(A)), 3.62 (s, 3H, CH3O(A)), 3.35-3.37 (m, 0.4H, H5′(B)), 3.32 (s, 3H, CH3N(A)), 3.29-3.32 (m, 1H, H5′(A), overlap with H2O), 3.23-3.27 (dd, 0.4H, J = 10.6 Hz, 2.6 Hz, H5′(B)), 3.15-3.19 (dd, 1H, J = 10.6 Hz, 2.6 Hz, H5′(A)), 3.03 (s, 1.2H, CH3N(B)); 13C NMR (DMSO-d6) δ 172.2, 172.1, 162.81, 162.78, 158.06, 158.03, 158.02, 150.5, 150.4, 144.6, 144.3, 140.2 (C6(B)), 140.1 (C6(A)), 135.4, 135.3, 135.2, 135.0, 130.7, 130.62, 130.56, 130.3, 130.2, 129.71 (DMTr), 129.69 (DMTr), 129.1, 128.1 (Py-CHA), 127.9 (Py-CHB), 127.8 (DMTr), 127.73 (DMTr), 127.70 (DMTr), 127.3 (Py), 127.14 (Py), 127.10 (Py), 126.73 (Py), 126.66 (DMTr), 126.1 (Py), 125.1 (Py), 125.0 (Py), 124.9 (Py), 124.5 (Py), 124.1, 124.0, 123.85 (Py), 123.76 (Py), 113.2 (DMTr), 113.1 (DMTr), 102.2 (C5(A)), 102.0 (C5(B)), 85.9, 85.7, 85.5 (C1′(B)), 85.2 (C4′(A)), 84.8 (C4′(B)), 83.2 (C1′(A)), 70.5 (C3′(A)), 69.3 (C3′(B)), 63.8 (C5′(A)), 63.7 (C5′(B)), 62.1 (C2′(B)), 59.0 (C2′(A)), 54.92 (CH3O(B)), 54.87 (CH3O(B)), 54.85 (CH3O(A)), 54.82 (CH3O(A)), 38.1 (CH2Py(A)), 37.8 (CH2-Py(B)), 34.2 (CH3N(A)), 31.4 (CH3N(B)).
2′-Amino-2′-deoxy-2′-N-methyl-2′-N-(pyren-1-yl-methyl)-3′-O-(N,N-diisopropylamino-2-cyanoethoxyphosphinyl)-5′-O-(4,4′-dimethoxytrityl)uridine (7Q)
Nucleoside 6Q (135 mg, 0.18 mmol) was co-evaporated with CH3CN (2 × 4 mL) and redissolved in anhydrous CH3CN (2.5 mL). To this was added DIPEA (153 μL, 0.87 mmol) and PCl-reagent (78 μL, 0.35 mmol). The reaction mixture was stirred at rt for 4h, whereupon it was cooled on an ice bath and abs. EtOH (3 mL) was added. The solvent was evaporated off and the resulting residue purified by silica gel column chromatography (0-40% EtOAc in petroleum ether, v/v; column built in 0.5 % Et3N) to afford nucleoside 6Q as a white foam (97 mg, 57 %). Rf: 0.3 (40% EtOAc in petroleum ether, v/v); MALDI-HRMS m/z 996.4046 ([M+Na]+, C57H60N5O8P·Na+, Calc. 996.4077); 31P NMR (CDCl3) δ 151.0, 149.8. The 31P NMR data are in general agreement with literature data.15
2′-Amino-2′-deoxy-2′-N-methyl-3′-O-(N,N-diisopropylamino-2-cyanoethoxyphosphinyl)-2′-N-(pyren-1-yl-carbonyl)-5′-O-(4,4′-dimethoxytrityl)uridine (7S)
Nucleoside 6S (219 mg, 0.28 mmol) was co-evaporated with anhydrous 1,2-dicholoroethane (2 × 2 mL) and redissolved in anhydrous CH2Cl2 (2 mL). To this was added DIPEA (58 μL, 0.33 mmol) followed by dropwise addition of PCl-reagent (74 μL, 0.33 mmol). After stirring at rt for 2h, CH2Cl2 (10 mL) was added and the reaction mixture stirred for additional 10 min whereupon the solvent was evaporated under reduced pressure. The resulting residue was purified by silica gel column chromatography (1st column: 0-40 % EtOAc in petroleum ether, v/v; 2nd column: 0-4 % MeOH in CH2Cl2, v/v) to afford a rotameric mixture of phosphoramidite 7S as a white foam (138 mg, 49 %). Rf: 0.8 (5% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 1010.3865 ([M+Na]+, C57H58N5O9P·Na+, Calc. 1010.3870); 31P NMR (CDCl3) δ 151.8, 151.2, 150.8.
2′-Amino-2′-deoxy-2′-N-methyl-3′-O-(N,N-diisopropylamino-2-cyanoethoxyphosphinyl)-2′-N-(pyren-1-yl-methylcarbonyl)-5′-O-(4,4′-dimethoxytrityl)uridine (7V)
Nucleoside 6V (0.30 g, 0.37 mmol) was co-evaporated with anhydrous 1,2-dicholoroethane (2 × 3 mL) and redissolved in anhydrous CH2Cl2 (4 mL). To this was added DIPEA (0.32 mL, 1.84 mmol) followed by dropwise addition of PCl-reagent (0.16 mL, 0.74 mmol). After stirring for 1.5 h at rt, CH2Cl2 (10 mL) was added and the reaction mixture stirred for additional 10 min whereupon the solvent was evaporated under reduced pressure. The resulting residue was purified by silica gel column chromatography (0-60 % EtOAc in petroleum ether, v/v) to afford phosphoramidite 7V as a bright yellow foam (153 mg, 42 %). Rf: 0.4 (60% EtOAc in petroleum ether, v/v); MALDI-HRMS m/z 1024.4037 ([M+Na]+, C58H60N5O9P·Na+, Calc. 1024.4026); 31P NMR (CDCl3) δ 150.6, 150.5.
Synthesis and purification of ONs
Synthesis of modified oligodeoxyribonucleotides (ONs) was performed on an DNA synthesizer using 0.2 μmol scale succinyl linked LCAA-CPG (long chain alkyl amine controlled pore glass) columns with a pore size of 500Å. Standard protocols for incorporation of DNA phosphoramidites were used. A ~50-fold molar excess of modified phosphoramidites in anhydrous acetonitrile (at 0.05 M) was used during hand-couplings (performed to conserve material) except with 4Z (~70-fold molar excess in anhydrous CH2Cl2, at 0.07M). Moreover, extended oxidation (45s) and coupling times were used (0.01 M 4,5-dicyanoimidazole as activator, 15 min for monomers V/W/Y, 35 min for monomer Z; 0.01 M 5-(bis-3,5-trifluoromethylphenyl)-1H-tetrazole [Activator 42], 15 min for monomers Q/S/V). Cleavage from solid support and removal of protecting groups was accomplished upon treatment with 32% aq. ammonia (55 °C, 24 h). Purification of all modified ONs was performed to minimum 80% purity using either of two methods: a) overall synthesis yield >80%: cleavage of DMT using 80% aq. AcOH, followed by precipitation from acetone (−18 °C for 12-16 h) and washing with acetone, or b) overall synthesis yield <80%: purification of ONs by RP-HPLC as described below, followed by detritylation and precipitation as outlined under “a”.
Purification of the crude ONs was performed on a HPLC system equipped with an XTerra MS C18 pre-column (10 μm, 7.8 × 10 mm) and a XTerra MS C18 column (10μm, 7.8 × 150 mm) using the representative gradient protocol depicted in Table S1. The identity of synthesized ONs was established through MALDI-MS/MS analysis (Table S2-S3) recorded in positive ions mode on a Quadrupole Time-Of-Flight Tandem Mass Spectrometer equipped with a MALDI source using anthranilic acid as a matrix, while purity (>80%) was verified by RP-HPLC running in analytical mode.
Thermal Denaturation Studies
Concentrations of ONs were estimated using the following extinction coefficients for DNA (OD/μmol): G (12.01), A (15.20), T (8.40), C (7.05); for RNA (OD/μmol): G (13.70), A (15.40), U (10.00), C (9.00); for fluorophores (OD/μmol): naphthalene (3.75)18, pyrene (22.4)10, and coronene (36.0).44 Each strand was thoroughly mixed and denatured by heating to 70-85 °C followed by cooling to the starting temperature of the experiment. Quartz optical cells with a path length of 1.0 cm were used. Thermal denaturation temperatures (Tm values [°C]) of duplexes (1.0 μM final concentration of each strand) were measured on a UV/VIS spectrophotometer equipped with a 12-cell Peltier temperature controller and determined as the maximum of the first derivative of the thermal denaturation curve (A260 vs. T) recorded in medium salt buffer (Tm buffer: 100 mM NaCl, 0.1 mM EDTA, and pH 7.0 adjusted with 10 mM Na2HPO4 and 5 mM Na2HPO4). The temperature of the denaturation experiments ranged from at least 15 °C below Tm to 20 °C above Tm (although not below 1°C). A temperature ramp of 0.5 °C/min was used in all experiments. Reported Tm-values are averages of two experiments within ± 1.0 °C.
Steady-state fluorescence emission spectra
Steady-state fluorescence emission spectra of ONs modified with pyrene-functionalized monomers Q/S/V/X/Y and the corresponding duplexes with complementary DNA/RNA targets were recorded in non-deoxygenated thermal denaturation buffer (each strand 1.0 μM) and obtained as an average of five scans using an excitation wavelength of λex = 350 nm, excitation slit 5.0 nm, emission slit 2.5 nm and a scan speed of 600 nm/min. Experiments were determined at 5 °C to ascertain maximal hybridization of probes to DNA/RNA targets. Solutions were heated to 80 °C followed by cooling to 5 °C over 10 min.
Supplementary Material
ACKNOWLEDGEMENT
P.J.H. appreciates support from Award Number R01 GM088697 from the National Institute of General Medical Sciences, National Institutes of Health; Institute of Translational Health Sciences (ITHS) (supported by grants UL1 RR025014, KL2 RR025015, and TL1 RR025016 from the NIH National Center for Research Resources); NIH Grant Number P20 RR016454 from the INBRE Program of the National Center for Research Resources; Idaho NSF EPSCoR and a Univ. Idaho Research Office and Research Council Seed Grant. P.N and T.B.J appreciate support from the The Danish National Research Foundation. We thank Dr. Lee Deobald and Prof. Andrzej J. Paszczynski (EBI Murdock Mass Spectrometry Center) for mass spectrometric analyses. Input from Todd Pankratz, Joanna Hawryluk and Johanna Root during preliminary phases of this study and scholarships from the National Science Foundation under award number 0648202 and the Department of Defense ASSURE (Awards to Stimulate and Support Undergraduate Research Experiences) Program supporting J.H and J.R. are appreciated.
Footnotes
Supporting information available: additional discussion regarding formation of cyclic N2′,O3′-hemiaminal ether (Scheme S1); representative RP-HPLC protocol (Table S1); MALDI-MS of ONs (Tables S2 and S3); representative thermal denaturation curves (Figures S1 and S2); discussion of RNA mismatch data for B2-series (Table S4 and Figure S3); absorption data (Figure S4 and Tables S5 and S6) and steady-state fluorescence emission spectra (Figure S5-S9) for modified ONs; additional discussion of fluorescence emission spectra; NMR spectra of compounds 2-7. This material is available free of charge via the Internet at http://pubs.acs.org/
REFERENCES
- (1).Bell NM, Micklefield J. ChemBioChem. 2009;10:2691–2703. doi: 10.1002/cbic.200900341. [DOI] [PubMed] [Google Scholar]
- (2).Duca M, Vekhoff P, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).(a) Bennett FC, Swayze EE. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]; (b) Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- (4).(a) Juskowiak B. Anal. Bioanal. Chem. 2011;399:3157–3176. doi: 10.1007/s00216-010-4304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dodd DW, Hudson RHE. Mini-Rev. Org. Chem. 2009;6:378–391. [Google Scholar]; (c) Asseline U. Curr. Org. Chem. 2006;10:491–518. [Google Scholar]
- (5).(a) Malinovskii VL, Wenger D, Haner R. Chem. Soc. Rev. 2010;39:410–422. doi: 10.1039/b910030j. [DOI] [PubMed] [Google Scholar]; (b) Filichev VV, Pedersen EB, Begley Tadhg P. Wiley Encyclopedia of Chemical Biology. WILEY-VCH Verlag GmbH & Co.; Weinheim: 2009. pp. 493–524. [Google Scholar]; (c) Teo YN, James N, Wilson JN, Kool ET. J. Am. Chem. Soc. 2009;131:3923–3933. doi: 10.1021/ja805502k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Østergaard ME, Hrdlicka PJ. Chem. Soc. Rev. 2011 doi: 10.1039/C1CS15014F. [Google Scholar]
- (7).(a) Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54:3607–3630. [Google Scholar]; (b) Obika S, Uneda T, Sugimoto T, Nanbu D, Minami T, Doi T, Imanishi T. Bioorg. Med. Chem. 2001;9:1001–1011. doi: 10.1016/s0968-0896(00)00325-4. [DOI] [PubMed] [Google Scholar]; (c) Kaur H, Babu BR, Maiti S. Chem. Rev. 2007;107:4672–4697. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- (8).Sørensen MD, Kværnø L, Bryld T, Håkansson AE, Verbeure B, Gaubert G, Herdewijn P, Wengel J. J. Am. Chem. Soc. 2002;124:2164–2176. doi: 10.1021/ja0168763. [DOI] [PubMed] [Google Scholar]
- (9).(a) Andersen NK, Wengel J, Hrdlicka PJ. Nucleosides Nucleotides Nucleic Acids. 2007;26:1415–1417. doi: 10.1080/15257770701539153. [DOI] [PubMed] [Google Scholar]; (b) Kumar TS, Madsen AS, Østergaard ME, Sau SP, Wengel J, Hrdlicka PJ. J. Org. Chem. 2009;74:1070–1081. doi: 10.1021/jo802037v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kumar TS, Madsen AS, Østergaard ME, Wengel J, Hrdlicka PJ. J. Org. Chem. 2008;73:7060–7066. doi: 10.1021/jo800551j. [DOI] [PubMed] [Google Scholar]
- (11).Kumar TS, Wengel J, Hrdlicka PJ. ChemBioChem. 2007;8:1122–1125. doi: 10.1002/cbic.200700144. [DOI] [PubMed] [Google Scholar]
- (12).Sau SP, Kumar TS, Hrdlicka PJ. Org. Biomol. Chem. 2010;8:2028–2036. doi: 10.1039/b923465a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kumar TS, Madsen AS, Wengel J, Hrdlicka PJ. J. Org. Chem. 2006;71:4188–4201. doi: 10.1021/jo060331f. [DOI] [PubMed] [Google Scholar]
- (14).Yamana K, Iwase R, Furutani S, Tsuchida H, Zako H, Yamaoka T, Murakami A. Nucleic Acids Res. 1999;27:2387–2392. doi: 10.1093/nar/27.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kalra N, Babu BR, Parmar VS, Wengel J. Org. Biomol. Chem. 2004;2:2885–2887. doi: 10.1039/B411626G. [DOI] [PubMed] [Google Scholar]
- (16).Nakamura M, Fukunaga Y, Sasa K, Ohtoshi Y, Kanaori K, Hayashi H, Nakano H, Yamana K. Nucleic Acids Res. 2005;33:5887–5895. doi: 10.1093/nar/gki889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yamana K, Ohashi Y, Nunota K, Kitamura M, Nakano H, Sangen OS. Tetrahedron Lett. 1991;32:6347–6350. [Google Scholar]
- (18).Oeda Y, Iijima Y, Taguchi H, Ohkubo A, Seio K, Sekine M. Org. Lett. 2009;11:5582–5585. doi: 10.1021/ol902142w. [DOI] [PubMed] [Google Scholar]
- (19).Sehgal RK, Kumar S. Org. Prep. Proced. Int. 1989;21:223–225. [Google Scholar]
- (20).Prepared as described in: Roy SK, Tang J-Y. Org. Process Res. Dev. 2000;4:170–171.
- (21).Yamana K, Aota R, Nakano H. Tetrahedron Lett. 1995;36:8427–8430. [Google Scholar]
- (22).Zemlicka J. Collect. Czech Chem. Commun. 1964;29:1734–1735. [Google Scholar]
- (23).Bair KW, Tuttle RL, Knick VC, Corey M, Mc Kee DD. J. Med. Chem. 1990;33:2385–2393. doi: 10.1021/jm00171a012. [DOI] [PubMed] [Google Scholar]
- (24).(a) Dale TJ, Rebek J. J. Am. Chem. Soc. 1990;33:2385–2393. [Google Scholar]; (b) Asseline E, Cheng E. Tetrahedron Lett. 2001;42:9005–9010. [Google Scholar]
- (25).Disappearance of 1H NMR signals from exchangeable protons upon D2O addition, in concert with NMR (1H, 13C, COSY and HSQC) and HRMS, ascertained the O2′/N2′-functionalized constitution of the reported nucleosides.
- (26).Ross BS, Springer RH, Tortorici Z, Dimock S. Nucleosides Nucleotides. 1997;16:1641–1643. [Google Scholar]
- (27).Al-Rawi S, Ahlborn C, Richert C. Org. Lett. 2005;7:1569–1572. doi: 10.1021/ol0502432. [DOI] [PubMed] [Google Scholar]
- (28).Abdel-Magid AF, Carson KG, Harris BD, Maryanoff CA, Shah RD. J. Org. Chem. 1996;61:3849–3862. doi: 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- (29).Sanghvi YS, Hoke GD, Freier SM, Zounes MC, Gonzalez C, Cummins L, Sasmor H, Cook PD. Nucleic Acids Res. 1993;21:3197–3203. doi: 10.1093/nar/21.14.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).(a) Printz M, Richert C. J. Comb. Chem. 2007;9:306–320. doi: 10.1021/cc060142a. [DOI] [PubMed] [Google Scholar]; (b) Printz M, Richert C. Chem. Eur. J. 2009;15:3390–3402. doi: 10.1002/chem.200801587. [DOI] [PubMed] [Google Scholar]
- (32).Christensen UB, Pedersen EB. Nucleic Acids Res. 2002;30:4918–4925. doi: 10.1093/nar/gkf624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bryld T, Højland T, Wengel J. Chem. Comm. 2004:1064–1065. doi: 10.1039/b402414a. [DOI] [PubMed] [Google Scholar]
- (34).Hendrix C, Devreese B, Rozenski J, van Aerschot A, de Bruyn A, van Beeumen J, Herdewijn P. Nucleic Acids Res. 1995;23:51–57. doi: 10.1093/nar/23.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Karla N, Parlato MC, Parmar VS, Wengel J. Bioorg. Med. Chem. Lett. 2006;16:3166–3169. doi: 10.1016/j.bmcl.2006.03.072. [DOI] [PubMed] [Google Scholar]
- (36).Korshun VA, Stetsenko DA, Gait MJ. J. Chem. Soc. Perkin Trans 1. 2002:1092–1104. [Google Scholar]
- (37).Dohno C, Saito I. ChemBioChem. 2005;6:1075–1081. doi: 10.1002/cbic.200400325. [DOI] [PubMed] [Google Scholar]
- (38).Dougherty G, Pilbrow JR. Int. J. Biochem. 1984;16:1179–1192. doi: 10.1016/0020-711x(84)90215-5. [DOI] [PubMed] [Google Scholar]
- (39).Manoharan M, Tivel KL, Zhao M, Nafisi K, Netzel TL. J. Phys. Chem. 1995;99:17461–17472. [Google Scholar]
- (40).Nakano S-I, Uotani Y, Uenishi K, Fujii M, Sugimoto N. J. Am. Chem. Soc. 2005;127:518–519. doi: 10.1021/ja045445s. [DOI] [PubMed] [Google Scholar]
- (41).Benvin AL, Creeger Y, Fisher GW, Ballou B, Wagonner AS, Armitage BA. J. Am. Chem. Soc. 2007;129:2025–2034. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Nakamura M, Murakami Y, Sasa K, Hayashi H, Yamana K. J. Am. Chem. Soc. 2008;130:6904–6905. doi: 10.1021/ja801054t. [DOI] [PubMed] [Google Scholar]
- (43).Sekine M, Oeda Y, Iijima Y, Taguchi H, Ohkubo A, Seio K. Org. Biomol. Chem. 2011;9:210–218. doi: 10.1039/c0ob00248h. [DOI] [PubMed] [Google Scholar]
- (44).Gupta P, Langkjaer N, Wengel J. Bioconj. Chem. 2010;21:513–520. doi: 10.1021/bc900421r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.