Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen capable of forming a biofilm under physiological conditions that contributes to its persistence despite long-term treatment with antibiotics. Here, we report that pathogenic P. aeruginosa strains PAO1 and PA14 are capable of infecting the roots of Arabidopsis and sweet basil (Ocimum basilicum), in vitro and in the soil, and are capable of causing plant mortality 7 d postinoculation. Before plant mortality, PAO1 and PA14 colonize the roots of Arabidopsis and sweet basil and form a biofilm as observed by scanning electron microscopy, phase contrast microscopy, and confocal scanning laser microscopy. Upon P. aeruginosa infection, sweet basil roots secrete rosmarinic acid (RA), a multifunctional caffeic acid ester that exhibits in vitro antibacterial activity against planktonic cells of both P. aeruginosa strains with a minimum inhibitory concentration of 3 μg mL-1. However, in our studies RA did not attain minimum inhibitory concentration levels in sweet basil's root exudates before P. aeruginosa formed a biofilm that resisted the microbicidal effects of RA and ultimately caused plant mortality. We further demonstrated that P. aeruginosa biofilms were resistant to RA treatment under in vivo and in vitro conditions. In contrast, induction of RA secretion by sweet basil roots and exogenous supplementation of Arabidopsis root exudates with RA before infection conferred resistance to P. aeruginosa. Under the latter conditions, confocal scanning laser microscopy revealed large clusters of dead P. aeruginosa on the root surface of Arabidopsis and sweet basil, and biofilm formation was not observed. Studies with quorum-sensing mutants PAO210 (ΔrhlI), PAO214 (ΔlasI), and PAO216 (ΔlasI ΔrhlI) demonstrated that all of the strains were pathogenic to Arabidopsis, which does not naturally secrete RA as a root exudate. However, PAO214 was the only pathogenic strain toward sweet basil, and PAO214 biofilm appeared comparable with biofilms formed by wild-type strains of P. aeruginosa. Our results collectively suggest that upon root colonization, P. aeruginosa forms a biofilm that confers resistance against root-secreted antibiotics.
Pseudomonas aeruginosa, a gram-negative bacterium commonly isolated from soil and water, is renowned for its nutritional and ecological versatility. As an opportunistic human pathogen, P. aeruginosa is a common cause of nosocomial infections and is responsible for persistent infections in immunocompromised individuals and for the chronic lung infections of patients with cystic fibrosis (Govan and Deretic, 1996). P. aeruginosa is also capable of causing serious infections in nonmammalian host species such as insects (Jander et al., 2000), nematodes (Mahajan-Miklos et al., 1999), and plants (Rahme et al., 1995; Silo-Suh et al., 2002). The effectiveness of this organism in causing infection is likely due to a suite of well-regulated virulence factors and defense mechanisms such as multidrug resistance pumps (Chuanchuen et al., 2001) and biofilm formation (Costerton et al., 1999).
Bacterial biofilms are defined as highly structured, surface-attached communities of cells encased within a self-produced extracellular polymeric matrix (Costerton et al., 1995). Most bacteria appear to form biofilms, including P. aeruginosa, and this multicellular mode of growth likely predominates in nature as a protective mechanism against hostile environmental conditions (Costerton et al., 1995; Costerton and Stewart, 2000). P. aeruginosa biofilm is a contributing factor in the persistent and incurable infections of patients with cystic fibrosis, as cells in the biofilm show a characteristically higher degree of resistance to host immune responses and antimicrobial treatments compared with planktonic cells (Costerton et al., 1999; Singh et al., 2000; Drenkard and Ausubel, 2002). It was originally hypothesized that the biofilm acted as a shield that protected the bacterial cells from harsh environmental conditions (Costerton et al., 1999). However, it is now believed that individual cells within the biofilm may have an altered metabolism that renders them more resistant to environmental conditions and prolonged antibiotic treatment. Moreover, the biofilm does not prevent the entrance and diffusion of antibiotics (Stewart, 2003; Walters et al., 2003). In contrast to these negative qualities of biofilm, Pseudomonas fluorescens has been reported to coat plant roots by forming a biofilm, which may protect roots against soil bacterial and fungal pathogens (O'Toole and Kolter, 1998a).
P. aeruginosa biofilm development proceeds through a series of programmed steps. The initial stages of biofilm formation require flagellar motility and type IV pili-mediated twitching for surface attachment and microcolony aggregation (O'Toole and Kolter, 1998a). As the bacterial cells continue to attach and form microcolonies, a mechanism of cell-cell signaling known as quorum sensing has been postulated to play an important role in the development of mature biofilm (Davies et al., 1998). Quorum-sensing systems in gram-negative bacteria use a population-dependent cell-cell signal, generally an acylated homo-Ser lactone molecule, to detect cell density. When the concentration of this autoinducer reaches a critical concentration, it activates a transcriptional regulator that induces specific target genes (Fuqua et al., 1994). P. aeruginosa contains two separate quorum-sensing systems, the las and rhl systems, that are responsible for the regulation of numerous genes (Pesci and Iglewski, 1999). Initial studies have suggested that mature biofilm formation in P. aeruginosa is dependent upon the las quorum-sensing system, but not the rhl system (Davies et al., 1998). Recent efforts have focused on developing methods of preventing or eradicating P. aeruginosa biofilm development to diminish its virulence in patients with cystic fibrosis (Singh et al., 2002).
Given that P. aeruginosa is a natural soil inhabitant and possible plant pathogen (Rahme et al., 1995; Silo-Suh et al., 2002), we explored the root-microbe interaction between this bacterium and two plant species: Arabidopsis and sweet basil (Ocimum basilicum). The area of soil surrounding a plant root represents a unique physical, biochemical, and ecological interface between the roots and the external environment, which is primarily influenced by the root system through chemicals secreted into the surrounding soil (Gleba et al., 1999; Nardi et al., 2000; Bais et al., 2001). We used Arabidopsis as a plant host because it has been shown to be susceptible to P. aeruginosa leaf infection (Rahme et al., 1995). Sweet basil was selected as our second plant system because in a previous communication we reported the isolation and functional characterization of rosmarinic acid (RA; α-o-caffeoyl-3-4-dihydroxyphenyllactic acid), an antimicrobial compound found in sweet basil root exudates and exhibiting potent bactericidal activity against P. aeruginosa (Bais et al., 2002). In the present study, we have developed an experimental system to study P. aeruginosa pathogenicity and biofilm formation by using plant roots as the host, and we explored the capabilities of plant roots to exude antimicrobial compounds into the rhizosphere.
RESULTS
Root Pathogenicity of P. aeruginosa
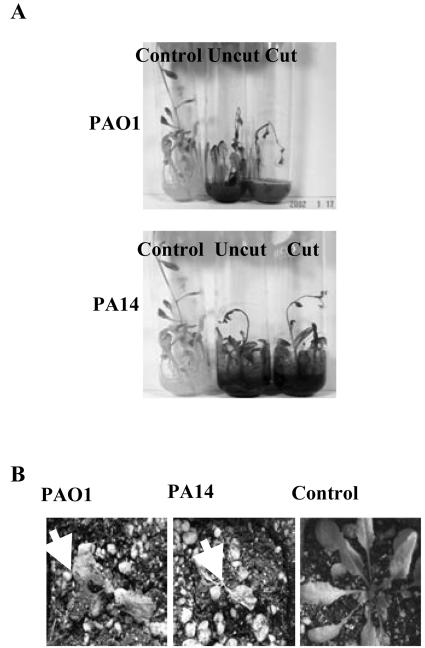
The root pathogenicity of P. aeruginosa strains PAO1 and PA14 was tested in vitro against two plant species: Arabidopsis and sweet basil. Both strains, when infiltrated into the liquid root media of Arabidopsis, caused characteristic disease-like symptoms such as black necrotic regions at the root tips. Arabidopsis infected with strains PAO1 and PA14 displayed progressively heightened symptoms of infection with bacterial cells initially infecting roots and fully developed leaves positioned near the base of the plant 2 to 3 d postinoculation, and spreading systemically to the top of the plant approximately 4 d postinoculation, with plant mortality occurring 7 d postinoculation (Fig. 1A). The degree of disease-like symptoms was unchanged in plants with severed root tips compared with uncut roots, despite the fact that bacterial plant pathogens typically require a wound or natural opening to penetrate tissue. In addition to in vitro studies, we tested the ability of strains PAO1 and PA14 to infect soil-grown Arabidopsis plants. Both strains caused extensive aerial tissue damage, leading to plant mortality approximately 7 d postinoculation when infiltrated into the soil immediately surrounding the root system (Fig. 1B). Both strains also caused plant mortality when infiltrated into attached leaves (data not shown).
Figure 1.
. Virulence of P. aeruginosa strains PAO1 and PA14 against Arabidopsis under in vitro and soil conditions. A, PAO1 and PA14 were infiltrated into the liquid media of cut and uncut plants, and disease symptoms and plant mortality were recorded after 7 d. B, Bacteria were also added to sterile soil of uncut plants and disease symptoms were again recorded after 7 d (arrows indicate aerial tissue damage leading to plant mortality).
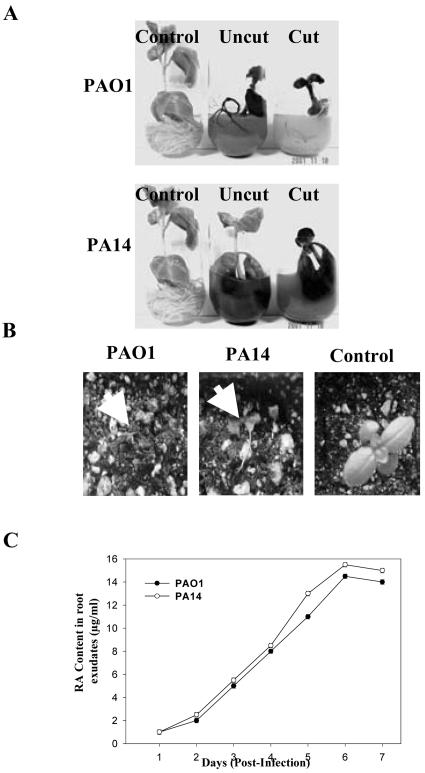
The addition of strains PAO1 and PA14 to the liquid media of in vitro-grown sweet basil plants caused disease-like symptoms similar to those in Arabidopsis, including black necrotic regions at the root tips and water-soaked lesions on leaf tissue, with plant mortality occurring 7 d postinoculation (Fig. 2A). This result was unexpected as we previously showed that RA secreted by sweet basil inhibited the growth of planktonic PAO1 and PA14 under in vitro conditions (Bais et al., 2002) and should therefore protect sweet basil from P. aeruginosa infection. Soil experiments revealed that both strains cause plant mortality after approximately 7 d postinoculation into the soil (Fig. 2B). Sweet basil leaves infiltrated with strains PA14 or PAO1 also caused plant mortality (data not shown).
Figure 2.
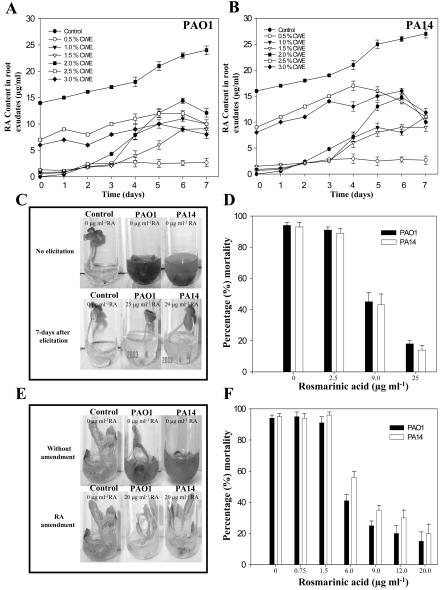
Virulence of P. aeruginosa strains PAO1 and PA14 against sweet basil under in vitro and soil conditions. A, PAO1 and PA14 were infiltrated into the liquid media of cut and uncut plants, and disease symptoms and plant mortality were recorded after 7 d. B, Bacteria were also added to sterile soil of uncut plants and disease symptoms were again recorded after 7 d (arrows indicate aerial tissue damage leading to plant mortality). C, Increased levels of RA (micrograms per milliliter) in the root exudates of sweet basil plants infected with P. aeruginosa was determined daily by HPLC analysis upon infection with PAO1 and PA14.
P. aeruginosa Infection Induces RA Secretion
To reconfirm that RA was secreted from the roots of sweet basil upon P. aeruginosa root infection, in vitro liquid media cultures containing infected sweet basil plants were collected daily for 7 d and were extracted with ethyl acetate. Bacterial cells were removed from the liquid media by centrifugation and filtering. Ethyl acetate extracts were separated by HPLC analysis, and the peak matching the retention time of commercially available RA and RA purified from prior studies (Bais et al., 2002) was collected and confirmed as RA by 1H NMR analysis. The levels of RA present in the root exudates of sweet basil infected with strain PA14 or PAO1 were quantified daily by HPLC analysis (Fig. 2C). RA was detected in the root exudates of infected plants 1 d postinfection, and the maximum concentration (14-15 μg mL-1) occurred 6 d postinfection (Fig. 2C). RA was not detected in the media of control plants (not infected with P. aeruginosa) or in media extracts from pure suspension cultures of PA14 or PAO1.
Antibacterial Activity of RA against PAO1 and PA14
The antibacterial activity of RA was tested against PAO1 and PA14 planktonic cells by the broth microdilution method in 96-well microtiter plates as described in “Materials and Methods.” The minimum inhibitory concentration (MIC) of RA was 3 μg mL-1 for both wild-type strains. The MICs in Murashige and Skoog basal media were comparable with MICs in cation-adjusted Mueller-Hinton broth (data not shown). Although sweet basil plants infected with PAO1 and PA14 secreted RA continuously during the first 6 d postinfection, PAO1 and PA14 planktonic cells were not initially killed because the MIC was not reached in the root exudates until 3 d postinfection (Fig. 2C).
Strains PAO1 and PA14 Form a Biofilm on Root Surfaces
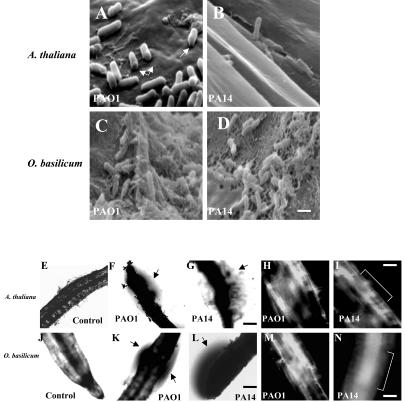
To visualize the cellular mode of attachment of strains PAO1 and PA14 to the root surfaces of Arabidopsis and sweet basil, roots were viewed by scanning electron microscopy (SEM). We observed P. aeruginosa cells attached perpendicularly and horizontally to the root cell walls of each plant species (Fig. 3, A-D). Similar modes of attachment were previously reported for strain PA14 on Arabidopsis leaves (Plotnikova et al., 2000). Bacterial cells from both strains oriented in either position appeared to be degrading and penetrating through the outermost layers of Arabidopsis root cell wall (Fig. 3, A and B). SEM also revealed that both wild-type strains colonized the root surface and produced biofilm-like communities on sweet basil (Fig. 3, C and D). Higher magnification of PAO1 and PA14 biofilms showed bacterial cells embedded within and connected together by an extracellular polymeric matrix (Fig. 3, C and D). Phase contrast microscopy also revealed that roots of Arabidopsis and sweet basil infected with PAO1 and PA14 were surrounded by phase-bright material suggestive of an extracellular matrix (Fig. 3, F, G, K, and L). Similar phase-bright material produced by P. aeruginosa was recently reported as indicative of a biofilm (Hogan and Kolter, 2002). Confocal scanning laser microscopy (CSLM) supported SEM and phase contrast microscopy, depicting an intact biofilm on the root surface of Arabidopsis and sweet basil produced by PAO1 and PA14 (Fig. 3, H, I, M, and N).
Figure 3.
SEM images of Arabidopsis roots infected with strains PAO1 (A) and PA14 (B) attaching perpendicularly to the root cell wall and forming a biofilm layer (arrows in A depict a perforation made by the bacterial strains). SEM of sweet basil roots with PAO1 (C) and PA14 (D) forming a mature biofilm 4 d postinoculation. Scale bar = 5 μm. Phase contrast and confocal images showing uninfected Arabidopsis roots (E) and roots infected with strains PAO1 (F and H) and PA14 (G and I); and uninfected sweet basil roots (J) and roots infected with strains PAO1 (K and M) and PA14 (L and N). Arrows indicate phase-bright material suggestive of a biofilm surrounding the roots; bracket indicates the root. Scale bar = 50 μm.
Effect of RA on P. aeruginosa Biofilm Formation
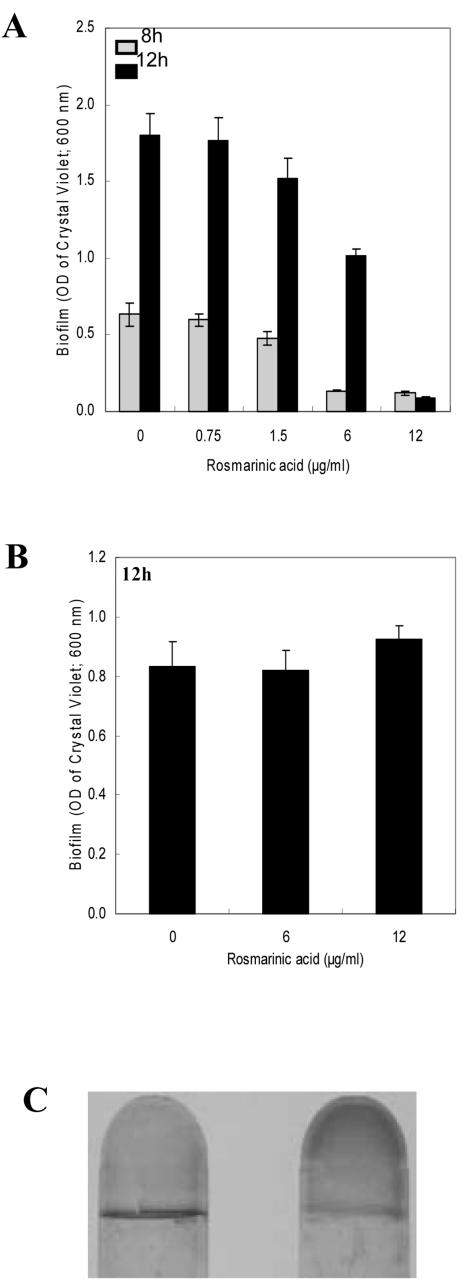
Because P. aeruginosa formed a biofilm on sweet basil root surfaces, we hypothesized that the biofilm may have rendered the bacterial communities resistant to RA; thus, we tested the effect of RA on biofilm development. Figure 4A reveals that sub-MIC levels of RA (0.75 and 1.5 μg mL-1) did not inhibit PA14 biofilm formation; however, it was observed that concentrations of RA above the MIC (6 and 12 μg mL-1) delayed or inhibited biofilm formation (Fig. 4A). RA was also assayed for its ability to disrupt or stop the development of PA14 preformed biofilms. We observed that RA at concentrations inhibiting planktonic growth and biofilm formation (6 and 12 μg mL-1) could not disrupt or prevent the development of preformed biofilms (8 h old; OD600 = 0.32) after 4 h of exposure (Fig. 4B). Interestingly, the biofilm that continued to form under the most elevated concentration of RA (12 μg mL-1) was found to coat the entire surface between the growth medium and the tube instead of producing the typical ring of growth at the air and liquid interface (Fig. 4C), suggesting that an anaerobic biofilm had developed under these conditions.
Figure 4.
Effect of varying concentrations of RA on initiation of biofilm formation by strain PA14 (A) and disruption of preformed PA14 biofilms (B). A, RA was added to the (Bushnell-Haas mineral salts medium supplemented with 0.2% [w/v] dextrose and 0.5% [w/v] tryptone [BDT medium]) from the onset of the incubation period. The range of RA assessed was above and below the MIC (0, 0.75, 1.5, 6.0, and 12 μg mL-1). Biofilm formation was quantified after 8 and 12 h of incubation. B, RA was added to preformed PA14 biofilms by replacing the BDT medium with fresh medium containing RA after 8 h of incubation. The concentrations of RA tested were 0, 6.0, and 12 μgmL-1. The biofilm was then allowed to develop for an additional 4 h (to 12 h) before quantification. Error bars represent ± sd, n = 3. C, Biofilm formed in absence (left) or presence of 12 μg mL-1 of RA (right) when RA was added to the growth medium 8 h after initiation of biofilm development.
Effect of Induction of RA Secretion and Exogenous Supplementation of RA on P. aeruginosa Infectivity
In vitro-grown seedlings of sweet basil were challenged with fungal cell wall elicitors (CWE) to induce root exudation of RA before P. aeruginosa infection (Bais et al., 2002). Elicitation with CWE from Phytophthora cinnamoni at 2% (v/v) induced RA secretion with concentrations ranging from 14.8 to 15.4 μg mL-1 during a 7-d time course (Fig. 5A, B). After 7 d of elicitation, sweet basil plants were subsequently infiltrated with PAO1 and PA14, but elicited plants were unaffected and did not succumb to infection by either strain (Fig. 5, C and D). Furthermore, the levels of RA induced by the CWEs (14.8-15.4 μg mL-1) increased to 24.9 to 28.6 μg mL-1 7 d postinoculation with PAO1 and PA14 (Fig. 5, A and B). Fungal CWE alone did not inhibit the growth of P. aeruginosa (data not shown), confirming the potent antipseudomonal properties of RA.
Figure 5.
A and B, Influence of P. cinnamoni CWE on RA exudation in in vitro-grown cultures of sweet basil. Plants were elicited with varying concentrations CWE for 7 d before inoculation with PAO1 and PA14 (see “Materials and Methods”; values are mean ± sd, n = 5). Day 0 of inoculation corresponds to d 7 postelicitation with CWEs. C, Reduced virulence of P. aeruginosa strains PAO1 and PA14 in sweet basil plants elicited with CWE. D, Mortality rates of elicited sweet basil plants inoculated with strains PAO1 and PA14. Values are mean ± sd, n = 5. E, Reduced virulence of strains PAO1 and PA14 in RA-supplemented Arabidopsis plants compared with an untreated control. F, Mortality rates of RA-supplemented (sub-MIC and above-MIC levels) Arabidopsis plants inoculated with strains PAO1 and PA14. Values are mean ± sd, n = 5.
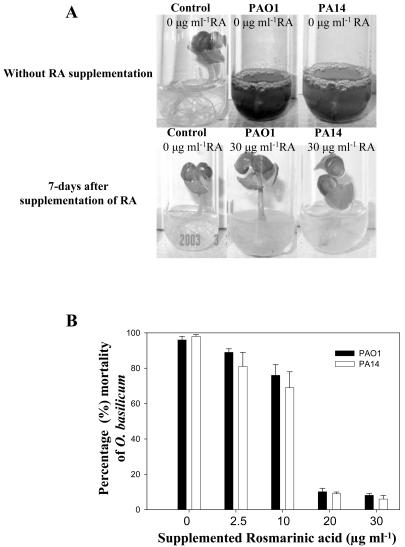
As described previously, Arabidopsis plants that are susceptible to P. aeruginosa infection do not produce or exude RA from their roots. Thus, we tested the effect of exogenously applied RA to the roots of in vitro-grown Arabidopsis plants. Based on the results described in the previous paragraph, our assumption was that exogenous RA would impede P. aeruginosa infection of Arabidopsis. We supplemented the root exudates of Arabidopsis with RA at sub-MIC levels (0.75-1.5 μg mL-1) and with higher-than-MIC levels (6.0-20.0 μg mL-1), and subsequently infiltrated the plants with PAO1 and PA14 as described previously. It was observed that concentrations above MIC levels (6-20.0 μg mL-1) resulted in reduced Arabidopsis mortality from PAO1 and PA14 (Fig. 5, E and F). However, sub-MIC levels of RA did not protect the treated plants against PAO1 and PA14 infections (Fig. 5F). Supplementation of sweet basil root exudates with RA (30 μg mL-1) also resulted in reduced plant mortality rates (Fig. 6, A and B).
Figure 6.
Effect of exogenous supplementation of RA to sweet basil root exudates. A, RA was added to sweet basil root exudates before infection with PAO1 and PA14 at varying concentrations (2.5-30 μg mL-1). Control plants were not supplemented with RA. Strains PAO1 and PA14 were less virulent upon supplementation with RA (30 μg mL-1). B, Mortality rates of sweet basil plants supplemented with RA (sub-MIC and above-MIC levels; 2.5-30 μg mL-1) and inoculated with strains PAO1 and PA14. B, Sweet basil plants were supplemented with RA at concentrations 10-fold greater than MIC levels. Values are mean ± sd, n = 5.
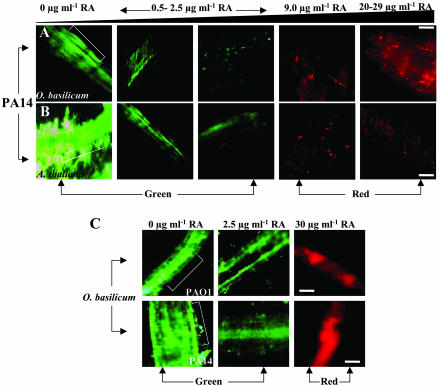
To analyze the interaction between P. aeruginosa and the roots of sweet basil and Arabidopsis that is likely mediated by RA induction and supplementation, we observed the roots of these species by CSLM 4 d after infection with PA14. CSLM results supported our previous data using phase contrast and SEM showing that PAO1 and PA14 form biofilms on the root surfaces of both plants (Fig. 7, A and B). We observed that an increase in exuded or added RA in sweet basil and Arabidopsis root rhizospheres was inversely proportional to the degree of biofilm formation and bacterial survival on the root surfaces (Fig. 7, A and B). Interestingly, large clusters of dead P. aeruginosa (PA14) were found on the root surfaces of the Arabidopsis and sweet basil with increased RA content (Fig. 7, A and B), supporting the potent bactericidal effect of RA. In contrast, Arabidopsis and sweet basil roots cocultured with sub-MIC levels of RA showed no signs of bacterial mortality, instead revealing an intact biofilm that ultimately led to plant mortality (Fig. 7, A and B). Similarly, exogenous supplementation of RA (30 μg mL-1) into sweet basil root exudates prevented PAO1 and PA14 biofilm formation, and clusters of dead bacteria were observed on the root surface. Thus, a definite correlation exists between increased sweet basil survival and diminished PAO1 and PA14 biofilm formation when RA concentrations are increased by induction with CWE or exogenous application of RA to sweet basil root exudates (Fig. 7C). Importantly, administration of RA above MIC levels (20 μg mL-1) 4 d after infection of Arabidopsis roots with PA14 did not prevent plant mortality (data not shown), indicating the inability of RA to affect the established biofilm communities.
Figure 7.
CSLM images of P. aeruginosa biofilms on in vitro-grown sweet basil (A) and Arabidopsis (B) roots. Bacterial viability was visualized by staining with a LIVE/DEAD BacLight Bacterial Viability kit (Molecular Probes, Eugene, OR): red areas indicate dead bacteria, and green areas indicate live bacteria (brackets represent root; scale bars = 50 μm. A, Influence of P. cinnamoni CWEs on sweet basil RA exudation and subsequent inoculation with PA14. Increasing concentrations of P. cinnamoni CWEs (2.0%-2.5%, v/v) resulted in PA14 mortality as shown in red. B, Effect of exogenous RA (0.75-20.0 μg mL-1) on PA14 biofilm formation on infected Arabidopsis roots. C, CSLM image of P. aeruginosa biofilm on in vitro-grown sweet basil roots supplemented with RA. C, Effect of exogenous RA (2.5 and 30 μg mL-1) on PAO1 and PA14 biofilm formation on infected sweet basil roots. Arabidopsis and sweet basil roots exhibited no autofluorescence.
Collectively, these results show that RA may account for reduced P. aeruginosa infectivity when MIC levels of this antibiotic are present before P. aeruginosa infection and/or before the development of a biofilm.
Role of lasI and rhlI Quorum-Sensing Systems in P. aeruginosa Plant Pathogenicity and RA Resistance
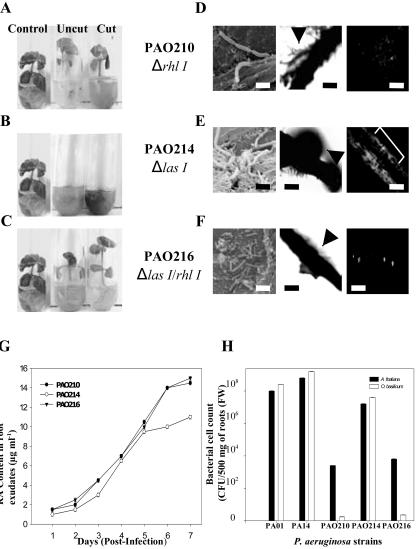
The above results strengthened our hypothesis that biofilm development contributed to P. aeruginosa resistance to RA. To further test this idea, we performed in vitro root pathogenicity assays with quorum-sensing mutants of strain PAO1: PAO210 (ΔrhlI), PAO214 (ΔlasI), and PAO216 (ΔlasIΔrhlI). Arabidopsis plants were susceptible to all quorum-sensing mutants, displaying similar symptoms of infection as observed when infected with PAO1 and PA14, and succumbing to infection 7 d postinoculation (data not shown). However, with PAO210 and PAO216, fewer bacterial cells were attached to the root surfaces, and biofilm development appeared to be diminished (data not shown). It should be reemphasized that Arabidopsis does not produce or secrete RA from its roots. SEM of Arabidopsis root surfaces infected with quorum-sensing mutants revealed perpendicular and horizontal attachment.
The ability of quorum-sensing mutants to infect the roots of sweet basil differed significantly from their ability to infect Arabidopsis. PAO214 (ΔlasI) was the only mutant to cause black necrotic regions at the root tips and water-soaked lesions on leaf tissue, and sweet basil mortality occurred approximately 7 d postinoculation, whereas mutants PAO210 and PAO216 had no adverse effects on plant growth (Fig. 8, A-C). SEM of sweet basil plant roots inoculated with quorum-sensing mutants revealed bacterial cells attached to the root surface and a similar mode of attachment to PAO1 and PA14 (Fig. 8, D-F). Notably, PAO214 appeared to form a nearly complete biofilm, whereas the biofilm formed by PAO210 and PAO216 appeared less developed with only minimal amounts of extracellular polymeric matrix visible compared with PAO1 and PA14 (Fig. 8, D-F). Phase contrast microscopy of sweet basil roots infected with quorum-sensing mutants revealed some phase-bright material surrounding the roots (Fig. 8, D-F). CSLM also supported SEM and phase contrast microscopy depicting an intact but incomplete biofilm on the root surface of Arabidopsis and sweet basil by PAO214 (data not shown for Arabidopsis; Fig. 8, D-F). Interestingly, all of the quorum-sensing mutant bacterial cells visualized by SEM appeared elongated and, in certain instances, showed cellular division (Fig. 8, D-F).
Figure 8.
Infection and mortality of sweet basil inoculated with three P. aeruginosa quorum-sensing mutants. A through C, Effect of P. aeruginosa quorum-sensing mutants in sweet basil infections assessed 7 d postinoculation. D through F, In vitro root pathogenicity in sweet basil: corresponding SEM (scale bar = 5 μm), phase contrast (scale bar = 50 μm), and CSLM images (scale bar = 50 μm). E, Strain PAO214 on sweet basil roots reveals a nearly complete biofilm compared with PAO210 and PAO216 (arrows indicate phase-bright material suggestive of a biofilm surrounding the roots; bracket indicates the root). G, RA content (micrograms per milliliter) in sweet basil root exudates infected with P. aeruginosa quorum-sensing mutants as determined daily by HPLC analysis. H, Bacterial cell counts on Arabidopsis and sweet basil roots 4 d postinfection with strains PAO1, PA14, and three quorum-sensing mutants. Average values were plotted (mean ± sd; n = 5) after inoculation with 105 bacteria per seedling. Five plants of each species were used per treatment.
HPLC analysis of root exudates after infection with the mutants showed an RA production (Fig. 8G) similar to that of sweet basil infected with PAO1 and PA14 (Fig. 2C). The three quorum-sensing mutants (planktonic cells) exhibited the same RA MICs (approximately 3 μg mL-1) as those observed with the wild-type strains (data not shown). RA induction and supplementation above MIC levels with sweet basil and Arabidopsis before infection with quorum-sensing mutants resulted in plant survival (data not shown). When we assessed the effect of RA on biofilm development by the quorum-sensing mutants, RA concentrations below MIC levels (0.75 μg mL-1) did not inhibit PAO214 (ΔlasI) biofilm formation (data not shown). Furthermore, similar to the results obtained with PA14, RA concentrations above MIC levels (6 and 12 μg mL-1) prevented PAO214 biofilm formation. In contrast, minimal biofilm formation was observed for PAO210 (ΔrhlI) and PAO216 (ΔlasI ΔrhlI) treated with RA above-MIC levels and in the control samples with no administration of RA, indicating that rhlI and lasIrhlI mutants have an impaired ability to form biofilm (data not shown). At higher-than-MIC levels (6 and 12 μg mL-1) and after 12 h of incubation, RA did not disturb or prevent the development of preformed biofilm in PAO214 (ΔlasI; data not shown). Mutants PAO210 (ΔrhlI) and PAO216 (ΔlasI ΔrhlI) produced minimal biofilm, and thus the disruptive effect of RA on these biofilms was not significant.
Correlation of Cell Counts of PA14 and Quorum-Sensing Mutants with Root Pathogenicity in Arabidopsis and Sweet Basil
To evaluate the number of bacterial cells associated with biofilm formation in planta, we analyzed the cell counts on the root surface of Arabidopsis and sweet basil on the 4th d after infection (Fig. 8H). Interestingly, the number of bacteria recovered from infected roots was relatively constant for a given strain. Strains PAO1, PA14, and PAO214 (ΔlasI) produced a high number of counts in planta on Arabidopsis (approximately 1.98 × 107 [±16%], 2.27 × 108 [±21%], and 1.26 × 107 [±12%]) and sweet basil (approximately 1.2 × 108 [±14%], 2.81 × 108 [±19%], and 2.1 × 107 [±15%]) 500 mg-1 of analyzed roots (Fig. 8H). There was a positive correlation between the bacterial cell counts on the root surface, the degree of biofilm formation, and pathogenicity of PAO1, PA14, and PAO214 (ΔlasI) on Arabidopsis and sweet basil. In Arabidopsis, fewer (approximately 104-fold) bacteria were typically obtained from roots infected with PAO210 (ΔrhlI; 2.86 × 103 [±12%]) and PAO216 (ΔlasI ΔrhlI; 1.75 × 104 [±18%]) compared with PAO1- and PA14-infected roots (Fig. 8H). In contrast, we observed a dramatic decrease (approximately 103-fold) in the bacterial count from sweet basil roots infected with PAO210 (ΔrhlI; 2.1 × 101 [±14%]) and PAO216 (ΔlasI ΔrhlI; 4.3 × 101 [±17%]) compared with Arabidopsis roots (Fig. 8H).
DISCUSSION
In the present study, we have found that P. aeruginosa clinical strains PAO1 and PA14 can infect the roots of two plant species: Arabidopsis and sweet basil. Infection of sweet basil was surprising, as this plant secretes the potent antimicrobial RA when challenged with P. aeruginosa (Bais et al., 2002). Plant infection and subsequent mortality due to P. aeruginosa was traced to the formation of a biofilm colonizing the root surface. This phenomenon is similar to P. aeruginosa biofilms that form on lung tissue and likely possess an altered metabolism that renders them resistant to antibiotic treatment in patients with cystic fibrosis (Costerton et al., 1999; Silo-Suh et al., 2002; Coleman et al., 2003). Although it has previously been reported that strains PAO1 and PA14 are capable of infecting Arabidopsis and lettuce (Lactuca sativa) leaves, respectively (Plotnikova et al., 2000; Rahme et al., 1995), this is the first report describing how these pathogens can use biofilm formation to evade the antimicrobial effects of root-secreted secondary compounds.
The pathogenicity of strains PAO1 and PA14 to the roots of Arabidopsis and sweet basil in vitro and in soil indicates that P. aeruginosa virulence in vitro and in soil are similar, and that our experimental system is a reliable method to further study the interaction between P. aeruginosa and plant roots. Because PAO1 and PA14 were capable of causing the mortality of Arabidopsis and sweet basil, we investigated the bacterial interactions with the roots using SEM, phase-contrast microscopy, and CSLM. As previously reported for P. aeruginosa attached to Arabidopsis leaves (Plotnikova et al., 2000), we observed PAO1 and PA14 cells attached perpendicularly to the root cell walls of Arabidopsis and sweet basil and forming small holes approximately the same diameter as the bacterial cells (Fig. 3). Furthermore, SEM, CSLM, and phase-contrast microscopy of root tissue confirmed the formation of biofilms on the root surface of both plant species (Fig. 3).
Biofilm-forming P. aeruginosa are often resistant to antibiotic treatment, and this mode of growth likely contributes to the persistent and often lethal infections in individuals stricken with cystic fibrosis (Costerton et al., 1999; Singh et al., 2000; Drenkard and Ausubel, 2002). Several mechanisms of P. aeruginosa's biofilm-mediated resistance to antibiotics have been postulated (Mah and O'Toole, 2001). Recently, Drenkard and Ausubel (2002) showed that antibiotic-resistant phenotypic variants of P. aeruginosa persist in vitro and in the lungs of patients with cystic fibrosis. A second hypothesis to account for biofilm resistance postulates that certain cells within the biofilm are metabolically less active, and thus exhibit less susceptibility to antibiotics compared with the metabolically active cells within the biofilm (Evans et al., 1990; Xu et al., 2000). A third possible mechanism suggests that the presence of the extracellular polymeric matrix retards the diffusion of antibiotics within the biofilm; however, antibiotics have shown various rates of diffusion through biofilm depending on the chemical nature of the compound and the thickness of the biofilm. Such results suggest that in some cases the antibiotic may freely penetrate the biofilm and that other factors such as altered metabolism inside the biofilm may account for resistance (Darouiche et al., 1994; Stewart, 1996). We believe a combination of the above-mentioned mechanisms likely contributes to the reduced activity of RA against biofilm compared with its activity against planktonic cells. Consistent with the third mechanism, it is notable that RA, a polyphenolic organic acid, is in addition likely to be repelled by the high density of negative charges on the alginate polymer that composes much of the P. aeruginosa biofilm. Finally, the intriguing observation that an apparently anaerobic biofilm mode of growth was selected by strain PA14 when elevated concentrations of RA were present is a further indication that the altered metabolism of anaerobic bacteria may be involved with resistance to RA (Yoon et al., 2002).
The MIC of RA for all the P. aeruginosa strains tested was 3 μg mL-1; however, P. aeruginosa planktonic cells were initially able to colonize the root surfaces because MIC levels of RA were not achieved in the root exudates of sweet basil in the first 3 d postinfection (Fig. 2C). These data suggest that because sub-MIC levels of RA do not inhibit planktonic growth or initial biofilm formation (Fig. 4A), P. aeruginosa was able to form an antibiotic-resistant biofilm on the sweet basil roots before greater-than-MIC levels of RA were present in the root exudates. Furthermore, once MIC levels were achieved in the root exudates, we observed that RA has no inhibitory effect on pre-established biofilm communities (Fig. 4B). In contrast, when RA was preinduced or supplemented in concentrations above MIC levels before adding the planktonic cells of P. aeruginosa, we did not observe plant mortality but instead found bacterial mortality due to RA's bactericidal activity. Furthermore, the degree of biofilm formation and root pathogenicity correlated with the bacterial cell counts on the root surface. Similar to our results, a recent study indicated that P. aeruginosa infection and biofilm development in cystic fibrosis airways actually occurs in an anaerobic environment (Yoon et al., 2002), possibly shedding light on the partially anaerobic conditions under which P. aeruginosa was able to form a biofilm on infected roots in vitro and in the soil. Thus, our results suggest that P. aeruginosa biofilm may be inherently resistant to root-secreted antibiotics such as RA, in contrast to planktonic cells.
Prior studies have shown that biofilms produced by quorum-sensing mutants of P. aeruginosa are not fully developed, leaving bacterial cells more susceptible to antimicrobials (Davies et al., 1998; Shih and Huang, 2002). However, Heydorn et al. (2002) reported that biofilm formation in a lasI mutant is nearly undistinguishable from wild-type biofilm, which is in accordance with our observations. Our studies with quorum-sensing mutants also indicate that virulence factors regulated by the rhl system may be important for the full expression of P. aeruginosa pathogenicity toward sweet basil.
The antimicrobial activity of RA against P. aeruginosa deserves further attention. In a recent communication, we reported that P. aeruginosa treated with RA showed nucleoid damage with an increase in spatial division and condensation of genetic material (Bais et al., 2002), suggesting that its bactericidal activity acts at the genetic level rather than merely affecting the cell wall or ion gradients due to plasmolysis. Additionally, we observed that P. aeruginosa cells treated with RA exhibited a short burst of prolific cellular division just before cell death (Bais et al., 2002). In the present study, quorum-sensing mutants that could not infect sweet basil also appeared elongated and divided after exposure to RA secreted from sweet basil roots. Although its mechanism of action is not yet fully understood, it seems likely that one of the catecholic functions on either extremity plays a part in RA effectiveness against P. aeruginosa. Studies on synthetic catecholic-containing β-lactams and pyoverdin derivatives point to catechol-forming iron complexes as important in facilitating membrane transport (Minnick et al., 1992; Hennard et al., 2001). Although the carboxylic acid function could also be important in this antibacterial effect, we hypothesize that structural and functional modifications of this group in RA may enhance its ability to kill cells contained within the alginate biofilm or decrease the MIC so that P. aeruginosa cells do not have time to initiate biofilm formation.
The effective antimicrobial activity of RA (MIC approximately 3 μg mL-1) against planktonic P. aeruginosa demonstrates a potentially new approach to antimicrobial discovery using the inducible bio-synthetic and secretory capabilities of plant root systems. Additionally, these findings indicate that our system can possibly be used as a model for studying the pathogenicity of P. aeruginosa using plant roots as the pathogen host. As shown here, it appears that P. aeruginosa strains that can infect animals and plants form characteristic biofilms when infecting plant roots and likely use this extracellular matrix to resist antimicrobial factors. Incidentally, P. aeruginosa biofilm inhibitors have been recently identified in human mucous secretions (Singh et al., 2002); thus, it might be possible to adapt our methods to screen root exudates for biofilm inhibitors. Such inhibitors might greatly enhance the effectiveness of known antibacterial agents used against P. aeruginosa infections in humans.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of wild-type Arabidopsis ecotype Columbia (Col-O) were obtained from Lehle Seeds (Round Rock, TX). Seeds of sweet basil (Ocimum basilicum) were obtained from Shepherd's Garden Seeds (Torrington, CT). Seeds were surface sterilized using commercial sodium hypochlorite (0.3%, v/v) for 10 to 12 min and were then washed four times in sterile double-distilled water. Surface-sterilized seeds were placed on static Murashige and Skoog (Murashige and Skoog, 1962) basal media in petri dishes for germination and were incubated in a growth chamber. Fifteen-day-old seedlings of each species were individually transferred to 50-mL culture tubes containing 5 mL of liquid Murashige and Skoog basal media. Plant cultures were maintained on an orbital platform shaker (Lab-Line Instruments, Melrose Park, IL) set at 90 rpm with a photoperiod of 16 h of light and 8 h of dark at 25°C ± 2°C.
Bacterial Strains and Culture Conditions
The following Pseudomonas aeruginosa strains were used in this study: PAO1 and PA14, both wild-type clinical isolates, were obtained from the laboratories of Frederick M. Ausubel and Herbert P. Schweizer, respectively. Quorum-sensing mutants PAO210 (ΔrhlI), PAO214 (ΔlasI), and PAO216 (ΔlasI ΔrhlI) were derived from PAO1 as previously described (Hoang et al., 2000). Freshly plated cells from frozen stock cultures were used for all experiments. All strains were plated on Luria-Bertani (LB) agar and were incubated at 37°C. Plated cells were suspended in 5 mL of LB broth for overnight growth at 37°C and were shaken at 250 rpm.
In Vitro Root Pathogenicity Assay
Twenty-five-day-old Arabidopsis and sweet basil plants were used for the in vitro root pathogenicity assays. P. aeruginosa strains were grown to OD600 = 0.3 to 0.4 and were added separately to the 5 mL of Murashige and Skoog media of each plant species to reach an initial OD600 = 0.02. Before the addition of the bacterial suspensions, root tips from one-half of the population of Arabidopsis and sweet basil were severed because plant pathogens require a wound or natural opening to penetrate the plant cell wall. Murashige and Skoog basal media (5 mL) without plant material was inoculated with the same volume of each bacterial strain tested. A noninfected plant control was maintained under the same conditions. All of the treatments and controls were incubated at 30°C in a controlled environment incubator shaker (New Brunswick Scientific, Edison, NJ) set at 30 rpm with a photoperiod of 16 h of light and 8 h of dark. Root tissues (500 mg of fresh weight basis) of Arabidopsis and sweet basil infected with PAO1, PA14, PAO210 (ΔrhlI), PAO214 (ΔlasI), and PAO216 (ΔlasI ΔrhlI) were rinsed with water and homogenized in 1 mL of saline (0.9% [w/v] sodium chloride) with a tissue grinder (Kontes, size C), and the suspension was serially diluted in saline and plated to determine bacterial cell counts as previously described (Rahme et al., 1995). Each experiment was conducted in triplicate.
Media Extraction, HPLC, and 1H NMR Analysis
Media extracts (5 mL) from all treatments were collected daily for 7 d. Extracts were centrifuged at 10,000g for 10 min at room temperature to recover supernatants. Collected supernatants were then passed through a 0.45-μm nylon syringe filter to remove cellular debris (Scientific Resources, Newton, MA) and were extracted with 3 mL of HPLC-grade ethyl acetate (Fisher Scientific, Pittsburgh) at room temperature for 24 h. The ethyl acetate extract was collected and concentrated under vacuum and was dissolved in 200 μL of HPLC-grade absolute methanol (Fisher Scientific). HPLC and 1H NMR conditions for RA analysis were followed as described previously (Bais et al., 2002). All extractions were conducted in triplicate, and the entire experiment was repeated twice.
Antibacterial Assays with RA
MICs of RA against planktonic cells of P. aeruginosa were determined by the broth microdilution method using an inoculum of approximately 1 × 105 cfu mL-1. Microtiter plates (96-well; Nalge Nunc International, Rochester, NY) were prepared with serial 2-fold dilutions of RA in cation-adjusted Mueller-Hinton broth (Difco Laboratories, Detroit). RA was added from a 1 mg mL-1 stock solution in dimethyl sulfoxide. The MIC was visually defined as the lowest concentration of an antibiotic that completely inhibited cell growth after incubation for 22 h at 37°C. All susceptibility trials were conducted in triplicate.
Biofilm Formation Assay
The effect of RA on biofilm formation by strains PA14, PAO210 (ΔrhlI), PAO214 (ΔlasI), and PAO216 (ΔlasI ΔrhlI) was investigated using a crystal violet assay as previously described (Déziel et al., 2001), except that polypropylene tubes were used instead of polystyrene tubes. Briefly, tubes containing 500 μL of a one-one-hundreth dilution of an overnight LB broth culture in BDT medium were incubated statically at 30°C. Biofilms were quantified by crystal violet staining followed by ethanol solubilization and OD measurement at 600 nm (O'Toole and Kolter, 1998b; Déziel et al., 2001). Two different treatments were tested. In the first experimental condition, RA was added in the BDT medium from the onset of the incubation period. The concentrations of RA assessed were above and below the MIC (0, 0.75, 1.5, 6.0, and 12 μg mL-1). The volume of added dimethyl sulfoxide was adjusted so that all tubes contained the same concentration of the solvent. Biofilm formation was quantified after 8 and 12 h of incubation. For the second assay, RA was added to preformed biofilms of PA14 by replacing the BDT medium with fresh medium containing RA after 8 h of incubation. The concentrations of RA tested were 0, 6.0, and 12 μg mL-1. The biofilm was then allowed to develop for an additional 4 h before the biofilm was quantified. All treatments were conducted in triplicate.
Leaf and Soil Pathogenicity Assay
Seeds of Arabidopsis and sweet basil were surface sterilized and germinated as described previously (see “Materials and Methods”). Fifteen-day-old seedlings of each species were transplanted from static Murashige and Skoog media to 10-cm black plastic pots containing 50 g (dry weight) of PM-O5 Arabidopsis-growing medium (Lehle Seeds). Plants were incubated in a growth chamber at 30°C with 12 h of light and were watered daily for 2 weeks before inoculation with bacteria. For leaf assays, P. aeruginosa strains were grown in LB at 37°C to OD600 = 0.2 to 0.3 and were diluted 1:100. Diluted suspensions were individually injected with the blunt end of a hypodermic needle into intact leaves of Arabidopsis and sweet basil at a dose of approximately 1 × 103 cfu/cm2 as previously described (Rahme et al., 1995). Infiltrated plants were incubated in a growth chamber at 30°C and 80% relative humidity with 16 h of light and 8 h of dark. For soil infiltration, 50 g of soil with Arabidopsis and sweet basil was flooded with 10 mL of bacterial suspension to give an inoculum concentration of 1 to 3 × 107 cfu g-1 of soil. Plants were incubated under identical conditions as those used for leaf infiltration assays.
Induction of RA Secretion and Exogenous Supplementation of RA
In vitro seedlings of sweet basil grown in 5 mL of Murashige and Skoog basal media in 40-mL culture tubes were elicited with fungal cell wall preparations. As previously reported, fungal CWE from Phytophthora cinnamoni were used to induce root secretion of RA by sweet basil before infection with P. aeruginosa (Bais et al., 2002). The fungal CWE were prepared and used according to Bais et al. (2002). Fungal CWE were administered at various volumes (1-3 mL) into 40-mL culture tubes containing 5 mL of Murashige and Skoog basal media. A time-course study of the influence of CWE on root exudation of RA was conducted by harvesting the media on a daily basis for 1 week; a nonelicited control was also harvested during the same period and was analyzed for RA content in the root exudates. RA content in the root exudates was analyzed by HPLC as described previously. Sweet basil plants were infected with PAO1 and PA14 7 d after the elicitation with fungal CWEs. The mortality rate for each plant was determined 7 d after infection with the P. aeruginosa strains. In the case of Arabidopsis, RA (0.75-20 μg mL-1) was added exogenously to 40-mL culture tubes containing 5 mL of Murashige and Skoog basal media in which the Arabidopsis plants were floating. Plants were subsequently infected with PAO1 and PA14, and were checked for mortality as described previously (see in vitro root pathogenicity assay). The infection assay with exogenous application of RA to sweet basil was similar as described for Arabidopsis except for the range of RA supplemented (2.5-30 μg mL-1). Each experiment was repeated twice with five replicates each.
Microscopy
Phase contrast images of P. aeruginosa-infected root tissues were captured with a 10× objective on an microscope (BX60; Olympus, Melville, NY) equipped with imaging software (CoolSnap; San Diego) as described previously (Hogan and Kolter, 2002). For SEM, segments of Arabidopsis and sweet basil roots were rinsed with water and were fixed in 4% (w/v) paraformaldehyde and passed through increasing concentrations of ethanol (30%, 50%, 70%, 96%, and 100% [w/v]). The fixed roots were dried in a Samdri-PVT-3B critical point drying apparatus, mounted on stubs, coated with a 12-nm layer of gold-palladium in a Hummer-II sputter coater, and visualized using a scanning electron microscope-FEI (505; Philips, Eindhoven, The Netherlands). Phase contrast microscopy and SEM were performed 4 d postinoculation. For observation of P. aeruginosa biofilm by CSLM, P. aeruginosa-colonized root tissues of sweet basil and Arabidopsis were rinsed with water and analyzed for fluorescence with a confocal laser microscope (Fluroview LGPS-2; Olympus) equipped with imaging software (CoolSnap). CSLM was performed 4 d postinoculation using a LIVE-DEAD BacLight Bacterial Viability kit (Molecular Probes) by incubating P. aeruginosa-colonized Arabidopsis and sweet basil roots at room temperature in the dark for 15 min, according to the manufacturer's manual. The samples were mounted with Citifluor antifading (Sigma-Aldrich, St. Louis) and were observed for fluorescence. Each experiment was repeated twice with three replicates each.
Acknowledgments
We thank Dr. Frank R. Stermitz for interpreting 1H NMR results and Andrea Remick for technical assistance.
This work was supported by the Colorado State University Agriculture Experiment Station (to J.M.V.), by a National Science Foundation-Faculty Early Career Development Award (CAREER; grant no. MCB 0093014 to J.M.V.), by the National Institutes of Health (grant no. GM56685 to H.P.S.), by the Department of Energy (grant no. DE-FG03-97ER20274 to R.F.), and by the Canadian Institutes of Health Research (to E.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027888.
References
- Bais HP, Loyola Vargas VM, Flores HE, Vivanco JM (2001) Root-specific metabolism: the biology and biochemistry of underground organs. In Vitro Cell Dev Biol Plant 37: 730-741 [Google Scholar]
- Bais HP, Walker TS, Schweizer HP, Vivanco JM (2002) Root-specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant Physiol Biochem 40: 983-995 [Google Scholar]
- Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkoff-Schweizer RR, Schweizer HP (2001) Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants over expressing MexCD-OprJ. Antimicrob Agents Chemother 45: 428-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman FT, Mueschenborn S, Meluleni G, Ray C, Carey VJ, Vargas SO, Cannon CL, Ausubel FM, Pier GB (2003) Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc Natl Acad Sci USA 100: 1949-1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49: 711-745 [DOI] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS (2000) Bacterial biofilms. In JP Nataro, MJ Blaser, S Cunningham-Rundles, eds, Persistent Bacterial Infections. American Society of Microbiologists, Washington, DC, pp 423-439
- Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318-1322 [DOI] [PubMed] [Google Scholar]
- Darouiche RO, Dhir A, Miller AJ, Landon GC, Raad II, Musher DM (1994) Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Disease 170: 720-723 [DOI] [PubMed] [Google Scholar]
- Davies GD, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295-298 [DOI] [PubMed] [Google Scholar]
- Déziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183: 1195-1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM (2002) Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416: 740-743 [DOI] [PubMed] [Google Scholar]
- Evans DJ, Brown MR, Allison DG, Gilbert P (1990) Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J Antimicrob Chemother 25: 585-591 [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba D, Borisjuk NV, Borisjuk LG, Kneer R, Poulev A, Skarzhinskaya M, Dushenkov S, Logendra S, Gleba YY, Raskin I (1999) Use of plant roots for phytoremediation and molecular farming. Proc Natl Acad Sci USA 96: 5973-5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Deretic V (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60: 539-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennard C, Truong QC, Desnottes JF, Paris JM, Moreau NJ, Abdallah MA (2001) Synthesis and activities of pyoverdin-quinolone adducts: a prospective approach to a specific therapy against Pseudomonas aeruginosa. J Med Chem 44: 2139-2151 [DOI] [PubMed] [Google Scholar]
- Heydorn A, Ersboll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S (2002) Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol 68: 2008-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Kutchma AJ, Becher A, Schweizer HP (2000) Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43: 59-72 [DOI] [PubMed] [Google Scholar]
- Hogan A, Kolter R (2002) Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296: 2229-2232 [DOI] [PubMed] [Google Scholar]
- Jander G, Rahme LG, Ausubel FM (2000) Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182: 3843-3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34-39 [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan M-W, Rahme LG, Ausubel FM (1999) Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96: 47-56 [DOI] [PubMed] [Google Scholar]
- Minnick AA, Mckee JA, Dolence EK, Miller MJ (1992) Iron transport-mediated antibacterial activity of and development of resistance to hydroxamate and catechol siderophore-carbacephalosporin conjugates. Antimicrob Agents Chemother 36: 840-850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tissue culture. Physiol Plant 15: 473-497 [Google Scholar]
- Nardi S, Concheri G, Pizzeghello D, Sturaro A, Rella R, Parvoli G (2000) Soil organic matter mobilization by root exudates. Chemosphere 5: 653-658 [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R, (1998a) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30: 295-304 [DOI] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R (1998b) Initiation of biofilm in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol 28: 449-461 [DOI] [PubMed] [Google Scholar]
- Pesci EC, Iglewski BH (1999) Molecular mechanisms of signalling in Pseudomonas aeruginosa. In G Dunny, SC Winans, eds, Cell-Cell Signaling in Bacteria. American Society of Microbiologists, Washington, DC, pp 147-155
- Plotnikova JM, Rahme LG, Ausubel FM (2000) Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol 124: 1766-1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Calderwood SB, Ausubel FM (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268: 1899-1902 [DOI] [PubMed] [Google Scholar]
- Shih P-C, Huang C-T (2002) Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J Antimicrob Chemother 49: 309-314 [DOI] [PubMed] [Google Scholar]
- Silo-Suh L, Suh S-J, Sokol PA, Ohman DE (2002) A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc Natl Acad Sci USA 99: 15699-15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Parsek MR, Greenberg EP, Welsh MJ (2002) A component of innate immunity prevents bacterial biofilm development. Nature 417: 552-555 [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407: 762-764 [DOI] [PubMed] [Google Scholar]
- Stewart PS (1996) Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother 40: 2517-2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS (2003) Diffusion in biofilms. J Bacteriol 185: 1485-1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC III, Roe F, Bugnicourt A, Franklin MJ Stewart PS (2003) Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47: 317-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu KD, McFeters GA, Stewart PS (2000) Biofilm resistance to antimicrobial agents. Microbiology 146: 547-549 [DOI] [PubMed] [Google Scholar]
- Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U et al. (2002) Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3: 593-603 [DOI] [PubMed] [Google Scholar]