Abstract
In higher plant plastids, ferredoxin (Fd) is the unique soluble electron carrier protein located in the stroma. Consequently, a wide variety of essential metabolic and signaling processes depend upon reduction by Fd. The currently available plant genomes of Arabidopsis and rice (Oryza sativa) contain several genes encoding putative Fds, although little is known about the proteins themselves. To establish whether this variety represents redundancy or specialized function, we have recombinantly expressed and purified the four conventional [2Fe-2S] Fd proteins encoded in the Arabidopsis genome and analyzed their physical and functional properties. Two proteins are leaf type Fds, having relatively low redox potentials and supporting a higher photosynthetic activity. One protein is a root type Fd, being more efficiently reduced under nonphotosynthetic conditions and supporting a higher activity of sulfite reduction. A further Fd has a remarkably positive redox potential and so, although redox active, is limited in redox partners to which it can donate electrons. Immunological analysis indicates that all four proteins are expressed in mature leaves. This holistic view demonstrates how varied and essential soluble electron transfer functions in higher plants are fulfilled through a diversity of Fd proteins.
Ferredoxin (Fd) is a soluble, low Mr protein that mediates transfer of one electron from a donor to an acceptor. The redox active center is a [2Fe-2S] cluster that confers a highly negative redox potential on the protein (-350 to -450 mV), making Fd a powerful reductant. The [2Fe-2S] cluster is ligated by four highly conserved Cys residues.
A broad spectrum of redox metabolism in higher plant plastids involves Fd. Although the name Fd was first used to describe a non-photosynthetic bacterial protein involved in nitrogen fixation (Mortenson et al., 1962), Fd is best known for a photosynthetic role: accepting electrons from photosystem I (PSI) and donating them to the enzyme Fd:NADP+ oxidoreductase (FNR) for photoreduction of NADP+ (Arnon, 1989). Donation of electrons by Fd has now been demonstrated to many other plastid enzymes essential for cellular processes, including nitrogen assimilation (nitrite reductase), sulfur assimilation (sulfite reductase [SiR]), amino acid synthesis (Glnoxoglutarate amino transferase), fatty acid synthesis (fatty acid desaturase), and redox regulation (Fd: thioredoxin reductase) (Knaff, 1996). In addition to PSI, Fd may be reduced by NADPH oxidation in a reversal of the FNR reaction (Suzuki et al., 1985). This enables Fd-dependent metabolism to continue under non-photosynthetic conditions, such as in root plastids.
Fds are present as multiple isoforms in many plants and algae (Bertini et al., 2002). In higher plants, those predominantly expressed in photosynthetic tissues can be crudely divided from those that are not on the basis of primary sequence (Wada et al., 1986). Work using maize (Zea mays) has exposed functional differences between these Fd types; the rate of light-dependent NADP+ reduction being higher with leaf type (photosynthetic) than root type (non-photosynthetic), and the root type being more efficiently reduced by NADPH than the leaf type (Hase et al., 1991b; Onda et al., 2000). In addition, root type Fd was found to mediate a more efficient electron flow from FNR to SiR, in a reconstruction of the root type sulfite reduction electron cascade (Yonekura-Sakakibara et al., 2000).
One Arabidopsis Fd gene has previously been cloned (Somers et al., 1990), and its expression was found to be light regulated and specific to chloroplast-containing tissues (Vorst et al., 1993). A second Arabidopsis Fd gene is up-regulated in response to nitrogen availability (Wang et al., 2000). However, the proteins involved remain uncharacterized. The availability of all Arabidopsis genomic Fd sequences (Arabidopsis Genome Initiative, 2000) provides the opportunity to study the entire complement of Fd proteins of one plant and to discover whether the capacity of Fd to perform such a wide variety of functions is reflected in differences between isoforms.
We have recombinantly expressed and purified all conventional [2Fe-2S] Fds encoded in the Arabidopsis genome, and we present here a detailed analysis of the physical and functional properties of the individual proteins, with confirmation of their expression in mature plants.
RESULTS
The Arabidopsis Genome Contains Four Genes Coding for Three Different Types of [2Fe-2S] Fd
Using the amino acid sequence of maize FdI, a National Center for Biotechnology Information BLAST2.0 search of the Arabidopsis genome (http://www.arabidopsis.org) identified six sequences with significant similarity. Of these, four were considered conventional Fds, because they shared more than 35% amino acid identity with maize FdI and conserved most structural features common to [2Fe-2S] Fd proteins from other higher plants (as represented within the 88 sequences compiled by Bertini et al. [2002]). The remaining two Fd-like proteins have conserved elements, such as the [2Fe-2S] cluster-binding Cys residues, but contain significant C-terminal extensions, which will probably impact greatly on protein structure and function because the C-terminus surrounds the redox center (Bertini et al., 2002). These proteins were therefore not included in this study, although accession numbers are included in “Materials and Methods.” Conventional Arabidopsis Fd (AtFd) genes were named AtFd1, AtFd2, AtFd3, and AtFd4 (see “Materials and Methods” for accession nos.). AtFd2 corresponds to the only Fd gene previously characterized, by Somers et al. (1990). Published expressed sequence tags and mRNA sequences confirm the predicted coding sequences of AtFd1, AtFd2, and AtFd3 but were unavailable for AtFd4 at the time of writing. The ChloroP program (http://www.cbs.dtu.dk/services/ChloroP/index.html) predicted highly probable chloroplast transit peptides for AtFd1, AtFd2, AtFd3, and AtFd4 (prediction scores of 0.573, 0.531, 0.577, and 0.569 with predicted transit peptides lengths of 69, 52, 49, and 49 amino acids, respectively). In Figure 1, predicted AtFd amino acid sequences are compared with known proteins typical of root, leaf, and cyanobacterial Fd types. On alignment (see Fig. 1A), AtFd1 and AtFd2 share many residues specific to leaf type Fds, and in support of this, light regulation of the AtFd2 gene has been reported (Vorst et al., 1993). On the basis of amino acid sequence comparison, AtFd3 encodes a typical root type Fd, and Wang et al. (2000) found expression of this gene was nitrate inducible, reminiscent of a root type Fd from maize (Matsumura et al., 1997). The AtFd4 protein differs from all well-studied wild-type Fds at several residues. Of particular interest are the Ser and Glu residues marked by a single star in Figure 1A, point mutation of which has been shown to greatly increase redox potential of the [2Fe-2S] cluster (Aliverti et al., 1995; Hurley et al., 1997; Taniguchi et al., 1997; Akashi et al., 1999).
Figure 1.
Comparison of AtFd amino acid sequences with characterized Fd proteins. A, Protein sequences typical of leaf, root, and cyanobacterial [2Fe-2S] Fds aligned with Arabidopsis Fds in ClustalW 1.8 (http://searchlauncher.bcm.tmc.edu/multi-align/multialign.html) and compared in BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX-_form.html). Transit peptides are shown in lowercase and mature proteins in capitals. All transit peptide cleavage sites are as resolved through chemical sequencing of mature N termini (spinach [Spinacia oleracea] and radish [Raphanus sativus] available on http://www.ncbi.nlm.nih.gov; see Hase et al. [1991a] for maize sequences), except those of Arabidopsis, rice, and sweet orange, which were estimated by comparison with known mature proteins. White on black residues are those conserved in more than nine of the sequences analyzed. B, Fd phylogenetic tree. An alignment of mature Fd amino acid sequences, lacking transit peptides, was generated in ClustalW 1.8 as for A. This alignment is presented as a Phylogram.
The phylogram of mature Fd proteins shown in Figure 1B illustrates further that AtFd1 and AtFd2 are leaf type Fds and AtFd3 is a root type Fd. AtFd4 is grouped close to root type Fds, but the long evolutionary distance makes it unclear whether AtFd4 shares a recent common ancestor with root type Fds.
DNA sequences coding for mature AtFd proteins were chemically synthesized, and the proteins were expressed recombinantly in Escherichia coli and then purified for detailed biochemical analysis. We consider the Chloro-P-predicted transit peptide cleavage site for AtFd1 unlikely because it includes residues highly conserved in mature Fds and that of AtFd3 unlikely because it includes seven residues before the conserved N terminus of many mature Fd proteins as determined by chemical sequencing. Therefore the first amino acid for recombinant expression was estimated by comparison with known mature Fd protein sequences and is given in Figure 1A as the first capital letter of AtFd proteins. In the case of AtFd3 and AtFd4, an additional Met residue was introduced as a transcription start site at the N terminus (see supplementary information for full nucleotide sequence details).
Purified Recombinant AtFd Proteins Vary in Surface Charge
Although all expressed AtFd sequences were similar in length and predicted mass (97 amino acids and 10.5 kD, 97 amino acids and 10.4 kD, 99 amino acids and 10.7 kD, and 100 amino acids and 10.9 kD for AtFd1, AtFd2, AtFd3, and AtFd4, respectively), there was wide variation in mobility during SDS-PAGE (Fig. 2A, left panel). Fd proteins contain a high proportion of negatively charged residues, reducing the SDS bound from the standard 1 to 1.4 molecules per peptide bond and resulting in migration to a position of higher perceived molecular mass (Huisman et al., 1978). The further relative migration of AtFd4 reflects the fewer negative residues in this molecule.
Figure 2.
Physical properties of AtFd proteins. A, Separation of purified mature recombinant proteins. Left panel, SDS-PAGE (1 μg protein lane-1); right panel, native gradient PAGE (20 μg lane-1). Lanes were 1, AtFd1; 2, AtFd2; 3, AtFd3; and 4, AtFd4. B, Absorption spectra: AtFd1, bold gray; AtFd2, bold black; AtFd3, light gray; and AtFd4, dark gray.
Fds are known to interact with electron transfer partners mainly through a network of salt bridges (Akashi et al., 1999; Kurisu et al., 2001). The variation in migration through the native gel shown in Figure 2A, right panel, is intriguing, because it allows a comparison of surface charge. AtFd1 and AtFd2 are the most mobile proteins through the native gradient gel. AtFd3 and AtFd4 migrate 90% and 76% as far, respectively. This indicates that the typical leaf type Fds, AtFd1 and AtFd2, have a more negative surface charge than the root type AtFd3, consistent with observations in maize (Kimata and Hase, 1989), and suggests AtFd4 has a much less negatively charged surface area.
The Spectrum of AtFd4 Contains a Red Shift
The spectra of all purified recombinant AtFd proteins, shown in Figure 2B, are typical of plant Fds, with the exception of a unique red shift in the spectrum of AtFd4 around 340 to 390 nm. We have cultivated AtFd4-expressing bacteria at 37°C, 30°C, and 25°C, which might result in alteration in peptide synthesis and cluster formation, and we observed no variation in the spectra of AtFd4 purified from such cells. We speculate that this red shift may be due to variation from the normal microenvironment of Fd [2Fe-2S] clusters and not a result of the artificial heterologous expression system.
Comparison of AtFd Redox Potential
Efficiency of electron transfer between Fd and its partner enzymes is dependent not only on protein-protein interaction but is also determined by the redox potential of the Fd [2Fe-2S] cluster relative to that of its partner enzyme. We therefore measured the redox potential of AtFd proteins by cyclic voltammetry. Figure 3A shows cyclic voltammograms of the current generated when Fd proteins interact with an electrode as its potential is decreased then increased. Calculated redox potentials (Fig. 3B) of AtFd1 and AtFd2 (-425 and -433 mV, respectively) are close to that of maize leaf Fd and considerably lower than the FAD moiety of maize leaf FNR, consistent with the physiological direction of electron flow in photosynthesis. The redox potential of AtFd3 is -337 mV, close to that of maize root type Fd, NADPH, and maize root FNR, and therefore favorable for efficient electron flow from catabolically generated NADPH to Fd, such as occurs in roots. At -152 mV, the redox potential of AtFd4 is remarkably high for a Fd molecule, implying that electron donation would only be efficient to a more restricted number of redox proteins with a similar or still higher redox potential. Other proteins containing plant type [2Fe-2S] clusters do have higher redox potentials than classical leaf and root type Fds, such as mitochondrial adrenodoxin (-250 mV according to Huang and Kimura [1983]), and some bacterial Riesk protein type [2Fe-2S] Fds (-150 mV according to Couture et al. [2001]). The greater current seen with the two leaf type AtFds in Figure 3A may be due to their more negative surface charge allowing increased interaction with the electrode, which is coated with positively charged poly-l-Lys.
Figure 3.
Redox potential. A, Cyclic voltammograms taken in 100 μm solutions of AtFd1 (bold gray), AtFd2 (bold black), AtFd3 (fine gray), and AtFd4 (fine black), versus a standard hydrogen electrode. Results are typical of three independent measurements. B, Comparison of calculated potentials with known measurements of NADPH and maize proteins: FdI, Matsumura et al. (1999); FdIII, Akashi et al. (1999); leaf FNR (L-FNR) and root FNR (R-FNR), Aliverti et al. (2001). sd was less than 5 mV in all cases.
Photosynthetic NADP+ Reduction
Actual measurements of electron transfer are presented in Figure 4, and average kinetic parameters for interaction of AtFds with electron transfer partners are recorded in Table I. We measured a significant difference in the affinity of root and leaf type Fds for electron transport between PSI and leaf type FNR. The leaf type Fds, AtFd1 and AtFd2, have a higher efficiency in mediating transfer of photosynthetic reductant, with Km values approximately 20 times lower than the root type AtFd3. AtFd4 showed no NADP+ photoreduction activity, presumably because its higher redox potential (Fig. 3B) prevented electron donation to leaf type FNR. We consider it likely that AtFd4 is reduced by PSI, because illumination of thylakoids resulted in a AtFd4 concentration-dependent decrease in OD422, consistent with reduction of the [2Fe-2S] cluster (results not shown).
Figure 4.
Electron transfer activity. A range of concentrations of AtFd1 (black circles), AtFd2 (white circles), AtFd3 (black squares), AtFd4 (white squares) were used to mediate electron flow: A, from photoreduced PSI to leaf FNR (L-FNR); B, from NADPH reduced L-FNR to cytochrome c; C, from NADPH reduced root FNR (R-FNR) to cytochrome c; D, from NADPH reduced R-FNR to SiR. Results are single data sets representative of three to four independent measurements.
Table I.
Kinetic parameters of various enzymes for Arabidopsis Fds
| Fd Species
|
Photosystem I
|
Leaf FNR
|
Root FNR
|
Sulfite Reductase
|
||||
|---|---|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Km | Vmax | Km | Vmax | |
| μm Fd ± sd | μmol NADPH μg-1 chlorophyll s-1 ± sd | μm Fd ± sd | μmol μmol-1 enzyme s-1 ± sd | μm Fd ± sd | μmol μmol-1 enzyme s-1 ± sd | μm Fd ± sd | μmol μmol-1 enzyme s-1 ± sd | |
| AtFd1 | 0.16 ± 0.07 | 0.020 ± 0.004 | 4.0 ± 0.5 | 61.6 ± 9. | 18.0 ± 5.6 | 197 ± 21 | 6.2 ± 0.97 | 7.5 ± 1.9 |
| AtFd2 | 0.18 ± 0.09 | 0.023 ± 0.007 | 2.8 ± 0.3 | 61.2 ± 16 | 19.0 ± 5.7 | 209 ± 39 | 5.2 ± 1.3 | 7.2 ± 0.8 |
| AtFd3 | 3.6 ± 0.8 | 0.025 ± 0.006 | 4.9 ± 1.4 | 79.4 ± 17 | 4.7 ± 0.5 | 231 ± 69 | 2.1 ± 0.33 | 13.7 ± 2.6 |
| AtFd4 | — | — | 27.4 ± 8.1 | 90.6 ± 14 | 14.7 ± 1.6 | 334 ± 78 | — | — |
Non-Photosynthetic Support of Redox Metabolism
Under non-photosynthetic conditions, Fd is reduced by FNR using NADPH, and this reaction was replicated in vitro with leaf type FNR (Fig. 4B) and with root type FNR (Fig. 4C). Leaf FNR has almost interchangeable kinetic parameters with leaf and root type Fds. However, root type FNR has a comparatively reduced affinity for leaf type Fds AtFd1 and AtFd2. These results are consistent with an efficient role for AtFd3 in transfer of non-photosynthetically derived reductant. AtFd4 shows a very high activity in both of these reactions, combined with a low affinity for both FNR molecules. In this assay, Fd reduces cytochrome c, and the relatively high redox potential of AtFd4 is no barrier to electron transfer, because cytochrome c has an even more positive redox potential of around +250 mV (Margalit and Schejter, 1973).
SiR is a Fd-dependent enzyme catalyzing reduction of sulfite to sulfide. Figure 4D shows sulfite reduction supported by a NADPH, root FNR, Fd reduction system. Root type AtFd3 is a more efficient electron donor than leaf type AtFd1 and AtFd2, having a higher activity for this reaction and a higher affinity for SiR. Probably due to its relatively high redox potential, AtFd4 is unable to support sulfite reduction in this system.
Expression in Mature Leaves and Roots
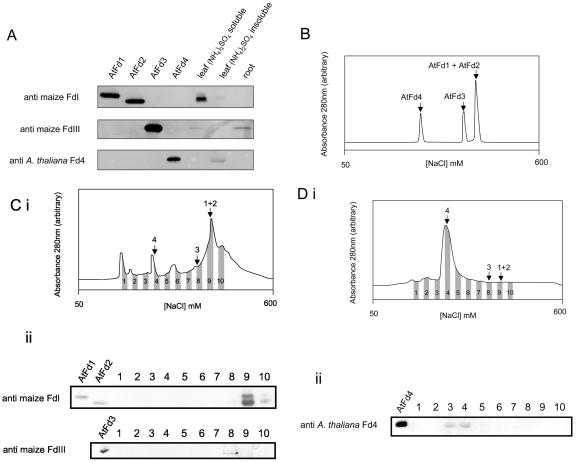
To confirm the presence of AtFd proteins in the plant, we performed immunodetection studies using antibodies specific to leaf and root type Fds. Neither antibody reacted efficiently with AtFd4, so we generated a new antibody to detect this protein. As model tissues, we chose mature Arabidopsis leaves and roots and prepared crude extracts from them. The leaf extract was fractionated into 70% saturated (NH4)2SO4 soluble and precipitated fractions. The antibody specific to leaf type Fds detects two bands corresponding to AtFd1 and AtFd2 mainly in the soluble fraction from leaves and not in roots, after SDS-PAGE (Fig. 5A). These leaf proteins also behave similarly to AtFd1 and AtFd2 during native PAGE (data not shown). To further confirm that the two proteins detected by the antibody specific to leaf type Fds are AtFd1 and AtFd2, we compared their chromatographic behaviors with those of recombinant AtFds using an anion exchange column. The two proteins are eluted at an identical NaCl concentration to recombinant AtFd1 and AtFd2 (Fig. 5, B and C).
Figure 5.
Detection of AtFd proteins in planta. A, Western blots using antibodies specifically reacting with leaf type Fds (raised against maize FdI), root type Fds (raised against maize FdIII), and AtFd4 (raised against AtFd4). Control lanes were loaded with 200 nmol of recombinant proteins, except for detection by anti-AtFd4, where 20 nmol of AtFd4 was used. Mature Arabidopsis leaf proteins were separated into fractions soluble and insoluble in 70% saturated (NH4)2SO4, and gels were loaded with the equivalent to 12 μg of total leaf protein, except when detecting with anti-AtFd4, where 90 μg was used. Twenty micrograms of Arabidopsis root protein was loaded. B, Elution profiles of recombinant AtFd1, AtFd2, AtFd3, and AtFd4 from an anion exchange column, over a salt gradient (based on conductivity of the eluent). C, i, Elution profile of Arabidopsis leaf proteins soluble in 70% saturated (NH4)2SO4. Arrows indicate relative elution positions of recombinant proteins: 1 + 2, AtFd1 and AtFd2; 3, AtFd3; 4, AtFD4. ii, Western blots to detect putative AtFd1 and AtFd2 after SDS-PAGE (top) and putative AtFd3 after gradient native PAGE (bottom). Two hundred nanograms of recombinant proteins was used as controls. D, i, Elution of Arabidopsis leaf proteins insoluble in 70% saturated (NH4)2SO4. Arrows as described above. ii, Labeled fractions were separated by SDS-PAGE and challenged with anti-AtFd in the western blot displayed. Twenty nanograms of recombinant AtFd4 was used as a control. All western blots are representative of two to three separate experiments.
The specific root type Fd antibody detected a band at a similar position to AtFd3, in the root extract and in the leaf (NH4)2SO4 soluble fraction (Fig. 5A). This assignment was also confirmed by elution profile over anion exchange chromatography, as described above (Fig. 5, B and C), and by comparison of migration over native PAGE (Fig. 5C, ii).
The antibody raised against AtFd4 is highly specific, 10 times less recombinant AtFd4 than for all other controls gave a strongly reacting band. Despite this specific, high titer of the antibody, the AtFd4 protein is so scarce in leaf tissues that 7.5 times more leaf protein was loaded onto the gel to detect a protein corresponding to recombinant AtFd4. The protein is detected in the (NH4)2SO4-precipitated fraction, but not in root protein extract. We also confirmed that this protein is eluted in the equivalent fraction to recombinant AtFd4 during anion exchange chromatography (Fig. 5, B and D). Precipitation of leaf AtFd4 in 70% saturated (NH4)2SO4 contrasts with the recombinant protein, although we have observed that a greater proportion of AtFd4 than other AtFds was precipitated by this treatment during purification of recombinant proteins from E. coli. Its entire precipitation may indicate a complex with another protein in vivo, which is still less soluble in solutions of (NH4)2SO4.
Densitometric comparison of leaf AtFd signals, obtained on the western blots, with standard curves of recombinant protein, blotted onto the same membrane, indicates that putative AtFd2 is most abundant and comprises around 90% of all leaf Fd, with putative AtFd1 and AtFd3 contributing around 7% and 3%, respectively, whereas the putative AtFd4 makes up just 0.05% of total leaf Fd. The possibility of Fd antibodies cross-reacting with the two Fd-like proteins in the Arabidopsis genome and leading to misidentification of AtFd proteins cannot be absolutely discounted. However, we consider it very unlikely given the number of bands detected, their behavior during PAGE and on anion exchange columns, and the high specificity of the antibodies, IgG reacting to conserved Fd sequences having being removed during antibody purification (see “Materials and Methods”).
DISCUSSION
We have characterized all conventional [2Fe-2S] Fd proteins encoded within the Arabidopsis genome, following recombinant expression and purification. Three basic types of protein are encoded, corresponding to classic leaf type Fds, a classic root type Fd, and a previously undescribed high redox potential type Fd.
Two leaf type Fds, AtFd1 and AtFd2, are present and, in all parameters so far measured, appear very similar. Both leaf type Fds have relatively negative redox potentials and have the highest affinity for NADP+ photoreduction of all of the Fd proteins measured. Using recombinant maize Fd proteins Hase et al. (1991b) also measured a higher efficiency of NADP+ photoreduction by leaf over that of root type Fd. A difference in physical properties between two leaf type Fds has only been described in maize (Kimata-Ariga et al., 2000), where the absence of an Asp residue (Fig. 1A, double star) reduces interaction with FNR, favoring partitioning to cyclic electron flow around PSI. This Asp residue is conserved between AtFd1 and AtFd2, and interaction with FNR molecules is very similar (Table I). Two proteins are detected in mature Arabidopsis leaves by a specific leaf type Fd antibody, and we conclude that they are AtFd1 and AtFd2. The only clue to a different function for AtFd1 and AtFd2 within the plant lies in the relative expression levels of these two proteins. Under these standard growth conditions, the putative AtFd2, comprises around 90% of the leaf Fd complement and so appears to be the major photosynthetic Fd. Whether the AtFd1, expressed at around 7% under these conditions, has a distinct physiological role remains unclear. Studies of expression pattern and protein-protein interaction should establish whether these two proteins have differential functions or represent redundancy.
AtFd3 is the only root type Fd in the Arabidopsis genome. This is in contrast to maize, where two separate isoforms have been described (Matsumura et al., 1997): one constitutively expressed and one nitrate induced. In agreement with measurements taken for maize root type Fd, AtFd3 has a more positive redox potential than leaf type Fds, favoring its reduction under non-photosynthetic conditions. This is reflected in measurements of electron transport, where AtFd3 has a much lower affinity for PSI than leaf type Fds. Confirming measurements made with maize proteins (Onda et al., 2000; Yonekura-Sakakibara et al., 2000), root type FNR has a higher affinity for AtFd3 than leaf type Fds during nonphotosynthetic Fd reduction, and AtFd3 more efficiently supports SiR activity after reduction by root type FNR. A protein was detected in mature Arabidopsis roots and leaves using a root type Fd-specific antibody, and we conclude it is AtFd3. There are previous reports of root and leaf type Fds being present in the leaf tissue of maize (Kimata and Hase, 1989), and the presence of root type in addition to leaf type Fds has been linked to the capacity for heterotrophic Fd reduction in tomato (Lycopersicon esculentum) fruit plastids (Aoki et al., 1998). Bearing in mind the low affinity of AtFd3 for photoreduction, it seems likely that this isoprotein is predominantly reduced heterotrophically. Root type FNR expression has been detected in maize leaves (Sakakibara, 2003), indicating that heterotrophic Fd reduction can occur in leaf tissues. Considering the superior support of sulfite reduction by AtFd3, its expression in leaves would be advantageous to the plant by increasing bioassimilatory and biosynthetic efficiency, particularly when photosynthesis may be limiting, such as during the dark or early in leaf development.
In addition to previously described Fd types, the Arabidopsis genome also contains a gene coding for AtFd4, a protein that shares some homology with root type Fds, but is evolutionarily distant. The AtFd4 sequence contains substitutions of two residues known to make a large contribution to the negative redox potential of Fds, and this probably contributes to its remarkably positive redox potential of around -150 mV, which likely prevents it from efficiently donating electrons to FNR or SiR. It is, however, redox active and donates electrons to cytochrome c in assays of leaf type and root type FNR with very high activity. A putative AtFd4 protein was detected at very low levels in mature leaves using an antibody specific for AtFd4, and its salting out behavior in (NH4)2SO4 indicates that it may be complexed to another protein(s) in vivo. Its low expression and our failure to detect it in roots may explain why, as yet, no expressed sequence tag is available. The number of redox proteins to which AtFd4 is capable of donating electrons is much more restricted than for other Fds, so it may have some kind of signaling function, perhaps when expressed in a specific cell type or under certain conditions. These questions would be best answered by a promoter expression study, and the identification of AtFd4-interacting proteins.
In conclusion, variation between Arabidopsis Fd sequences has been shown in this study to confer different functional characteristics on the individual proteins. We can now begin to see how different Fd isoproteins have the capacity to support a wide range of physiological roles under varying conditions. Studies of protein-protein interaction and expression pattern will further clarify the physiological roles of separate Fd proteins.
MATERIALS AND METHODS
Gene Construction, Recombinant Expression, and Purification
Arabidopsis Fd genomic sequences were identified by a BLAST search of the Arabidopsis Information Resource (http://www.arabidopsis.org/Blast/) using maize (Zea mays) FdI amino acid sequence. Stop codons were confirmed by comparison with cDNAs, where available, and cleavage sites of chloroplast transit peptides were estimated by comparison with known mature sequences. Sixteen oligonucleotides (oligos) of approximately 30 bp, corresponding to the coding and template strands of each mature amino acid sequence, were designed with increased A/T ratio in the first 60 bp from the 5′ end to increase expression level in Escherichia coli (Matsumura et al., 1999) and NcoI and SmaI overhangs at the 5′ and 3′ ends, respectively. All coding oligo pairs overlapped by 9 bp at 5′ ends to ensure specific annealing of consecutive DNA strands in the gene sequence. Figure 6 shows the design strategy for the coding and template oligo pair at the 5′ end of AtFd2. Entire sequences for all oligos are given in the supplementary information. All oligos, except the terminal 5′ ends, were phosphorylated using a kit (TaKaRa, Otsu, Shiga, Japan). Coding oligo pairs were denatured at 90°C for 3 min, cooled to 4°C, and then annealed by incubation at 75°C for 5 min followed by cooling to 20°C over 3 h. Consecutive DNA strands were stepwise ligated (T4DNA ligase kit, TaKaRa) and then gel purified to produce sequences coding for entire proteins. These sequences were then ligated into a modified version of the pTrc99A-1 vector, the proteins expressed in E. coli JM105, and then purified essentially as described by Matsumura et al. (1999). Fd concentration was estimated by spectroscopy using a molar extinction coefficient of 9.68 mm-1 cm-1 at 422 nm (Tagawa and Arnon, 1968).
Figure 6.
Design of oligonucleotides for Fd gene synthesis. Sequences designed for the terminal coding and template oligonucleotide (oligo) pair at the 5′ end of the AtFd2 gene are shown. White on black nucleotides are those altered from genomic sequence. Full sequences of all oligos are given in supplementary information.
Physical Analysis
Spectra were measured on a UV-2500PC spectrophotometer (Shimadzu, Tokyo). Redox potential was calculated as the midpoint of cyclic voltammetric measurements made at room temperature versus a standard hydrogen electrode at pH 7.5 as described by Taniguchi et al. (1997).
Enzymatic Analysis
Photoreduction of NADP+ was measured at 340 nm in a final volume of 1 mL containing: 50 mm HEPES-NaOH, pH 7.5, 100 mm NaCl, 1 mm MgCl2, 0.2 mm NADP+, 10 μg of chlorophyll spinach (Spinacia oleracea) thylakoid membranes and 0.01 to 40 μm Fd. The assay was started by illuminating the cuvette with an LH7 red light source (Hansatech, Kings Lynn, UK), perpendicular to the light path of the spectrophotometer. Measurements of NADPH-dependent Fd reduction by FNR were performed as described by Onda et al. (2000). Sulfite reduction was measured using recombinant maize SiR supported by a Fd:root-FNR reduction system, as described by Yonekura-Sakakibara et al. (2000).
Plant Extracts
Mature leaves and roots from healthy 3-week-old wild-type Arabidopsis plants (Columbia ecotype) were purchased from Green Gold Biosystem (Okayama, Japan). The plants were grown under a 16-h-light (light intensity, 70 μmol m-2 s-1)/8-h-dark regime and treated with fertilizer from Hyponex, Corp. Ltd (Tokyo) with final NH4 and NO3 concentrations of 1.4 and 0.64 mm, respectively. Proteins were extracted into 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm MgCl2, 1 mm EDTA, 5% (w/v) polyclar A, 5 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail (Nacalai tesque, Kyoto). The extract was desalted, and remaining micelles and thylakoids were precipitated at 30% saturated (NH4)2SO4. A crude Fd fraction was generated by precipitating the majority of proteins at 70% saturated (NH4)2SO4. Precipitated proteins were re-suspended, and both fractions were dialyzed against 50 mm Tris HCl, pH 7.5, and 1 mm MgCl2.
Electrophoresis
Native gradient PAGE was performed as described by Kimata and Hase (1989). Samples for SDS-PAGE were denatured at 95°C in 2% (w/v) SDS and then separated on a 15% (w/v) gel essentially as described by Onda et al. (2000) followed by staining with Coomassie Brilliant Blue.
Antibody Purification and Western Blotting
Two milligrams of maize FdI, maize FdIII (purified as described by Matsumura et al. [1999]), and AtFd4 were separately used to produce polyclonal antisera in rabbits. Common epitopes between AtFd proteins were removed by incubation of 1 mL of antisera for 3 h at 4°C in 9 mL of 2.5% (w/v) bovine serum albumin with 1 mg of each nonspecific AtFd (e.g. antisera raised against AtFd4 was incubated with AtFd1, AtFd2, and AtFd3), which had been denatured and immobilized on polyvinylidene difluoride (PVDF) membrane. Antibodies were then purified against the AtFd of interest by incubating for 3 h at 4°C with 1 mg of the protein, denatured, and immobilized on PVDF membrane. The membrane was washed extensively before stripping off bound IgG in 0.2 m Gly, pH 2, followed by immediate neutralization to pH 7.5 through addition of 1 m Tris-HCl, pH 8.8. Proteins were separated by SDS-PAGE and blotted to PVDF membrane followed by immunodetection essentially according to the methods of Onda et al. (2000). Primary antibody incubation was performed at equivalent antisera dilutions of anti-maize FdI (1:200), anti-maize FdIII (1:200), and anti-AtFd4 (1:80).
Anion Exchange Chromatography
An ÄKTA prime chromatographic system was used in combination with a RESOURCE Q 6-mL anion exchange column (both from Amersham Biosciences, Tokyo). Proteins were loaded on the column in 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, and 1 mm MgCl2. A mix of 50 μm each recombinant AtFd was used, and peak identity was confirmed by western blotting. Leaf proteins were loaded as the equivalent to 100 mg of total leaf protein. After extensive washing with the same buffer, elution was performed over a 100-mL gradient from 0.05 to 1 m NaCl.
Accession Numbers
Gene bank accession numbers for Fd genes are AtFd1 (At1g10960), AtFd2 (At1g60950), AtFd3 (At2g27510), AtFd4 (At5g10000), and Arabidopsis Fd-like protein encoding genes (At1g32550, At4g14890). Accession numbers for proteins are, rice (Oryza sativa) leaf (BAA06436), rice root (BAA06456), maize I (P27787), maize II (T01170), maize III (P27788), radish (Raphanus sativus) leaf (JX0088), radish root (P1493), spinach I (P00221), spinach II (P00224), and sweet orange (S62722).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining permissions will be the responsibility of the requestor.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.032755.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15G50320).
References
- Akashi T, Matsumura T, Ideguchi T, Iwakiri K, Kawakatsu T, Taniguchi I, Hase T (1999) Comparison of the electrostatic binding sites on the surface of ferredoxin for two ferredoxin-dependent enzymes, ferredoxin-NADP(+) reductase and sulfite reductase. J Biol Chem 274: 29399-29405 [DOI] [PubMed] [Google Scholar]
- Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G (2001) Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry 40: 14501-14508 [DOI] [PubMed] [Google Scholar]
- Aliverti A, Hagen WR, Zanetti G (1995) Direct electrochemistry and EPR spectroscopy of spinach ferredoxin mutants with modified electron transfer properties. FEBS Lett 368: 220-224 [DOI] [PubMed] [Google Scholar]
- Aoki K, Yamamoto M, Wada K (1998) Photosynthetic and heterotrophic ferredoxin isoproteins are colocalized in fruit plastids of tomato. Plant Physiol 118: 439-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1989) The discovery of ferredoxin: the photosynthetic path. Trends Biochem Sci 13: 30-33 [DOI] [PubMed] [Google Scholar]
- Bertini I, Luchinat C, Provenzani A, Rosato A, Vasos PR (2002) Browsing gene banks for Fe2S2 ferredoxins and structural modelling of 88 plant-type sequences: an analysis of fold and function. Proteins 46: 110-127 [DOI] [PubMed] [Google Scholar]
- Couture MM, Colbert CL, Babini E, Rosell FI, Mauk AG, Bolin JT, Eltis LD (2001) Characterization of BphF, a Rieske-type ferredoxin with a low reduction potential. Biochemistry 40: 84-92 [DOI] [PubMed] [Google Scholar]
- Hase T, Kimata Y, Matsumura T, Sakakibara H (1991a) Molecular cloning and differential expression of the maize ferredoxin family. Plant Physiol 96: 77-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T, Mizutani S, Mukohata Y (1991b) Expression of maize ferredoxin cDNA in Escherichia coli: comparison of photosynthetic and nonphotosynthetic ferredoxin isoproteins and their chimeric molecule. Plant Physiol 97: 1395-1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YY, Kimura T (1983) Reduction potential and thermodynamic parameters of adrenodoxin by the use of an anaerobic thin-layer electrode. Anal Biochem 133: 385-393 [DOI] [PubMed] [Google Scholar]
- Huisman JG, Moorman AF, Verkley FN (1978) In vitro synthesis of chloroplast ferredoxin as a high molecular weight precursor in a cell-free protein synthesizing system from wheat germs. Biochem Biophys Res Commun 82: 1121-1131 [DOI] [PubMed] [Google Scholar]
- Hurley JK, Weber-Main AM, Stankovich MT, Benning MM, Thoden JB, Vanhooke JL, Holden HM, Chae YK, Xia B, Cheng H et al. (1997) Structure-function relationships in anabaena ferredoxin: correlations between x-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants. Biochemistry 36: 11100-11117 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Hase T (1989) Localization of ferredoxin isoproteins in mesophyll and bundle sheath cells in maize leaf. Plant Physiol 89: 1193-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata-Ariga Y, Matsumura T, Kada S, Fujimoto H, Fujita Y, Endo T, Mano J, Sato F, Hase T (2000) Differential electron flow around photo-system I by two C(4)-photosynthetic-cell-specific Ferredoxins. EMBO J 19: 5041-5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaff DB (1996) Ferredoxin and ferredoxin dependent enzymes. In DR Ort, CF Yocum, eds, Oxygenic Photosynthesis: The Light Reactions. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 333-361
- Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T (2001) Structure of the electron transfer complex between ferredoxin and ferredoxin-NADP(+) reductase. Nat Struct Biol 8: 117-121 [DOI] [PubMed] [Google Scholar]
- Margalit R, Schejter A (1973) Cytochrome c: a thermodynamic study of the relationships among oxidation state, ion-binding and structural parameters: 1. The effects of temperature, pH and electrostatic media on the standard redox potential of cytochrome. Eur J Biochem 32: 492-499 [DOI] [PubMed] [Google Scholar]
- Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Murata H, Takao T, Shimonishi Y, Hase T (1999) Complementary DNA cloning and characterization of ferredoxin localized in bundle-sheath cells of maize leaves. Plant Physiol 119: 481-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T, Sakakibara H, Nakano R, Kimata Y, Sugiyama T, Hase T (1997) A nitrate-inducible ferredoxin in maize roots: genomic organization and differential expression of two nonphotosynthetic ferredoxin isoproteins. Plant Physiol 114: 653-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortenson LE, Valentine RC, Carnaham JE (1962) An electron transport factor from Clostridium pasteuranium. Biochem Biophys Res Commun 7: 448-552 [DOI] [PubMed] [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T (2000) Differential interaction of maize root ferredoxin:NADP(+) oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiol 123: 1037-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H (2003) Differential response of genes for ferredoxin and ferredoxin:NADP+ oxidoreductase to nitrate and light in maize leaves. J Plant Physiol 160: 65-70 [DOI] [PubMed] [Google Scholar]
- Somers DE, Caspar T, Quail PH (1990) Isolation and characterization of a ferredoxin gene from Arabidopsis thaliana. Plant Physiol 93: 572-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Oaks A, Jacquot JP, Vidal J, Gadal P (1985) An electron transport system in maize roots for reaction of glutamate synthase and nitrite reductase. Plant Physiol 78: 374-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa Y, Arnon DI (1968) Oxidation-reduction potentials and stoichiometry of electron transfer in ferredoxins. Biochim Biophys Acta 153: 602-613 [DOI] [PubMed] [Google Scholar]
- Taniguchi I, Miyahara A, Iwakiri K, Hirakawa Y, Hayashi Y, Nishiyama K, Akashi T, Hase T (1997) Electrochemical study of biological functions of particular evolutionary conserved amino acid residues using mutated molecules of maize ferredoxin. Chem Lett 1977: 929-930 [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T (2000) Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol 122: 887-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorst O, van Dam F, Weisbeek P, Smeekens S (1993) Light-regulated expression of the Arabidopsis thaliana ferredoxin A gene involves both transcriptional and post-transcriptional processes. Plant J 3: 793-803 [DOI] [PubMed] [Google Scholar]
- Wada K, Onda M, Matsubara H (1986) Ferredoxin isolated from plant non-photosynthetic tissues: purification and characterization. Plant Cell Physiol 27: 407-415 [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]