Abstract
Pectins are major components of primary plant cell walls and the seed mucilage of Arabidopsis. Despite progress in the structural elucidation of pectins, only very few enzymes participating in or regulating their synthesis have been identified. A first candidate gene involved in the synthesis of pectinaceous rhamnogalacturonan I is RHM2, a putative plant ortholog to NDP-rhamnose biosynthetic enzymes in bacteria. Expression studies with a promoter β-glucuronidase construct and reverse transcription PCR data show that RHM2 is expressed ubiquitously. Rhm2 T-DNA insertion mutant lines were identified using a reverse genetics approach. Analysis of the rhm2 seeds by various staining methods and chemical analysis of the mucilage revealed a strong reduction of rhamnogalacturonan I in the mucilage and a decrease of its molecular weight. In addition, scanning electron microscopy of the seed surface indicated a distorted testa morphology, illustrating not only a structural but also a developmental role for RGI or rhamnose metabolism in proper testa formation.
Pectins are a major component of the cell walls of higher plants (Carpita and Gibeaut, 1993). They are particularly abundant in the primary cell wall, i.e. the wall of growing cells, and the middle lamella. Pectins are thought to be responsible for cell to cell adhesion (Stephenson and Hawes, 1994), determination of wall porosity (Baron-Epel et al., 1988), and, at least in part, for wall strength (O'Neill et al., 2001). The pectic polysaccharides encompass mainly three classes of polysaccharides, homogalacturonan (HGA), rhamnogalacturonan I (RGI), and rhamnogalacturonan II (RGII; Mohnen, 1999). Common to all of these polysaccharides is the presence of d-GalUA (GalA). HGA is a homopolymer consisting of α 1,4-linked GalA, which is often partially methylesterified. RGII is a complex heteropolysaccharide consisting of a HGA backbone decorated with various structurally complex side chains composed of 11 different monosaccharides (O'Neill et al., 2001). RGII can occur as a dimer via a borate ester, which contributes to the tensile strength of the wall (O'Neill et al., 2001). RGI is a heteropolysaccharide consisting of the repeating disaccharide unit (1,2) α-l-rhamnosyl-(1,4) α-GalA. Numerous side chains can be attached to the rhamnosyl residues of the RGI backbone. These include galactans (β-1,4-linked) or arabinans (α-1,5-linked), both of which can be further decorated with arabinosyl residues or side chains.
Although much is known about the structure of RGI, its function in plant cell growth and development is less well characterized. Recent results indicate that RGI side chains are developmentally regulated (Willats et al., 1999), which was shown by labeling cell walls with specific antibodies raised against these side chains. Another approach to understanding the function of RGI facilitates the expression of fungal glycosylhydrolases in planta altering RGI structure. Expression of an endo-β-1,4 galactanase in potato (Solanum tuberosum) tubers resulted in an RGI structure with a 70% reduction of galactan side chains (Sorensen et al., 2000). However, no morphological or developmental phenotype was observed in the mutant plant under the growth conditions examined. However severe morphological phenotypes, such as death of stolons and discoloring of the leaves was observed when expressing an endo-α-1,5 arabinanase in potatoes (Skjot et al., 2002). When the enzyme was genetically engineered to be retained in the endoplasmic reticulum instead of being secreted into the apoplast, the plant morphology remained normal despite a 50% reduction in arabinan side chains. Finally, expression of an RGI lyase in potatoes resulted in an unusual fragmentation of RGI (Oomen et al., 2002), but only a minor phenotype in the potato tubers was observed.
Another approach to gain insight into the function of pectins is to modulate their synthesis to produce an altered pectin structure. In general, it is assumed that activated sugar substrates, the nucleotide sugars, are used by various glycosyltransferases to form the pectic polysaccharides. Of the 412 putative glycosyltransferases identified in the completed genome sequence of Arabidopsis (Henrissat et al., 2001), it has been estimated that at least 53 glycosyltransferases are required to assemble pectic polysaccharides (Mohnen, 1999). Assigning function to glycosyltransferases and hence identifying their involvement in pectin synthesis has been difficult mainly due to unknown substrate specificities and the multitude of candidate genes. Only two glycosyltransferase genes involved in the synthesis of pectins have been identified so far: a putative glycosyltransferase (quasimodo), whose knock-out line exhibited a 15% reduction of GalA in its wall (Bouton et al., 2002), and a putative glucuronosyltransferase (Iwai et al., 2002), whose inactivation resulted in an altered RGII structure and an increase in cell separation. There are 11 different monosaccharides present in pectins, and thus it is expected that at least 11 different nucleotide sugars are involved in pectin biosynthesis. To accommodate the need for such a diversity of nucleotide sugars, plants possess an extensive nucleotide sugar interconversion pathway for the de novo synthesis of those nucleotide sugars (Feingold and Avigad, 1980). In addition, plants have salvage pathways, in which monosaccharides are activated through phosphokinases and are subsequently converted to nucleotide sugars through pyrophosphorylases. This has been shown for many but not all sugars (Feingold and Avigad, 1980). Thus far, only one of the genes involved in the nucleotide sugar interconversion pathway has been shown to be involved in the synthesis of pectins, specifically RGII, a GDP-Man-4,6-dehydratase (Bonin et al., 1997). Inactivation of this enzyme (MUR1) led to an altered RGII structure resulting in a lower affinity for borate and thus a reduction in RGII dimerization (O'Neill et al., 2001). This led to a decrease in the tensile strength of the stem. Other enzymes involved in the nucleotide sugar conversion pathway may be identified through gene sequence comparisons. Because the orthologous genes from bacteria are relatively well characterized and the candidate genes found in the Arabidopsis genome cluster into distinct gene families (Reiter and Vanzin, 2001), a function can easily be predicted. However, to date, no nucleotide sugar converting enzyme has been shown to be involved in RGI synthesis. Here, we present evidence that an enzyme from Arabidopsis predicted to be involved in nucleotide sugar synthesis influences the synthesis of RGI.
RESULTS
Identification and Attributes of RHM2
Nowadays, reverse genetics is a useful tool for identifying plant cell wall-related enzymes (Perrin et al., 2001). Therefore, hidden Markov models (HMMs) were extracted from the Pfam protein family database (Bateman et al., 2002) using enzymatic keywords (“epimerase” and “dehydratase”) to identify candidate genes possibly involved in the synthesis of NDP-rhamnose in plants. These HMMs from the Pfam database were used to search for Arabidopsis proteins with HMMsearch from the HMMer software package (Eddy, 1998). The Pfam database generally contains high-quality HMMs based on alignments of multiple related proteins, enabling the user to identify even remotely homologous sequences (Eddy, 1998) as opposed to simple BLAST searches. Especially PF01370 proved to be very useful, because one earlier described nucleotide sugar-converting enzyme (Doerman and Benning, 1998) and its putative paralogs were among the highest scoring hits. Among the identified genes was a small gene family comprising three members that could be aligned with the full PF01370 HMM, corresponding to 315 amino acids in these three genes. These alignments produced an E value below 1 × 10-107. The “alignment” part of the HMMsearch results showed approximately 140 exact matches against highly and less highly conserved amino acids. Moreover, of the remaining 175 amino acids, roughly 100 residues were identified as positives by the program. Together these data indicate a good similarity of this gene family to the conserved epimerase/dehydratase motive. A National Center for Biotechnology Information conserved domain analysis (Marchler-Bauer et al., 2003) indicated a bipartite structure of these genes. The N-terminal part of the proteins showed highest similarity to the rfbB dehydratase conserved domain (accession no. COG1088), a domain characteristic for proteins involved in the first step of bacterial dTDP-rhamnose synthesis. In bacteria, three separate enzymes encompassing a dehydratase, an epimerase, and a reductase are responsible for conversion of dTDP-Glc into the corresponding rhamnose precursor (Giraud and Naismith, 2000). A closer look at the plant gene sequences revealed a modification of the extended signature found in nucleotide cofactor (e.g. FAD and NAD(P))-binding Rossmann folds (Wierenga et al., 1985; Kleiger and Eisenberg, 2002) in the N-terminal domain of these proteins. In addition, the signature sequences YXXXK and TXE involved in the catalytic mechanism of epimerases and dehydratases such as bacterial dTDP-rhamnose dehydratase (Allard et al., 2001) can be assigned (Fig. 1A).
Figure 1.
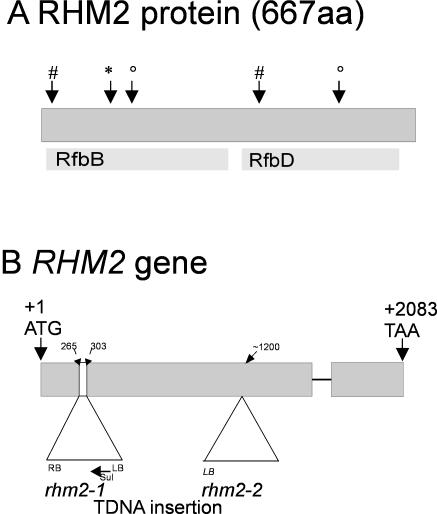
Structures of the RHM2 gene and the RHM2 protein. A, Results of a National Center for Biotechnology Information conserved domain analysis of RHM2 showing only the most significant hit for each domain. RfbB is annotated as dTDP-d-Glc 4,6-dehydratase, RfbD as dTDP-4-dehydrorhamnose reductase. #, Rossmann fold signature BIXGXXGXXAXXXAXB or BIXGXXGXXGXXXAXB (B is a hydrophobic amino acid); *, the conserved residues important for catalysis (TXE); °, the signature important for catalysis (YXXXK, 161-165). B, RHM2 gene structure and insertion of the T-DNAs (numbers indicate the insertion of the T-DNA); +1 indicates the A of the start codon ATG; +2,083 the A of the stop codon; sul, sulfadiazine resistance gene; LB/RB, left/right T-DNA border; thin line, single intron.
The C-terminal part of these proteins shows highest similarity to the rfbD reductase (COG1091) domain, characteristic of the last step in bacterial dTDP-rhamnose synthesis. This small gene family also shows a conserved extended nucleotide cofactor-binding site where only the last hydrophobic amino acid is missing. Moreover, a conserved YXXXK loop was identified. All family members were predicted to exclude any transmembrane domains by the transmembrane prediction program TMHMM (Krogh et al., 2001). Tentative subcellular localization prediction by TargetP (Emanuelsson et al., 2000) or Prot-Comp (http://www.softberry.com), a prediction software trained on plant proteins, did not suggest targeting to any specialized compartment (data not shown).
Recently, Reiter and Vanzin (2001) have also identified this gene family using a BLAST-based approach using bacterial genes as template. There, the genes were termed RHM1 to RHM3 for rhamnose biosynthetic enzymes. We chose RHM2 for further characterization as a representative candidate of this gene family, because all functional predictions were similar for the whole family. Because only 5′-truncated versions of RHM2 cDNA were available in public databases, the full-length coding region was PCR amplified using primers specific for the predicted full-length coding sequence, proving the validity of the predicted full-length gene model.
RHM Proteins Are Common in Plants
The expressed sequence tag (EST) clustering database SPUTNIK (Rudd et al., 2003) was queried with the RHM2 sequence to explore whether RHM proteins are present in other plant species. Best scoring EST clusters (accession nos. are given in the end of this paper) were aligned with either the N-terminal dehydratase (Fig. 2) or the C-terminal epimerase/reductase domain of RHM2. For both domains, very similar sequences were identified in legumes, solanaceous plants, and members of the graminae. For soybean (Glycine max), potato, wheat (Triticum aestivum), and barley (Hordeum vulgare), high-scoring EST clusters spanning parts of both domains of RHM2 could be identified, indicative of a conservation of the two domain protein architecture in plants in contrast to bacteria.
Figure 2.
Sequence alignment of RmlB from Salmonella with the dehydratase domain of Arabidopsis RHM2 and other plant orthologs. The alignment was produced with ESPript (Gouet et al., 1999). Totally conserved residues are highlighted in inverted black. Boxed residues are conserved. The extended motive indicative of a Rossman fold is represented as a bar below the sequence alignment. A star below the sequence alignment indicates residues important for catalysis. (The accession nos. for the Sputnik EST clusters are given in the accession no. section.)
RHM2 Is Expressed Ubiquitously
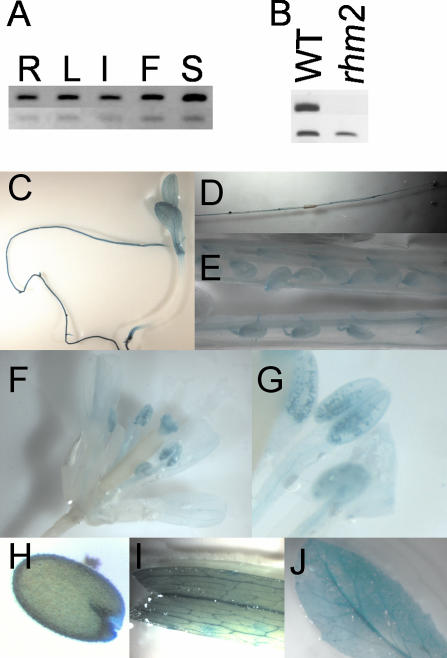
RHM2 transcript abundance in several tissues was investigated using reverse transcription (RT)-PCR to assess the possible function of RHM2. Expression was detectable in all examined tissues and was therefore judged to be ubiquitous (Fig. 3A). A reporter gene construct was prepared to localize RHM2 expression in more detail. The 2.2-kb region spanning the 5′ promoter region of RHM2 including the C-terminal part of the previous predicted gene was fused to the β-glucuronidase (GUS) reporter gene (Jefferson et al., 1987) and transformed into Arabidopsis. Fifteen plant lines were assessed, and expression patterns of GUS activity were visualized histochemically (Fig. 3, C-J). Expression in the young seedling was strongest in the roots, the vascular tissue of the leaves, and parts of the stem. In mature plants, strong expression was detected in the roots, in the flowers, and in the siliques. In the flowers, strongest expression was observed in the vascular tissue of petals, in the pollen grains, and on top of the stigma. In the siliques, strongest expression was obtained at the base of the silique, in the vascular tissue, in the funiculi, and in the developing seed coat, which even persisted in mature dried seeds. Weaker, less uniform staining was detectable in the vascular tissue of mature rosette leaves and in the stem. Taken together, these results are consistent with the RT-PCR data, indicating a ubiquitous expression of RHM2.
Figure 3.
Expression analysis of RHM2. A, Tissue specificity: RT-PCR was performed with specific primers for RHM2 (top lane, 33 PCR cycles) and APT1 (control; bottom lane, 29 PCR cycles) on roots (R), leaves (L), stems (I), flowers (F), and siliques (S) of WT plants. B, Mutant analysis: RT-PCR on siliques of rhm2-1 and WT plants with RHM2-specific primers spanning the insertion site of the T-DNA. C to J, GUS analysis of plant tissues. C to G, Incubation for 2 h. H to J, Incubation overnight. C, Seedling; D, root; E, silique; F, flower; G, stamens; H, mature seed; I, cauline leaf; J, rosette leaf.
Expression of the whole extended RHM gene family was assessed by querying the MPSS database (http://www.dbi.udel.edu/mpss) for Arabidopsis transcripts. A rather ubiquitous expression of the whole family was observable with preferential expression of RHM3 in the root and highest expression of RHM1 in the flower.
Identification of Arabidopsis Lines with T-DNA Insertions in RHM2
An rhm2 Arabidopsis mutant line (rhm2-1) was identified within the GABI-KAT collection when looking for insertion lines in the RHM gene family. Heterozygous plants showed a segregation of 3:1 when grown on sulfadiazine (χ2 = 0.0054; P = 0.941, n = 988) indicative of a single T-DNA insertion. Similar results were obtained by Southern-blot analysis (data not shown). PCR-based analysis of the progeny showed that only those plants that were homozygous for the T-DNA insertion had an observable phenotype in their seeds. Sequencing of the PCR-amplified right and left border products indicated that the T-DNA was inserted in the first exon between bp 265 and 307, resulting in a deletion of approximately 40 bp in the genomic DNA (Fig. 1B).
The effect of the T-DNA insertion on mRNA expression level was studied by RT and PCR amplification using primers spanning the putative insertion site of the T-DNA in the mRNA. Whereas a clear signal was visible in WT plants, no signal was obtained in the mutant, indicative of an alteration of RHM2 mRNA (Fig. 3B).
A second allele was obtained from the SALK collection, which will henceforth be referred to as rhm2-2. In this line, the T-DNA insertion was in the second domain of RHM2, thus potentially representing a weaker allele than rhm2-1 (Fig. 1B).
Rhm2 Mutant Phenotype
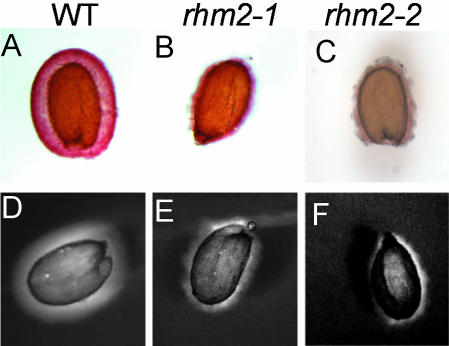
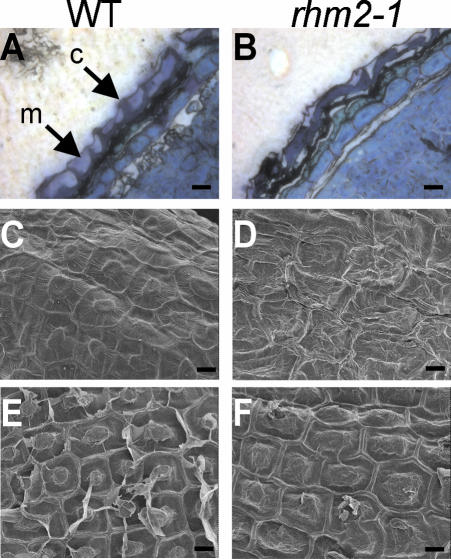
Cell wall material from different tissues of WT and rhm2-1 plants was prepared, and its monosaccharide composition was determined. However, no remarkable alteration in wall composition was detected in leaf material from mature plants or roots from plants grown on Murashige and Skoog agar. Moreover, no obvious growth, developmental, or morphological phenotype could be observed when rhm2 plants were grown in phytotrons or in the greenhouse. However, when mature seeds were stained for pectin with ruthenium red, a pectin-staining dye (Hanke and Northcote, 1975), only little irregularly patterned staining could be observed in both rhm2-1 and rhm2-2 seeds, whereas wild-type (WT) seeds showed intense regularly spherical staining (Fig. 4, A-C). To investigate the possibility of a compensatory up-regulation of other wall polysaccharides in the seed mucilage of rhm2, imbibed seeds were placed into an aqueous solution of high Mr blue dextran. WT seeds showed a larger more regular zone not penetrable by blue dextran, whereas again in both rhm2 mutants, this zone was more irregular and smaller in size, indicative of a loss of penetration barrier in the mutant (Fig. 4, D-F). Because these stainings revealed similar effects both for rhm2-1 and rhm2-2, the following analyses were conducted on rhm2-1 only. Because the structural integrity of the seed coat is affected in some mucilage mutants (Western et al., 2001), the structure of the rhm2-1 seed coat was investigated by sectioning the seeds and subsequent staining with toluidine blue, a polychromatic dye (O'Brien et al., 1964). These analyses revealed that the columellae (Fig. 5B) of rhm2-1 seeds were flattened and distorted and mucilage was reduced when compared with sections of WT seeds (Fig. 5A). These findings were further corroborated by examination of the whole seed surface structure by scanning electron microscopy (SEM). When WT seeds are not imbibed, they exhibit a regular pattern of elevated columellae within each epidermal cell, separated from their neighboring cells by clearly visible radial cell walls (Fig. 5C). In contrast, the rhm2-1 seeds show a rather distorted structure where the columella is not easily distinguishable and radial cell walls are no longer clearly visible (Fig. 5D) due to the distortion of the rest of the epidermal cells, which harbor the mucilage (Western et al., 2000).
Figure 4.
Histochemical analysis of WT (A and D), rhm2-1 (B and E), and rhm2-2 (C and F) seeds. A to C, Staining of imbibed seeds with ruthenium red. D to F, Staining of imbibed seeds with blue dextran (2,000-kD) solution.
Figure 5.
Microscopical analysis of WT (A, C, and E) and rhm2-1 (B, D, and F) seeds. A and B, Sections of mature seeds fixed under conditions preserving the mucilage, stained with toluidine blue (c, columella; m, mucilage). C and D, SEM of the seed surface (testa) of dry untreated seeds. E and F, SEM of imbibed seeds after air-drying; the mucilage is removed. Scale bar in A through F = 10 μm.
WT seeds exhibit a release of material around the columellae after imbibition, and as a result, deep valleys around the columella become visible. In imbibed rhm2-1 seeds, the valleys around the columella seem to be shallower and the columella seems to be more flattened (Fig. 5, E and F). These SEM pictures confirm that both mutant and WT seeds did excrete mucilage in accordance with the observed staining patterns with ruthenium red. Taken together, these results suggest that the mutant still has a mucilage remnant, which can be released upon hydration but that does not seem to be sufficient for proper seed coat development.
Analysis of Seed Mucilage
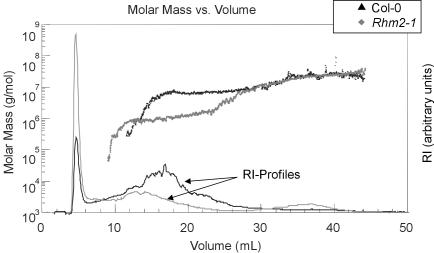
The monosaccharide composition of extractable mucilage of rhm2-1 and WT seeds was determined. The seed mucilage is largely composed of unsubstituted RGI (Goto, 1985; Penfield et al., 2001), and the most abundant sugars present in both WT and rhm2-1 seed mucilage were rhamnose and uronic acid (Table I). However, only approximately 35% and approximately 45% of the rhamnose amount obtained from the WT was released from the rhm2-1 and rhm2-2 mutant seeds, respectively. This is in agreement with the observed collapsed structure in mature seeds, where the testa cells seem to contain less mucilage. The ratio of rhamnose to GalUA of 1:1 remained approximately constant (Table I). Solubilized mucilage was subjected to field flow fractionation (FFF) and Mr determination by multi-angle laser light scattering (MALLS)-differential refractive index. The advantage of the FFF technique over size exclusion chromatography is its extremely high upper working limit in terms of Mr and hydrodynamic volume of polymers and the lack of sensitivity to adsorption (Roessner and Kulicke, 1994). Significant differences in the elution profiles and in the molar mass distributions of rhm2-1 and WT mucilage were obtained (Fig. 6). Rhm2-1 mucilage has a molecular mass average of 8.8 × 105 g mol-1, whereas WT mucilage has a much larger molecular mass average of 5.2 × 106 g mol-1. Simultaneous elution of molecules with different molecular masses indicates differences in the molecular conformation of WT and rhm2-1 mucilage. The gyration radius that was obtained from light-scattering data constitutes in rhm2-1 mucilage only half of that of WT (averages of 75 and 152 nm, respectively). Only rhm2-1 mucilage shows a significant second peak (15 mass%) with a molecular mass average of 2 × 107 g mol-1, which might represent RGI aggregates due to the used method. In addition, approximately 35% of rhm2-1 mucilage elutes within the first 2 to 3 min, whereas only approximately 10% of the WT sample elutes in this range. Because similar amounts based on uronic acid content of equally treated samples were injected, this early eluting material probably contains also small Mr pectins and not only salt, further indicating a substantial higher portion of smaller pectic oligoand polysaccharides in rhm2-1.
Table I.
Monosaccharide composition of solubilized mucilage
Mucilage was solubilized and its monosaccharide composition was determined. The values are the average of five independent experiments.
| WT | Rhm2-1 | |
|---|---|---|
| μg mg−1treated seeds | ||
| Galactose | 1.6 ± 0.4 | 1.4 ± 0.4 |
| Mannose | 0.9 ± 0.1 | 0.9 ± 0.1 |
| Xylose | 1.2 ± 0.1 | 1.3 ± 0.1 |
| Arabinose | 1.1 ± 0.1 | 1.0 ± 0.2 |
| Fucose | 1.2 ± 0.1 | 1.2 ± 0.1 |
| Rhamnosea | 8.9 ± 2.3 | 2.9 ± 0.4 |
| Glucose | 3.8 ± 1.4 | 2.9 ± 1.3 |
| Uronic Acidsb | 9.9 ± 3.0 | 4.8 ± 0.5 |
a Significance at 1% level. b Significance at 5% level.
Figure 6.
Molar mass versus FFF elution volume for mucilage extracted from WT and rhm2-1 seeds. The molar mass distribution obtained by MALLS for WT (black dots) and rhm2-1 (gray dots) are shown. Refractive index (RI) profiles indicating polymer abundance in arbitrary units for WT (black) and rhm2-1 (gray) are indicated by solid lines.
Because the seed mucilage may also play a role in seed germination under water-limiting conditions (Penfield et al., 2001), germination rate of mutant and WT seeds was investigated by addition of various concentrations of PEG-8000. However, no significant difference in germination rate was observed (data not shown).
DISCUSSION
Choosing a reverse genetics approach to study plant cell wall synthesis and function, several Arabidopsis alleles of rhm2 were identified and characterized, strongly indicating that a disruption of RHM2 is responsible for the observed seed coat phenotype. First, the observed phenotype was linked to a homozygous insertion in the RHM2 gene in both rhm2-1 and rhm2-2. It has been shown that there is only a single T-DNA insertion in the genome of the rhm2-1 plants by segregation analysis and Southern blotting. Second, an RNA interference approach (Waterhouse et al., 2001; Wesley et al., 2001) down-regulating the RHM2 transcript resulted in a similar seed phenotype as the rhm2 mutants (B. Usadel and M. Pauly, unpublished data). Finally, mum4, a seed coat mutant identified in a forward genetic screen also showed a reduction in mucilage and developmental effects (Western et al., 2001). Recently, mum4 has been mapped to the RHM2 locus (Western et al., 2004).
RT-PCR analysis of rhm2-1 with primers spanning the T-DNA insertion failed to amplify a product, indicative of an altered transcript. Because the T-DNA is inserted in the N-terminal domain of RHM2, rhm2-1 could represent a close to null allele, even though absence of a general phenotype might be explained by a weak allele.
Mucilages in general are very abundant and have been implicated for hydration regulation and as glue for substrate adhesion (Frey-Wyssling, 1976). Moreover Arabidopsis seed mucilage has been shown to be involved in germination efficiency (Penfield et al., 2001). However, a defect in RHM2 leading to a reduced amount of extractable seed mucilage had no significant effect on germination rate under water-limiting conditions. This is in contrast to myb61 seeds where no mucilage was extractable and a decrease in germination rate was observed (Penfield et al., 2001). These findings could indicate that small amounts of mucilage may be sufficient to assist seed germination under reduced water potential or that a loss of MYB61 function leads to additional defects. In this context, it is noteworthy that RHM2 transcript is up-regulated in response to abscisic acid (Hoth et al., 2002), a plant hormone known to be involved in response to water status.
The deposition of mucilage is rather complex. Mucilage and seed coat development have only been recently studied in greater detail (Western et al., 2000; Windsor et al., 2000). The mucilage secretory cell development has been sectioned into five stages, where different sets of genes seem to be necessary for proper seed coat development underlining the vital role of the mucilage during seed development (Western et al., 2001). The reduction of rhamnose and thus RGI in the seed coat provides a new tool for dissecting the signaling pathway in the establishment of the seed coat in Arabidopsis.
Despite the strong effect in the mucilage, rhm2 did not show any other morphological or cell wall-related phenotype, even though RHM2 seems to be expressed in other plant organs as judged by GUS staining and RT-PCR. Even though GUS staining could be prone to artifacts in some plant parts (e.g. vascular tissue; Martin et al., 1992), RHM2 expression was also shown to be ubiquitous by semiquantitative RT-PCR. A likely explanation therefore is that the loss of RHM2 transcript is compensated through functional transcripts of the other RHM-gene family members, which also seem to be expressed ubiquitously. Apparently, there is a preferential expression of RHM2 in the reproductive organs, where pectins also play a role in proper development (Rhee and Somerville, 1998).
Rhm2 is impaired in RGI biosynthesis, and because at least the first domain of RHM2 exhibits a high sequence similarity to bacterial rhamnose synthesis genes, it seems possible that RHM2 actually encodes an enzyme that catalyzes the conversion of NDP-Glc to NDP-rhamnose as discussed previously by Reiter and Vanzin (2001). This is further corroborated by heterologous expression of a RHM2 paralog, RHM1, in Escherichia coli, which resulted in an enzyme fraction that converts UDP-d-Glc into a product that upon hydrolysis comigrates with rhamnose upon thin-layer chromatography plate separation (M. Mølhøj and W.-D. Reiter, personal communication). Together, these data suggest that RHM2 might be involved in NDP-rhamnose biosynthesis, although further experiments are necessary to unambiguously demonstrate its activity. Rhamnose, in its terminal form, also occurs in cell wall arabinogalactanproteins (AGPs; Fincher et al., 1983), and therefore these structures could also be altered if rhamnose becomes limiting due to lack of RHM2. However, linkage analysis of the Arabidopsis seed mucilage has revealed the presence of AGPs but not any terminal rhamnosyl residues (Penfield et al., 2001) rendering a role for RHM2 in mucilage AGP synthesis unlikely.
Moreover, rhamnose-containing oligosaccharides, exuded from germinating seeds, have been implicated in hypocotyl growth (Yokotani-Tomita et al., 1998). These could also account for sugars such as fucose in the mucilage. Reduced rhamnose supply could also change the abundance of these. If these oligosaccharides would have an impact on seed development, they can be responsible for the observed seed phenotype. The seed mucilage provides a useful model system to study pectin biosynthesis because of its high content of relatively pure unsubstituted RGI (Penfield et al., 2001) and the expendability of the mucilage under laboratory conditions as suggested earlier (Western et al., 2000). The data shown here provide evidence that limiting substrate availability seems to be involved in the determination of polysaccharide chain length, as indicated by the different Mr of rhm2-1 RGI. Thus, rhm2, as the first mutant known to be involved in RGI synthesis represents a novel tool for dissecting the regulation of pectin synthesis. For example, this mutant could be used to identify novel mutants, which suppress the developmental but not the biochemical phenotype or vice versa in a suppressor screen. Other genes affected by the reduction and lack of RGI could be discovered by looking for potential interaction partners of RHM2. Such data would give further insights into the synthesis of RGI, its regulation, and its function.
MATERIALS AND METHODS
Plant Material
A T-DNA insertion line (rhm2-1) was obtained from the GABI-KAT consortium (Max-Planck-Institut, Cologne, Germany). The line was generated as follows: Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) harboring plasmid pAC161 (for the sequence of pAC161, see Gen-Bank/EMBL accession no. AJ537514) was used for plant transformation based on the floral dip method of Clough and Bent (1998). Selection of T1-plants in the greenhouse was performed essentially as described by Hadi et al. (2002) Initial segregation analysis was performed using 100 surface sterilized T2 progeny seeds on Murashige and Skoog agar plates containing 7.5 mg L-1 sulfadiazine. Segregation analysis was repeated on the confirmed rhm2-1 line with approximately 1,000 seeds. A second T-DNA insertion line (rhm2-2) was obtained from the Salk Institute for Genomic Analysis Laboratory Collection (Alonso et al., 2003) via the Nottingham Arabidopsis Stock Centre. All Arabidopsis plants were grown in environmental chambers on soil under standard conditions (120 μmol m-2 s-1, 60% humidity, 20°C, 16 h light/8 h dark).
WT plants were of the Columbia-0 ecotype, and plants carrying the rhm2 mutations had the Colombia-0 background.
RNA Extraction and Semiquantitative PCR
Total RNA from 7-week-old plant organs was extracted with guanidinium hydrochloride as previously described (Logemann et al., 1987). The extracted RNA (approximately 4 μg) was treated with 1 unit of DNase (Roche Diagnostics, Mannheim, Germany) and subsequently with reverse transcriptase from Moloney murine leukemia virus (Qbiogene, Heidelberg) as recommended by the manufacturer. The cDNA content of all RT reactions was normalized by amplifying the APT1 transcript, a constitutive control (Moffatt et al., 1994). An aliquot (0.5 μL) of this normalized cDNA was used for semiquantitative PCR. As a control, APT1 transcript was amplified using previously described primers (Orsel et al., 2002). For amplification of RHM2, the primers RTRA (5′-GTTTCAAGCAACACGGTCC-3′) and RTRB (5′-GGTACATACATTGTCAAATTCTCTCAAG-3′) were used. To investigate the influence of the T-DNA insertion RHM2, cDNA was amplified with the T-DNA spanning primers RHMHA (5′-CAGATTTCAAGGATGGATGATAC-3′) and RHMHB (5′-AGAGTAAGGATTCGTCGGTAACAG-3′).
Molecular Analysis
The GABI-KAT collection was screened by gene code for RHM2 insertions. A putative insertion line was confirmed by PCR with the T-DNA left border-specific primer GLB (5′-GGGAATGGCGAAATCAAGGCATCG-3′) and an RHM2-specific primer (GRHM2, 5′-ATTTCTCCTTTGTGAAGAAYAACC-3′). The resulting PCR product was sequenced with a nested T-DNA-specific primer (G8409, 5′-ATATTGACCATCATACTCATTGC-3′)
T-DNA copy number was determined by Southern-blot analysis using the product of the PCR reaction with the T-DNA-specific primers G8418 (5′-GCAATGAGTATGATGGTCAATATG-3′) and G10706 (5′-GAACCCTAATTCCCTTATCTGGG-3′) as a probe.
The genotype of T2 plants and later generations was determined by a PCR-based approach using either the gene-specific primers RHMHA and RHMHB or one gene-specific primer (RHMHB) and the T-DNA-specific primer G8409.
The integrity of the right border was checked by using the genomic DNA of the mutant as a template for a PCR reaction with the gene-specific primer RHMHA and the T-DNA right border-specific primer G2588 (5′-CGCCAGGGTTTTCCCAGTCACGACG-3′). The resulting PCR product was cloned using the TOPO TA cloning kit (Invitrogen, Karlsruhe, Germany), and two independent clones were partially sequenced using M13 standard primers.
A full-length clone of RHM2 was prepared by using cDNA prepared as described above as template for a PCR reaction with the RHM2-specific primers RFF (5′-ATGGATGATACTACGTATAAGCC-3′) and RFB (5′-TTAGGTTCTCTTGTTTGGTTCAAA-3′). The resulting PCR product was cloned using the TOPO TA cloning kit (Invitrogen) and sequenced.
The Salk collection was queried with the gene code of RHM2 (At1g53500), and a putative insertion line (SALK 139223) was ordered from the Nottingham Stock collection. A homozygous insertion line was identified with the gene-specific primers RTRA and RTRB and the T-DNA-specific primer Lba1 (5′-TGGTTCACGTAGTGGGCCATCG-3′).
Construction and Expression of pRHM2::GUS Fusions
A 2,272-bp sequence upstream of the predicted ATG of RHM2 was PCR amplified with the primers 5′-CTAGTCTAGAGACCTCCACCAAGAGTTCCA-3′ (XbaI) and 5′-GCGCGGATCCTCCTTGAAATCTGCAA-3′ (BamHI). The PCR product was cloned as a BamHI/SpeI fragment into the pBI101.1 vector (Jefferson et al., 1987) in front of a promoterless GUS gene. Plants of the Columbia ecotype were transformed by A. tumefaciens-mediated transformation (Clough and Bent, 1998) and selected by screening for T-DNA-encoded kanamycin resistance. Fifteen transformants for the construct were obtained, and GUS activity was visualized by submerging plant organs in 50 mm sodium phosphate, pH 7.2, containing 0.05% (w/v) 5-bromo-4-chloro-3-indolyl β-d-GlcUA, 10 mm ethylene diamine tetraacetic acid, and 0.1% (v/v) Triton X-100 (Jefferson et al., 1987). Tissues were incubated at 37°C for 30 min up to 16 h depending on staining intensity. Tissues were cleared of chlorophyll by passing through a graded ethanol series.
Seed Analysis
SEM was performed on imbibed and dry mature seeds. The SEM investigations were performed by a JSM 6330 F (JEOL, Tokyo) at an acceleration voltage of 5 kV. Thin layers of about 4 nm of platinum were deposited by sputtering onto the surfaces of the seed to avoid electrical charging during SEM investigations. The SEM images were taken by a digital image processing and recording system.
Resin Embedding for Bright-Field Microscopy
Mature seeds were fixed overnight in an aqueous solution of formaldehyde-acetic acid (5% [v/v] formalin, 5% [v/v] acetic acid, and 67% [v/v] ethanol) at 4°C. The sample was dehydrated and infiltrated with LR-White (London Resin Co., Berkshire, UK) according to the manufacturer's suggestions. For microscopy, 1-μm sections were cut with a glass knife on a Reichert-Jung Ultracut E microtom (Leica, Bensheim, Germany) and mounted on charged glass slides. The sections were stained with 1% (w/v) toluidine blue O in 100 mm sodium phosphate (pH 7.2) for 1 h and washed several times with distilled water. Pictures were visualized with a BX41 microscope (Olympus, Hamburg, Germany) connected to a digital SPOT 2.2.1 camera (Visitron Systems, Puchheim, Germany).
Mucilage Analysis
Mucilage was extracted from 2- to 4-week-old seeds (approximately 5 mg) by suspending them in 1 mL of 0.2% (w/v) ammonium oxalate and shaking for 2 h at 30°C. The released mucilage material was lyophilized and dissolved in water. The content of uronic acids was determined using the m-hydroxybiphenyl colorimetric uronic acid assay (Blumenkrantz and Asboe-Hansen, 1973). The content of neutral monosaccharides was determined by gas chromatography-mass spectrometry analysis of the derivatized alditol acetates (Albersheim et al., 1967).
For Mr determination, dissolved mucilage was concentrated via an Amicon filter (Mr cut off 10,000; Millipore, Bedford, MA), and the solvent was transferred to 0.1 m sodium nitrate/0.05% (w/v) sodiumazide, which represents the eluant for the flow FFF-MALLS-refractive index device (Wyatt, 1993). After centrifugation (14,000g, 5 min), 250 μL (200 μg) of the sample was injected into a symmetrical flow FFF instrument (F-1000, regenerated cellulose membrane, cut off 10 kD; FFFractionation Inc., Salt Lake City, UT). It was then separated by the interaction of a channel flow (1 mL min-1) and a linear cross flow gradient (0-5 min, 1.5 mL min-1; 35-50 min, 0.2 mL min-1) according to the diffusion coefficients of the various molecules. The light-scattering detection was performed with a multi-angle DAWN DSP laser photometer (He-Ne-laser, WTC, Santa Barbara, CA), whereas for the detection of the concentration, an Optilab DSP Interferometric Refractometer (WTC) was used. The data were processed by using the ASTRA software from WTC (v4.73). The extrapolation was carried out by Debye (second order) or by random coil because it allowed a more accurate fit.
Seed Staining
Ruthenium red staining of seeds was performed by placing the seeds in a solution of 0.01% (w/v) ruthenium red and shaking them for 10 min. For dextran blue staining, seeds were first imbibed in water and then covered with a 1% (w/v) aqueous solution of blue dextran (average Mr 2,000,000) for 5 min. Seeds were visualized using a MZ FZ III binocular (Leica) with a mounted SPOT 2.2.1 CCD camera (Visitron Systems).
Identification of Candidate Genes
Proteins potentially involved in the nucleotide sugar pathway were identified as follows. The Pfam HMMs (release 6.6) and the hmmer software package (release 2.2g) were downloaded and installed locally. The Pfam HMMs were indexed with hmmindex, and interesting HMMs were extracted via hmmfetch. These were searched against a flat file containing all predicted Arabidopsis proteins in fasta format which had been downloaded from The Institute for Genomic Research Web site (http://www.tigr.org).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Accession Numbers
The full-length coding sequence of RHM2 has been deposited as AJ565874 at the EMBL Nucleotide Sequence Database. The Sputnik accession numbers for putative RHM protein homologs from soybean (Glycine max), potato (Solanum tuberosum), maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare), and Pinus taeda were: C_BI787599, C_BG88611, C_BM660359, C_BJ268533, S_BI779431, and C_BE241380 for the N-terminal dehydratase like domain, respectively, and C_BM085701, C_BG593445, C_AW018105, C_BJ268533, C_AV916302, and C_BG275831 for the C-terminal domain.
Acknowledgments
We thank Dr. Kim Larsen and Claudia Beyer (Max Planck Institute of Molecular Plant Physiology) for excellent technical assistance; Drs. Alisdair Fernie and Leonard Krall (Max Planck Institute of Molecular Plant Physiology) for critical reading of the manuscript; Dr. Manfred Pinnow (Fraunhofer Institute for Applied Biopolymers, Golm, Germany) for SEM; Drs. Michael Mølhøj and Wolf-Dieter Reiter (University of Connecticut, Storrs); and Drs. Tamara Western and George Haughn (University of British Columbia, Canada) for communicating unpublished results. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutant and the Nottingham Arabidopsis Stock Centre for providing seeds.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.034314.
References
- Albersheim P, Nevins DJ, English PD, Karr A (1967) A method for the analysis of sugars in plant cell wall polysaccharides by gas-liquid chromatography. Carbohydr Res 5: 340-345 [Google Scholar]
- Allard ST, Giraud MF, Whitfield C, Graninger M, Messner P, Naismith JH (2001) The crystal structure of dTDP-d-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J Mol Biol 307: 283-295 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657 [DOI] [PubMed] [Google Scholar]
- Baron-Epel O, Gharyal PK, Schindler M (1988) Pectins as mediators of wall porosity in soybean cells. Planta 175: 389-395 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths- Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30: 276-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G (1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484-489 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD (1997) The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-d-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-l-fucose. Proc Natl Acad Sci USA 94: 2085-2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Hofte H, Truong HN (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577-2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1-30 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Dormann P, Benning C (1998) The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J 13: 641-652 [DOI] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755-763 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005-1016 [DOI] [PubMed] [Google Scholar]
- Feingold DS, Avigad, G (1980) Sugar nucleotide transformations in plants. In PK Stumpf, EE Conn, eds, Carbohydrates: Structure and Function. Academic Press, New York, pp 101-170
- Fincher GB, Stone BA, Clarke AE (1983) Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol 34: 47-70 [Google Scholar]
- Frey-Wyssling A (1976) The Plant Cell Wall. In Encyclopedia of Plant Anatomy, Ed 3. Gebrueder Borntraeger, Berlin
- Giraud MF, Naismith JH (2000) The rhamnose pathway. Curr Opin Struct Biol 10: 687-696 [DOI] [PubMed] [Google Scholar]
- Goto N (1985) A mucilage polysaccharide secreted from testa of Arabidopsis thaliana. Arabidopsis Inf Serv 22: 1-4 [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305-308 [DOI] [PubMed] [Google Scholar]
- Hadi MZK, Wendeler E, Reiss B (2002) Simple and versatile selection of Arabidopsis transformants. Plant Cell Rep 21: 130-135 [Google Scholar]
- Hanke DE, Northcote DH (1975) Molecular visualization of pectin and DNA by ruthenium red. Biopolymers 14: 1-17 [DOI] [PubMed] [Google Scholar]
- Henrissat B, Coutinho PM, Davies GJ (2001) A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol 47: 55-72 [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891-4900 [DOI] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S (2002) A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA 99: 16319-16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Eisenberg D (2002) GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H. O hydrogen bonds and van der waals interactions. J Mol Biol 323: 69-76 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383-396 [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567-580 [DOI] [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163: 16-20 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ et al. (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31: 383-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Wöhner RV, Hummel S, Willmitzer L, Frommer WB (1992) The GUS reporter system as a tool to study plant gene expression. In SR Gallagher, ed, GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. Academic Press, San Diego, pp 23-39
- Moffatt BA, McWhinnie EA, Agarwal SK, Schaff DA (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143: 211-216 [DOI] [PubMed] [Google Scholar]
- Mohnen D (1999) Biosynthesis of pectins and galactomannans. In D Barton, K Nakanishi, O Meth-Cohn, BM Pinto, eds, Comprehensive Natural Products Chemistry, Vol 3. Carbohydrates and Their Derivatives including Tannins, Cellulose, and Related Lignins. Elsevier, Oxford, pp 497-527
- O'Brien TP, Feder N, McCully ME (1964) Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59: 367-373 [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846-849 [DOI] [PubMed] [Google Scholar]
- Oomen RJ, Doeswijk-Voragen CH, Bush MS, Vincken JP, Borkhardt B, van den Broek LA, Corsar J, Ulvskov P, Voragen AG, McCann MC et al. (2002) In muro fragmentation of the rhamnogalacturonan I backbone in potato (Solanum tuberosum L.) results in a reduction and altered location of the galactan and arabinan side-chains and abnormal periderm development. Plant J 30: 403-413 [DOI] [PubMed] [Google Scholar]
- Orsel M, Krapp A, Daniel-Vedele F (2002) Analysis of the NRT2 nitrate transporter family in Arabidopsis: structure and gene expression. Plant Physiol 129: 886-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777-2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Wilkerson C, Keegstra K (2001) Golgi enzymes that synthesize plant cell wall polysaccharides: finding and evaluating candidates in the genomic era. Plant Mol Biol 47: 115-130 [PubMed] [Google Scholar]
- Reiter WD, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47: 95-113 [PubMed] [Google Scholar]
- Rhee SY, Somerville CR (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79-88 [DOI] [PubMed] [Google Scholar]
- Roessner D, Kulicke W-M (1994) On-line coupling of flow field-flow fractionation and multi-angle laser light scattering. J Chromatogr A 687: 249-258 [Google Scholar]
- Rudd S, Mewes HW, Mayer KF (2003) Sputnik: a database platform for comparative plant genomics. Nucleic Acids Res 31: 128-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjot M, Pauly M, Bush MS, Borkhardt B, McCann MC, Ulvskov P (2002) Direct interference with rhamnogalacturonan I biosynthesis in Golgi vesicles. Plant Physiol 129: 95-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SO, Pauly M, Bush M, Skjot M, McCann MC, Borkhardt B, Ulvskov P (2000) Pectin engineering: modification of potato pectin by in vivo expression of an endo-1,4-β-d-galactanase. Proc Natl Acad Sci USA 97: 7639-7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson MB, Hawes MC (1994) Correlation of pectin methylesterase activity in root caps of pea with root border cell separation. Plant Physiol 106: 739-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411: 834-842 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581-590 [DOI] [PubMed] [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998-1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALAZ, TRANSPARENT TESTA GLABRA1 and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga RK, de Mayer MC, Hol WG (1985) Interaction of pyrophosphate moieties with alpha-helixes in dinucleotide binding proteins. Biochemistry 24: 1346-1357 [Google Scholar]
- Willats WG, Steele-King CG, Marcus SE, Knox JP (1999) Side chains of pectic polysaccharides are regulated in relation to cell proliferation and cell differentiation. Plant J 20: 619-628 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM (2000) Arabidopsis seed coat development: morphological differentiation of the outer integument. Plant J 22: 483-493 [DOI] [PubMed] [Google Scholar]
- Wyatt PJ (1993) Light scattering and the absolute characterization of macromolecules. Anal Chim Acta 272: 1-40 [Google Scholar]
- Yokotani-Tomita K, Goto N, Kosemura S, Yamamura S, Hasegawa K (1998) Growth-promoting allelopathic substance exuded from germinating Arabidopsis thaliana seeds. Phytochemistry 47: 1-2a [DOI] [PubMed] [Google Scholar]