Abstract
In Arabidopsis, asparagine (Asn) synthetase is encoded by a small gene family (ASN1, ASN2, and ASN3). It has been shown that ASN1 and ASN2 exhibit reciprocal gene expression patterns toward light and metabolites. Moreover, changes in total free Asn levels parallel the expression of ASN1, but not ASN2. In this study, we show that ASN2 expression correlates with ammonium metabolism. We demonstrate that the light induction of ASN2 is ammonium dependent. The addition and removal of ammonium exerted fast and reciprocal effects on the levels of ASN2 mRNA, specifically under light-grown conditions. NaCl and cold stress increased cellular free ammonium and ASN2 mRNA levels in a coordinated manner, suggesting that the effects of stress on ASN2 expression may be mediated via accumulation of ammonium. The correlation between ASN2 and cellular ammonium metabolism was further demonstrated by analysis of ASN2 transgenic plants. When plants were grown on Murashige and Skoog medium containing 50 mm ammonium, ASN2 overexpressors accumulated less endogenous ammonium compared with the wild-type Colombia-0 and ASN2 underexpressors. When plants were subjected to high-light irradiance, ammonium levels built up. Under such conditions, ASN2 underexpressors accumulated more endogenous ammonium than the wild-type Colombia-0 and ASN2 overexpressors. These results support the notion that ASN2 is closely correlated to ammonium metabolism in higher plants.
To systematically study the differential physiological roles of different members in the asparagines synthetase (AS; the enzyme catalyzing the biosynthesis of asparagines) gene family, we previously cloned all the members of AS gene in the model plant Arabidopsis (ASN1, ASN2, and ASN3). Phylogenetic analysis revealed that although ASN1 clustered with all dicot AS genes, ASN2 and ASN3 are more closely related to monocot AS genes (Lam et al., 1998). ASN1 and ASN2 were shown to be reciprocally regulated by light and metabolites in Arabidopsis (Lam et al., 1998). Based on the phylogenetic data and gene expression data, it was suggested that ASN1 and ASN2 may play very different physiological roles in plant nitrogen metabolism (Lam et al., 1998).
The expression level of ASN1 is tightly correlated with free Asn levels (Lam et al., 1994, 1998, 2003). In wild-type Arabidopsis, ASN1 and free Asn increase in dark-adapted plants, consistent with the idea that Asn is an important nitrogen carrier, especially under carbon-limiting conditions. In transgenic plants overexpressing the ASN1 gene, free Asn levels in source and sink tissues as well as in the phloem increase accordingly. Total amino acid pools in seeds of the ASN1 overexpressors are enhanced quantitatively and qualitatively (Lam et al., 2003). These findings suggest that ASN1 plays a major role in nitrogen assimilation, regulating nitrogen transport and storage during seed development. These data are also consistent with the notion that free Asn is an important nitrogen carrier for long-range transport and storage in higher plants (Lea and Miflin, 1980; Sieciechowicz et al., 1988; Lea et al., 1990).
By contrast, studies have shown that although expressed at lower levels, the expression of ASN2 is reciprocal to that of ASN1. The discovery of a light-induced ASN2 gene was initially paradoxical because free Asn does not accumulate to a high level in light-grown plants (Lam et al., 1998). It was proposed that ASN2 may play a role other than in primary nitrogen assimilation.
Previous physiological studies suggest that there may be some correlations between Asn accumulation and ammonium metabolism, especially under light conditions. For instance, it was shown in 1922 that when plants were grown in light with high levels of exogenous ammonium, levels of Asn accumulated (Prianischnikow, 1922). On the other hand, under high-light intensity, photorespiration occurs in C3 plants and leads to an increased production of ammonium (Magalhaes and Wilcox, 1984; Givan et al., 1988; Kozaki and Takeba, 1996; Wingler et al., 2000). Under such circumstances, Asn can account for up to 7% of the total nitrogen (Ta et al., 1984; Sieciechowicz et al., 1988). It has been proposed that Asn may play a role in ammonium detoxification (Kanamori and Matsumoto, 1974; Givan, 1979; Stewart, 1979) and may act as an alternative nitrogen donor during photorespiration (Ta et al., 1984, 1985; Ta and Joy, 1986).
Ammonium accumulation may also occur when plants are under abiotic and biotic stresses. For instance, an increase of ammonium level was observed in tomato (Lyocopersicon esculentum) plants subjected to water and salinity stresses (Feng and Barker, 1993). On the other hand, when tomato was infected with root-knot nematode, foliar accumulation of ammonium was observed (Barker, 1999a). In fact, ammonium accumulation can be generally viewed as an index of stress in plants (Barker, 1999b).
Interestingly, although water and salinity stresses lead to ammonium accumulation, these stresses also enhance cellular free Asn levels. For example, when soybean (Glycine max) was subjected to severe water stress, 54% loss of leaf protein was balanced by a gain in the free amino acids in which 41% accumulated as Pro and Asn (Fukutoku and Yamada, 2002). Prolonged salt treatment also resulted in an increase of Asn and other amino acids in source tissues of Coleus blumei. The accumulation of Asn was at least partially due to de novo synthesis (Gilbert et al., 1998). It was also reported that genes encoding Asn synthetase in maize (Zea mays) are up-regulated by salt and heavy metal stresses (Chevalier et al., 1996).
The above findings suggest a correlation between Asn biosynthesis, ammonium accumulation, and stress conditions in higher plants. However, the possible regulation of AS genes in relation to ammonium metabolism remains unexplored. Because multiple AS isozymes (ASN1, ASN2, and ASN3) exist in the plant and AS activities are proven to be difficult to assay in vitro (Kern and Chrispeels, 1978; Joy et al., 1983; Huber and Streeter, 1985; Sieciechowicz et al., 1988), we addressed the question using molecular and transgenic approaches. In this report, we provided in vivo evidence showing that ASN2 gene expression correlates with changes in ammonium metabolism in Arabidopsis.
RESULTS
Ammonium Is Required for Light Induction of ASN2
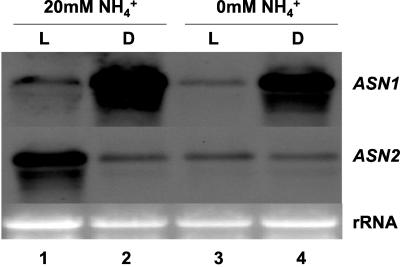
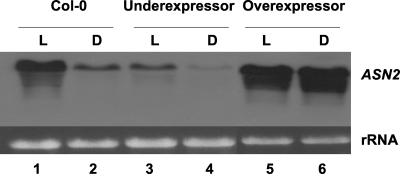
It was shown that supplementation of exogenous ammonium leads to Asn accumulation in plants (Prianischnikow, 1922). Therefore, we examined the regulation of ASN gene expression in Arabidopsis in response to light and ammonium treatment. In previous studies, we demonstrated that light induces ASN2 expression and represses ASN1 expression in plants grown in Murashige and Skoog media (Lam et al., 1998). Here, we show that the light regulation of ASN2, but not ASN1 is dependent on the presence of ammonium in the media (Fig. 1). When the plants were grown on regular Murashige and Skoog medium (20 mm ammonium/40 mm nitrate) for 48 h under continuous light (Fig. 1, lane 1), ASN2 mRNA level was increased compared with plants that had been dark adapted for 48 h (Fig. 1, compare lanes 1 and 2). This result is consistent with previous findings on light induction of ASN2 gene expression (Lam et al., 1998). However, if ammonium was removed from the growth medium, no light induction of ASN2 could be observed (Fig. 1, compare lanes 3 and 4). By contrast, the presence of ammonium has no effect on the light repression of ASN1 gene expression (Fig. 1).
Figure 1.
Ammonium controls the light induction of ASN2. Twelve-day-old seedlings grown on Murashige and Skoog agar plates under a regular day/light cycle (16 h of light and 8 h of dark) were transferred to new Murashige and Skoog medium containing 20 mm (Lanes 1 & 2) or no (Lanes 3 & 4) ammonium. NaNO3 was added to ammonium-free medium to maintain an equal molar concentration of nitrogen resources. Plants were subsequently treated under continuous light (L) or continuous dark (D) conditions for 48 h. Total RNA was extracted from the seedlings as described in “Materials and Methods.” An aliquot of 15 μg of total RNA from each line was loaded onto each lane. Northern-blot analysis was performed as described in “Materials and Methods.”
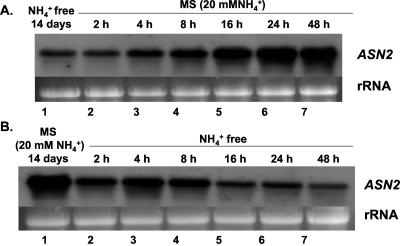
We further investigated the kinetics with which ammonium effects on ASN2 mRNA accumulation. For this, plants were first grown on ammonium-free Murashige and Skoog medium for 14 d before being transferred to growth medium containing 20 mm ammonium (Fig. 2A). Induction of ASN2 mRNA levels was first observed 4 h after the switch to the ammonium-containing media (Fig. 2A, lane 3). Maximal induction of ASN2 mRNA was observed after 24 h (Fig. 2A, lane 7). A reciprocal experiment was performed by first growing the plants on regular Murashige and Skoog medium containing 20 mm ammonium before transferring the plants to ammonium-free Murashige and Skoog medium (Fig. 2B). An initial decrease of ASN2 mRNA was observed 2 h after the transfer (Fig. 2B, lane 2), and maximal repression was observed after 16 h (Fig. 2B, lane 7). The reduction of ASN2 mRNA in ammonium-free medium was not due to nitrogen deprivation because additional nitrate was added to the ammonium-free medium to maintain an equal nitrogen supply. The above data further supports that ASN2 gene expression is strongly dependent on the presence of ammonium, especially under light-grown conditions.
Figure 2.
Ammonium controls the light induction of ASN2. Seedlings were germinated and grown under a regular day/light cycle on Murashige and Skoog agar plates containing no (A) or 20 mm (B) ammonium for 14 d before being transferred to Murashige and Skoog agar plates containing 20 mm (A) and no (B) ammonium respectively, and grown under continuous light. NaNO3 was added to ammonium-free medium to maintain an equal molar concentration of nitrogen resource. Samples were collected at time 0, 2, 4, 8, 16, 24, and 48 h (lanes 1-7, respectively) after treatments. Total RNA was extracted from the seedlings as described in “Materials and Methods.” An aliquot of 15 μg of total RNA from each line was loaded onto each lane. Northern-blot analysis was performed as described in “Materials and Methods.”
Stresses Increased Steady-State mRNA Level of ASN2 and Cellular Ammonium Contents
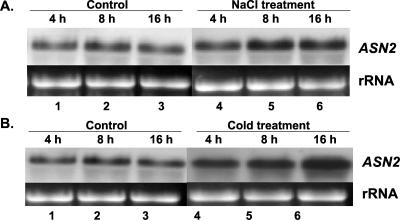
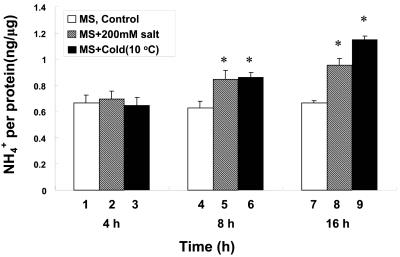
The ASN2 gene of Arabidopsis has been shown to group phylogenetically with the maize AS gene (Lam et al., 1998). Because the maize AS gene was found to be induced by stresses such as NaCl (Chevalier et al., 1996), we tested the effects of NaCl and cold stress on the levels of ASN2 mRNA in Arabidopsis. Plants were first grown on regular Murashige and Skoog medium before being subjected to NaCl and cold treatments (Fig. 3). Leaf samples were collected at 4, 8, and 16 h after each treatment. Northern-blot analysis using total RNA extracted from the leaf samples showed that ASN2 mRNA levels were increased at time points 8 and 16 h for NaCl (Fig. 3A, lanes 5 and 6) and cold (Fig. 3B, lanes 5 and 6) treatments. Ammonium content in the same batch of leaf samples was also determined (Fig. 4). A significant increase of ammonium content was observed at time points 8 and 16 h for NaCl (Fig. 4, lanes 5 and 8) and cold (Fig. 4, lanes 6 and 9) treatments, matching the induction of the ASN2 mRNA levels (Fig. 3). In control plants without stress treatments, no increase of ASN2 mRNA (Fig. 3, A, lanes 1-3, and B, lanes 1-3) or ammonium content (Fig. 4, lanes 1, 4, and 7) was observed. These findings suggest that the induction of ASN2 expression by NaCl and cold stress may be mediated through accumulation of ammonium under these stress conditions. This hypothesis is further supported by our data that the ASN2 gene is specifically responsive to ammonium treatments (Figs. 1 and 2).
Figure 3.
NaCl and cold stresses increase the levels of ASN2 mRNA. The effects of NaCl (200 mm; A) and cold (10°C; B) on the levels of ASN2 mRNA were tested. Twelve-day-old seedlings grown on Murashige and Skoog agar plates under a regular day/light cycle were transferred to new growth medium (A) or were subjected to new growth temperature (B) for 4 (lane 4), 8 (lane 5), and 16 h (lane 6). Samples of control plants without stress treatments were also collected at same time points (lanes 1-3). Total RNA was extracted from the seedlings as described in “Materials and Methods.” An aliquot of 15 μg of total RNA from each line was loaded onto each lane. Northern-blot analysis was performed as described in “Materials and Methods.”
Figure 4.
Endogenous free ammonium content increases under stress treatments. Samples were harvested as described in Figure 3. Free ammonium content was assayed as described in “Materials and Methods.” Each bar represents an average of five samples. Error bars = ses. The data was analyzed by one-way analysis of variance (ANOVA) followed by lsd test. An asterisk indicates significant difference when compared NaCl-(lanes 2, 5, and 8) and cold-(lanes 3, 6, and 9) treated samples to untreated controls (lanes 1, 4, and 7), with a P value less than 0.05.
Overexpression or Underexpression of ASN2 Alters Ammonium Content in Arabidopsis
To further investigate the possible relation between ASN2 and ammonium metabolism, transgenic Arabidopsis lines overexpressing or underexpressing the ASN2 gene were produced. In ASN2 underexpressors, the coding region of ASN2 was expressed in antisense orientation under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The level of ASN2 mRNA was found to decrease, especially in light-grown conditions (Fig. 5, lane 3). In ASN2 overexpressors, the coding region of ASN2 was placed in sense orientation under the control of the CaMV 35S promoter. Native ASN2 is expressed at higher levels in the light compared with dark (Fig. 5, lanes 1 and 2). By contrast, in ASN2-overexpressing transgenic plants, constitutive expression of ASN2 was observed and the light control of ASN2 was deregulated (Fig. 5, lanes 5 and 6). Figure 5 showed the results of one typical antisense (lanes 3 and 4) and one typical sense (lanes 5 and 6) lines. Other antisense and sense lines used in the research exhibited similar ASN2 expression patterns (data not shown).
Figure 5.
Northern-blot analysis of ASN2 transgenic lines. Twelve-day-old seedlings of the wild-type Col-0 (lanes 1 and 2), the ASN2-underexpressing line 401-A4-c1 (lanes 3 and 4), and the ASN2-overexpressing line 402-A1-b1 (lanes 5 and 6) grown on Murashige and Skoog agar plates under a regular day/light cycle were subsequently treated under continuous light (L) or continuous dark (D) conditions for 48 h. Total RNA was extracted from the seedlings as described in “Materials and Methods.” An aliquot of 15 μg of total RNA from each line was loaded onto each lane. Northern-blot analysis was performed as described in “Materials and Methods.”
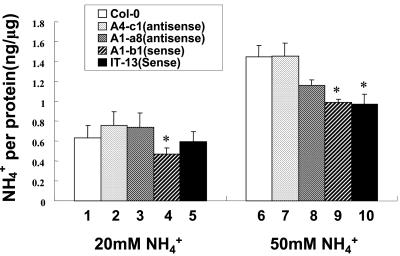
To test if the misexpression of ASN2 in transgenic plants has effects on the level of endogenous ammonium, two sets of experiments were performed. When the ammonium concentration in the growth medium was increased from 20 mm (in regular Murashige and Skoog medium) to 50 mm, endogenous ammonium content started to increase accordingly in all lines (Fig. 6). However, the overexpressing lines exhibited lower endogenous ammonium contents compared with wild type when grown on Murashige and Skoog medium containing 50 mm ammonium (Fig. 6, lanes 9 and 10) compared with the wild-type (Colombia) Col-0 (Fig. 6, lane 6). One ASN2-overexpressing line also showed a slight decrease in ammonium content even at the lower concentration of ammonium (Fig. 6, lane 4) compared with wild type. On the other hand, no significant difference was observed in ammonium content when comparing the underexpressing lines and Col-0 grown on 50 mm ammonium (Fig, 6, lanes 7 and 8).
Figure 6.
Endogenous free ammonium content decreases in ASN2-overexpressing transgenic lines grown on ammonium-containing medium. Ten-day-old seedlings of the wild-type Col-0 (lanes 1 and 6), ASN2-underexpressing lines 401-A4-c1 (lanes 2 and 7) and 401-A1-a8 (lanes 3 and 6), and ASN2-overexpressing lines 402-A1-b1 (lanes 4 and 9) and 402-IT-13 (lanes 5 and 10) grown under a regular day/light cycle on regular Murashige and Skoog (containing 20 mm ammonium) agar plates were transferred to Murashige and Skoog medium supplemented with 20 (lanes 1-5) or 50 mm (lanes 6-10) exogenous ammonium. Free ammonium content was assayed as described in “Materials and Methods.” Each bar represents an average of five samples. Error bars = ses. The data was analyzed by one-way ANOVA followed by lsd test. An asterisk indicates significant difference with a P value less than 0.05.
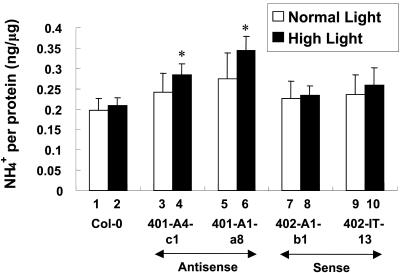
To directly increase the internal pool of ammonium without external supplements, the transgenic lines were subjected to high-light irradiance that may increase the levels of photorespiratory ammonium (Magalhaes and Wilcox, 1984; Givan et al., 1988; Kozaki and Takeba, 1996; Wingler et al., 2000). Under high-light conditions, ASN2 underexpressors accumulated significantly higher levels of cellular ammonium (Fig. 7, lanes 4 and 6) when compared with Col-0 (Fig. 7, lane 2). Such differences were not significant when the comparison was made under low-light irradiance (Fig. 7, lanes 1, 3, and 5). No significant difference was found between the overexpressing lines (Fig. 7, lanes 7-9) and Col-0 (Fig. 7, lanes 1 and 2).
Figure 7.
Endogenous free ammonium content increases in ASN2 antisense transgenic lines grown under high-light irradiance. Twelve-day-old seedlings of the wild-type Col-0 (lanes 1 and 2), ASN2-underexpressing lines 401-A4-c1 (lanes 3 and 4) and 401-A1-a8 (lanes 5 and 6), and ASN2-overexpressing lines 402-A1-b1 (lanes 7 and 8) and 402-IT-13 (lanes 9 and 10) grown under a regular day/light cycle at low light irradiance (35 μE) on regular Murashige and Skoog agar plates were exposed to continuous light at low (35 μE; lanes 1, 3, 5, 7, and 9) or high (150 μE; lanes 2, 4, 6, 8, and 10) for 72 h. Free ammonium contents were assayed as described in “Materials and Methods.” Each bar represents an average of five samples. Error bars = ses. The data was analyzed by one-way ANOVA followed by lsd test. An asterisk indicates significant difference with a P value less than 0.05.
The loss-of-function and gain-of-function approaches using transgenic lines described above thus showed that ASN2 overexpressors accumulated lower levels of ammonium, whereas the ASN2 underexpressors accumulated higher levels of ammonium. This data suggests that ASN2 gene expression levels correlate with changes in ammonium metabolism in Arabidopsis.
DISCUSSION
Drastic changes in the levels of free Asn under different physiological and environmental conditions have suggested that Asn may play an important role in various aspects of plant physiology and metabolism (Sieciechowicz et al., 1988). In general, AS enzymes were found to be encoded by a small gene family in various species of legume and nonlegume plants (for example, Davis and King, 1993; Waterhouse et al., 1996; Hughes et al., 1997; Lam et al., 1998). Expression of AS genes is under the control of light, metabolites, postharvesting conditions, and stress treatments (Tsai and Coruzzi, 1990, 1991; Davis and King, 1993; Lam et al., 1994, 1998; Chevalier et al., 1996). These complex modes of regulation of AS gene expression support the notion that AS genes may play different roles in various aspects of plant physiology.
In this study, we found that the expression of ASN2 is correlated with ammonium metabolism. We showed that when plants were grown on Murashige and Skoog media, the light induction of ASN2 is ammonium dependent. Time kinetics studies of ASN2 expression revealed a rapid and reciprocal control of ASN2 by ammonium supplementation or deprivation (Fig. 2). To our knowledge, it is the first report on ammonium induction of an AS gene. In addition to ammonium, we showed that stresses such as salinity and cold also increase ASN2 mRNA levels, and that these stresses correlate with increases in internal ammonium (Figs. 3 and 4). The effect of stresses on the induction of AS genes was shown in maize previously, but the relationship to cellular ammonium accumulation was not addressed (Chevalier et al., 1996).
Interestingly, there is a close relationship between ammonium accumulation and stress conditions. For instance, ammonium accumulates under various biotic and abiotic stresses (Feng and Barker, 1993; Barker, 1999a, 1999b). Under stress conditions, the photosynthetic rate will drop significantly and hence the carbon skeleton reaching the sink tissues may become limiting (Farrar, 1981; Kerr et al., 1985; Baysdorfer et al., 1998). Nitrogen assimilation and protein synthesis will decrease and catabolism of amino acids will increase to provide the carbon skeleton to sustain respiration and other metabolic processes (Saglio and Pradet, 1980; Journet et al., 1986; Brouquisse et al., 1991; Dieuaide et al., 1992, 1993; Baysdorfer et al., 1998). Extensive protein degradation leads to a drastic increase of internal ammonium content (Feng and Barker, 1992). To save valuable nitrogen resources, it is important for plants to recapture the ammonium otherwise lost via protein degradation. Nitrogen resources in the form of ammonium will be mainly recaptured by the enzymes Gln synthetase-Glu oxoglutarate aminotransferase that function in a cycle. We have shown that ASN2 is induced by ammonium treatments (Figs. 1 and 2) and by stress treatments that cause ammonium accumulation (Figs. 3 and 4). These findings suggest that the physiological role of ASN2 may be related, directly or indirectly, to the recapturing of lost nitrogen resources under stress conditions. By contrast, the role of ASN1 may be related to primary nitrogen assimilation and transport.
To further investigate the possible interaction between ASN2 and ammonium metabolism, transgenic Arabidopsis lines overexpressing or underexpressing the ASN2 gene were constructed (Fig. 5). Under high concentrations of exogenous ammonium supplies, endogenous ammonium accumulates in the treated plants (Fig. 6). However, ASN2 overexpressors exhibit a significantly lower level of endogenous ammonium under such conditions compared with wild type. Although ammonium is an important intermediate in nitrogen assimilation, high ammonium concentration is toxic to plants (Vines and Wedding, 1960; Fangmeier et al., 1994; Gerendas et al., 1997). Previous studies showed that Asn accumulates under high levels of exogenous ammonium (Prianischnikow, 1922) and that Asn may play a role in ammonium detoxification (Kanamori and Matsumoto, 1974; Givan, 1979; Stewart, 1979). Together with our finding that overexpression of ASN2 can reduce levels of endogenous ammonium, it reaffirms our hypothesis that ASN2 plays an important role in ammonium metabolism in Arabidopsis.
Under high-light irradiance, photorespiration is enhanced and leads to an increase in the loss of fixed carbon and fixed nitrogen as carbon dioxide and ammonium, respectively (Magalhaes and Wilcox, 1984; Givan et al., 1988; Kozaki and Takeba, 1996; Wingler et al., 2000). In accordance with this, we observed a significant increase of ammonium in wild-type plants grown under high-light irradiance compared with plants that received lower light irradiance (Fig. 7). However, ASN2 underexpressors showed a significant increase in levels of ammonium, especially when grown under high-light irradiance conditions. The data of our loss-of-function experiment suggests a possible relationship between the level of ASN2 gene expression and the level of ammonium loss via the photorespiratory pathway. It has been reported that Asn may act as an alternative nitrogen donor in the photorespiratory pathway in pea (Pisum sativum; Ta et al., 1984, 1985; Ta and Joy, 1986).
Whether ASN2 directly or indirectly involved in ammonium metabolism is still unclear. Some studies suggested that monocot AS enzymes may use ammonium as the direct substrate (Oaks and Ross, 1984; Oaks and Hirel, 1985). The protein encoded by the Arabidopsis ASN2 gene is actually more closely related to monocot AS proteins than to the ASN1 protein that clusters with dicot AS enzymes in a phylogeny tree (Lam et al., 1998). However, only a separate detailed enzyme kinetic studies of the purified ASN2 protein can give the ultimate answer. In planta studies of the AS activities in wild-type and transgenic plants has proven to be difficult due to instability of the enzyme, presence of inhibitors, and asparaginase activities (Kern and Chrispeels, 1978; Joy et al., 1983; Huber and Streeter, 1985; Sieciechowicz et al., 1988). As the transgenic results suggest that ASN2 may be playing a special role in ammonia assimilation, we will attempt to validate this hypothesis in future biochemical studies of the ASN1 and ASN2 enzymes using a variety of heterologous expression systems.
MATERIALS AND METHODS
Plant Growth and Stress Treatments
Seeds were surface sterilized by bleach and washed with autoclaved distilled water before sowing on Murashige and Skoog (Murashige and Skoog, 1962) basal medium (Invitrogen, Carlsbad, CA; pH adjusted to 5.7 with KOH) containing 3% (w/v) Suc and 0.9% (w/v) bactoagar. After being imbibed at 4°C in dark for 2 d, the agar plates were transferred to environmentally controlled growth chambers with regular day/light cycle (16 h of light at 22°C and 8 h of dark at 20°C).
In experiments testing the ammonium dependence of light induction of ASN2, 12-d-old seedlings were transferred to new Murashige and Skoog medium containing 20 mm ammonium (concentration in regular Murashige and Skoog medium) or ammonium-free Murashige and Skoog medium that contained the same molar amount of nitrogen (supplied in form of NaNO3). The plates were exposed to 48 h continuous light or continuous dark conditions before harvesting. In experiments showing the kinetics of ammonium effects on ASN2 gene expression, 14-d-old seedlings originally grown in regular day/light cycle on Murashige and Skoog medium containing no or 20 mm ammonia were transferred to Murashige and Skoog medium containing 20 mm and no ammonia, respectively. The seedlings were allowed to grow under continuous light, and samples were harvested at 2, 4, 8, 16, 24, and 48 h after treatments.
In NaCl stress experiments, the seedlings were transferred to Murashige and Skoog medium supplemented with 200 mm NaCl. In cold stress experiments, the seedlings were transferred to regular Murashige and Skoog medium but were grown in a 10°C growth chamber. The samples were harvested at 4, 8, and 16 h after treatment. All treatments were performed during the regular day/light cycle.
To test the endogenous ammonium contents in ASN2 transgenic lines when grown in medium supplemented with different concentrations of exogenous ammonium, 10-d-old seedlings were transferred to Murashige and Skoog medium containing 20 or 50 mm ammonium and were allowed to grow under regular day/light cycle for 5 d before harvesting.
To test the accumulation of ammonium under different light intensities, seeds were allowed to germinate on Murashige and Skoog medium containing 1% (w/v) Suc. Twelve-day-old seedlings grown under a regular day/light cycle (at in irradiance of 35 μE) were divided into two sets. The two sets of seedlings were exposed continuous low irradiance (35 μE) or high irradiance (150 μE) for 72 h before harvesting.
Construction of ASN2 Transgenic Lines
The ASN2 cDNA (Lam et al., 1998) was subcloned into a binary vector (Brears et al., 1993) and expressed under the control of the CaMV 35S promoter in sense or antisense orientation. Binary vector constructions were transformed into the disarmed Agrobacterium tumefaciens GV3101/pMP90 (Koncz and Schell, 1986) by electroporation. Vacuum infiltration in planta transformation (Bechtold and Pelletier, 1993) was performed using a modified protocol (Lam et al., 2003). The transformed plants were allowed to continue to grow and set seeds. Positive transformants were identified as green seedlings grown on Murashige and Skoog agar plates supplemented with 50 μg mL-1 kanamycin. Single-locus insertions were scored (using the chi-square test) by observing a 3:1 ratio (kanamycin resistant versus kanamycin sensitive) in T2 progenies resulted from self-pollination of T1 parents. Homologous T3 progenies were used for subsequent assays.
RNA Analysis and Ammonium Assay
For RNA analysis, samples were freshly collected and frozen immediately in liquid nitrogen before grinding with a precooled mortar and pestle. Total RNA was extracted using a modified phenol extraction protocol (Jackson and Larkins, 1976). Digoxygenin-labeled RNA probes were generated by a commercial kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Northern-blot analysis was performed by hybridizing RNA blots with labeled probes at 68°C in 50% (v/v) formamide hybridization solution for at least 16 h. Posthybridization washing and chemiluminescent detection were carried out as described in the manufacturer's instructions. All northern analyses were conducted in biological replicates of two to three times. A sample northern blot was shown for each experimental design.
For free ammonium assays, plant tissues were harvested and frozen immediately in liquid nitrogen. Each sample was ground in 150 μL of protein grinding buffer (50 mm Tris-HCl, pH 8.0, 10 mm imidazole, and 0.5% [w/v] β-mercaptoethanol; Oliveira et al., 2002). The samples were vortexed vigorously and spun for 15 min at maximum speed in a microfuge. The upper layer was transferred to a new microtube, of which 50 μL was saved for the protein assay (Bradford, 1976). Free ammonium was assayed in the remaining sample using a commercial kit (Boehringer Mannheim, Mannheim, Germany).
Acknowledgments
We thank Iris Tong and Lee Wong for their professional assistance in generating the transgenic plants. We also appreciate the technical help of Kwan-Meí Yam in some molecular and biochemical analysis. We also acknowledge the support provided by the Hong Kong University Grant Council Area of Excellence on Plant and Fungal Biotechnology Center.
This work was supported by the Hong Kong Research Grant Council (earmarked grant no. CUHK4292/98M to H.-M.L) and by the U.S. Department of Energy (grant no. DEFG01-92-20071 to G.M.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.033126.
References
- Barker AV (1999a) Ammonium accumulation and ethylene evolution by tomato infected with root-knot nematode and grown under different regimes of plant nutrition. Commun Soil Sci Plant Anal 30: 175-182 [Google Scholar]
- Barker AV (1999b) Foliar ammonium accumulation as an index of stress in plants. Commun Soil Sci Plant Anal 30: 167-174 [Google Scholar]
- Baysdorfer C, Warmbrodt RD, Van der Woude WJ (1998) Mechanism of starvation tolerance in Pearl millet. Plant Physiol 88: 1381-1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1993) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In J Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Humana Press, Totowa, NJ pp 259-266 [DOI] [PubMed]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Brears T, Liu C, Knight TJ, Coruzzi GM (1993) Ectopic overexpression of asparagine synthetase in transgenic tobacco. Plant Physiol 103: 1285-1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, James F, Raymond P, Pradet A (1991) Study of glucose starvation in excised maize root tips. Plant Physiol 96: 619-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Just D, Raymond P (1996) Metabolic regulation of asparagine synthetase gene expression in maize (Zea mays L.) root tips. Plant J 9: 1-11 [DOI] [PubMed] [Google Scholar]
- Davis KM, King GA (1993) Isolation and characterization of a cDNA clone for a harvest induced asparagine synthetase from Asparagus officinalis L. Plant Physiol 102: 1337-1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Brouquisse R, Pradet A, Raymond P (1992) Increased fatty acid β-oxidation after glucose starvation in maize root tips. Plant Physiol 99: 595-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Couee l, Pradet A, Raymond P (1993) Effects of glucose starvation on the oxidation of fatty acids by maize root tip mitochondria and peroxisomes: evidence for mitochondrial fatty acid β-oxidation and acyl-CoA dehydrogenase activity in a higher plant. Biochem J 296: 199-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangmeier A, Hadwiger-Fangmeier A, Van der Eerden L, H-J. J (1994) Effects of atmospheric ammonia on vegetation: a review. Environ Pollut 86: 43-82 [DOI] [PubMed] [Google Scholar]
- Farrar JF (1981) Respiration rate of barley roots: its relation to growth, substrate supply and the illumination of the shoot. Ann Bot 48: 53-63 [Google Scholar]
- Feng J, Barker A (1992) Ethylene evolution and ammonium accumulation by tomato plants with various nitrogen forms and regimes of acidity: Part I. J Plant Nutr 15: 2457-2469 [Google Scholar]
- Feng J, Barker AV (1993) Ethylene evolution and ammonium accumulation by tomato plants under water and salinity stresses. J Plant Nutr 15: 2471-2490 [Google Scholar]
- Fukutoku Y, Yamada Y (2002) Sources of proline-nitrogen in water-stressed soybean (Glycine max): fate of nitrogen-15-labeled protein. Physiol Plant 61: 622-628 [Google Scholar]
- Gerendas J, Zhu Z, Bendixen R, Ratcliffe RG, Sattelmacher B (1997) Physiological and biochemical processes related to ammonium toxicity in higher plants. Z Pflanzenernaehr Bodenkd 160: 239-251 [Google Scholar]
- Gilbert GA, Gadush MV, Wilson C, Madore MA (1998) Amino acid accumulation in sink and source tissues of Coleus blumei (benth) during salinity stress. J Exp Bot 49: 107-114 [Google Scholar]
- Givan CV (1979) Metabolic detoxification of ammonia in tissues of higher plants. Phytochemistry 18: 375-382 [Google Scholar]
- Givan CV, Joy KW, Kleczkowski LA (1988) A decade of photorespiratory nitrogen recycling. Trends Biol Sci 13: 433-437 [DOI] [PubMed] [Google Scholar]
- Huber TA, Streeter JG (1985) Purification and properties of asparagine synthetase from soybean root nodules. Plant Sci 42: 9-17 [Google Scholar]
- Hughes CA, Beard HS, Matthews BF (1997) Molecular cloning and expression of two cDNA encoding asparagine synthetase in soybean. Plant Mol Biol 33: 301-311 [DOI] [PubMed] [Google Scholar]
- Jackson AO, Larkins BA (1976) Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol 57: 5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, Bligny R, Douce R (1986) Biochemical changes during sucrose deprivation in higher plant cells. J Biol Chem 261: 3193-3199 [PubMed] [Google Scholar]
- Joy KW, Ireland RJ, Lea PJ (1983) Asparagine synthesis in pea leaves, and the occurrence of an asparagine synthetase inhibitor. Plant Physiol 73: 165-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T, Matsumoto H (1974) Asparagine biosynthesis by Oryza sativa seedlings. Phytochemistry 13: 1407-1412 [Google Scholar]
- Kern R, Chrispeels MJ (1978) Influence of the axis in the enzymes of protein and amide metabolism in the cotyledons of mung bean seedlings. Plant Physiol 62: 815-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr PS, Rufty TW, Huber SC (1985) Changes in non-structural carbohydrates in different parts of soybean (Glycine max L. Merr.) plants during a light/dark cycle and in extended darkness. Plant Physiol 78: 576-581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383-396 [Google Scholar]
- Kozaki A, Takeba G (1996) Photorespiration protects C3 plants from photooxidation. Nature 384: 557-560 [Google Scholar]
- Lam H-M, Hsieh M-H, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345-353 [DOI] [PubMed] [Google Scholar]
- Lam H-M, Peng S, Coruzzi G (1994) Metabolic control of asparagine synthetase gene expression in Arabidopsis thaliana. Plant Physiol 106: 1347-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Wong P, Chan H-K, Yam K-M, Chen L, Chow C-M, Coruzzi GM (2003) Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol 132: 926-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Miflin BJ (1980) Transport and metabolism of asparagine and other nitrogen compounds within the plant. In PK Stumpt, EE Conn, eds, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 569-607 [Google Scholar]
- Lea PJ, Robinson SA, Stewart GR (1990) The enzymology and metabolism of glutamine, glutamate, and asparagine. In BJ Miflin, PJ Lea, eds, The Biochemistry of Plants: Amino Acids and Derivatives, Vol 16. Academic Press, New York, pp 121-159 [Google Scholar]
- Magalhaes JR, Wilcox GE (1984) Ammonium toxicity development in tomato plants relative to nitrogen form and light intensity. J Plant Nutr 7: 1477-1496 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for the growth and bioassay with tobacco tissue culture. Physiol Plant 15: 473-497 [Google Scholar]
- Oaks A, Hirel B (1985) Nitrogen metabolism in roots. Annu Rev Plant Physiol 36: 345-365 [Google Scholar]
- Oaks A, Ross D (1984) Asparagine synthetase in Zea mays. Can J Bot 62: 68-73 [Google Scholar]
- Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM (2002) Overexpression of cytosolic glutamine synthetase: relation to nitrogen, light, and photorespiration. Plant Physiol 129: 1170-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prianischnikow D (1922) Uber den aufbau un abbau des asparagins in den pflanzen Ber. Bot Ges 40: 242-248 [Google Scholar]
- Saglio P, Pradet A (1980) Soluble sugars, respiration, and energy charge during aging of excised maize root tips. Plant Physiol 66: 516-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieciechowicz KA, Joy KW, Ireland RJ (1988) The metabolism of asparagine in plants. Phytochemistry 27: 663-671 [Google Scholar]
- Stewart CR (1979) The effect of ammonium, glutamine, methionine sulfoxamine and azaserine on asparagine synthetase in soybean leaves. Plant Sci Lett 14: 269 [Google Scholar]
- Ta TC, Joy KW (1986) Separation of amino acid and amide nitrogen from plant extracts for 15N analysis. Anal Biochem 154: 564-569 [DOI] [PubMed] [Google Scholar]
- Ta TC, Joy KW, Ireland RJ (1984) Amino acid metabolism in pea leaves: utilization of nitrogen from amide and amino groups of [15N]-asparagine. Plant Physiol 74: 822-826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta TC, Joy KW, Ireland RJ (1985) Role of asparagine in the photorespiratory nitrogen metabolism of pea leaves. Plant Physiol 78: 334-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM (1990) Dark-induced and organ-specific expression of two asparagine synthetase genes in Pisum sativum. EMBO J 9: 323-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM (1991) Light represses transcription of asparagine synthetase genes in photosynthetic and nonphotosynthetic organs of plants. Mol Cell Biol 11: 4966-4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vines HM, Wedding RT (1960) Some effects of ammonia on plant metabolism and a possible mechanism for ammonia toxicity. Plant Physiol 35: 820-825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RN, Smyth AJ, Massonneau A, Prosser IM, Clarkson DT (1996) Molecular cloning and characterization of asparagine synthetase from Lotus japonicus: dynamics of asparagine synthesis in N-sufficient conditions. Plant Mol Biol 30: 883-897 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration: metabolic pathways and their role in stress protection. Phil Trans Royal Soc London Series B Biol Sci 355: 1517-1529 [DOI] [PMC free article] [PubMed] [Google Scholar]