Abstract
We have developed a fluorescence imaging-based approach to detect expression of tumor marker genes in pancreatic cancer cells using molecular beacons (MBs). MBs are short hairpin oligonucleotide probes that bind to specific oligonucleotide sequences and produce fluorescent signals. MBs targeting transcripts of two tumor marker genes, mutant K-ras and survivin, were synthesized and their specificity in detection of the expression of those genes in pancreatic cancer cells was examined. We found that K-ras MBs differentially bind to mutant K-ras mRNAs, resulting in strong fluorescent signals in pancreatic cancer cells with specific mutant K-ras genes but not in normal cells or cancer cells expressing either wild type or a different mutation of the K-ras gene. Additionally, MBs targeting survivin mRNA produced a bright fluorescent signal specifically in pancreatic cancer cells. We also demonstrated that MBs labeled with different fluorophores could detect survivin and mutant K-ras mRNAs simultaneously in single cancer cells. Furthermore, we showed that survivin and K-ras MBs have a high specificity in identifying cancer cells on frozen sections of pancreatic cancer tissues. In conclusion, molecular beacon-based imaging of expression of tumor marker genes has potential for the development of novel approaches for the detection of pancreatic cancer cells.
Keywords: molecular imaging, molecular beacon, K-ras mutation, survivin, in situ gene expression detection and pancreatic cancer cells
INTRODUCTION
Development of human cancer is a multistage process that involves a series of genetic alterations in oncogenes and tumor suppressor genes, and abnormalities in the level of gene expression that provide growth advantage and metastatic potential to the cells.1–3 Methods that allow us to detect the alteration of mRNA transcripts, either mutations or changes in the level of gene expression are promising approaches for identifying cancer cells. In this study, we developed a molecular beacon imaging technology for the detection of expression of tumor marker genes in pancreatic cancer cells. Molecular beacons (MBs) are dual labeled anti-sense oligonucleotide probes with a fluorophore at one end and a quencher at the other. MBs form a hairpin structure in the absence of a complimentary target such that fluorescence of the fluorophore is quenched. Upon hybridization with the target mRNAs that are expressed specifically in cells, the stem of MBs opens up, leading to fluorescence in the cells.4 A major advantage of the stem-loop probes is that they can recognize their targets with a higher specificity than linear probes. Properly designed MBs could discriminate between targets that differ by as little as a single nucleotide.5–7 MBs have been used in a variety of applications including DNA mutation detection, real-time enzymatic cleavage assay, protein–DNA interaction, real-time monitoring of PCR, analyzing loss of heterozygosity and mRNA detection in living cells.8–15 However, applications of this technology for detecting the expression of tumor marker genes in cancer cells and tissues have yet to be developed.
It is well known that the K-ras oncogene is one of the most attractive molecular markers for pancreatic cancer.16–18 Point mutations of the K-ras gene are found in over 90% of pancreatic carcinomas.16,17 Further, most of these mutations are concentrated at codon 12, which makes design and synthesis of mutation-specific MBs feasible. Recently, the genes of an inhibitor of apoptosis (IAP) protein family have been characterized. IAPs are able to inhibit the cascade of the apoptotic pathway by inhibiting activation of caspases.19,20 Survivin, a member of the IAP family, is normally expressed during fetal development but not in most normal adult tissues. However, a high level of survivin is detected in many human cancers, suggesting that survivin may be a specific marker for many common tumor types.21,22 Recent studies demonstrated the presence of survivin in 77 to 83% of pancreatic duct cell adenocarcinomas and 56% of intraductal papillary-mucinous tumors.23,24 Expression of survivin could be detected in all stages of pancreatic duct cell carcinoma including the early stage of neoplastic transition. However, survivin was not detected in pancreatic tissues obtained from normal subjects and patients with chronic pancreatitis.23 Absence of survivin expression in normal pancreas and pancreatic tissue of chronic pancreatitis makes it an ideal molecular marker for pancreatic cancer cells.
In this study, we examined the feasibility of detecting pancreatic cancer cells using MBs targeting mutant K-ras and survivin mRNAs. We showed that the MB-based molecular imaging approach is a simple and efficient method for the detection of mRNA transcripts in cancer cells. We also demonstrate that K-ras and survivin MBs are able to produce specific fluorescent signals in pancreatic cancer cells as well as in frozen tissue sections of pancreatic cancer. These findings suggest that MB-based imaging has potential for the development of novel detection methods for pancreatic cancer.
METHODS
Human tumor and normal cell lines
Pancreatic cancer cell lines PANC-1, Capan-2, MIA PaCa-2 and BXPC-3 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The primary normal human dermal fibroblast cell line HDF was purchased from the Emory University Skin Disease Center (Atlanta, GA). BXPC-3 and PANC-1 cell lines were cultured in RPMI-1640 medium and MIA PaCa-2 cells were cultured in DMEM medium. The Capan-2 cell line was cultured in McCOY’5A medium. All the above media were purchased from Mediatech, Herndon, VA and supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT), 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. The HDF cell line was maintained in DMEM medium with 20% FBS.
Human normal and pancreatic cancer tissues
The paired pancreatic cancer and normal tissues were collected by pathologists at Emory University from pancreatic cancer patients during surgery to remove the tumor, according to an approved IRB protocol. The tissues were frozen immediately in liquid nitrogen and stored in a −80°C freezer until further study.
Detection of K-ras mutation
Genomic DNA samples from pancreatic cancer cell lines and tissues were isolated using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Since human pancreatic cancer tissues consisted of a mixture of normal and cancer cells, we used a mutant-enriched PCR to determine the presence and type of mutant K-ras gene in the pancreatic cancer tissues.25
One microgram of DNA samples were amplified by PCR using a K-ras exon 1 primer pair designed to enrich the mutant K-ras gene. K-ras primers were: K-ras forward, 5′-GAGAGAATTCTATAAACTTG-TGGTAGTTGGACCT-3′ and K-ras reverse, 5′-GA-GAGAATTCATCTGTATCAAAGAATGGTC-3′. The bold region of each primer codes for an EcoR 1 site inserted for cloning the PCR products. The underlined base contained a G to C mutation that created a BstN1 restriction site (5′-CCTGG-3′) if a wild type K-ras (GGT) exon 1 region was amplified. When the K-ras gene was mutated at codon 12 from wild type GGT to GAT, GTT or TGT, this abolished the BstN1 restriction site. Therefore, after BstN1 enzyme digestion, PCR fragments derived from the wild type gene were cut and lost their binding sequences for the K-ras forward primer. As a result, only PCR products containing a mutant K-ras gene could be further amplified by the K-ras primer pair. Following the second PCR amplification, PCR fragments were further digested by BstN1 and EcoR 1. The resulting fragments were purified by gel electrophoresis and cloned into pcDNA 3 (′) plasmid (Invitrogen, Carisbad, CA). The plasmids were then transformed into Top 10 E. coli competent cells (Invitrogen) and positive clones were selected. After PCR amplification of the K-ras fragments from the selected clones, PCR products were sequenced in the DNA Sequence Core Facility at Emory University.
Design and synthesis of MBs for detection of mRNAs
The designs of MBs targeting mutant K-ras and survivin mRNAs are shown in Table 1. For each MB design, the sequence of one arm of the stem and the loop region of the MBs was complementary to a segment of its target gene. In Table 1, the underlined bases were those added to form a stem. K-ras MB1, K-ras MB2, survivin MB and GAPDH MB were synthesized by MWG-Biotech Inc. (High Point, NC). A Cy3, Texas-red or FITC fluorophore was conjugated to the 5′-end of each oligonucleotide and a quencher, 4-(4′-dimethylamino phenylazo) benzoic acid (Dabcyl), was linked to its 3′-end.
Table 1.
Design of molecular beacons for detection of e and survivin genes and corresponding target sequences xpression of mutant K-ras
| MBs | Target sequences | Design of the MBs |
|---|---|---|
| K-ras MB1 | K-ras codon 12 GGT to GAT mutation | 5′-Cy 3-CCTACGCCATCAGCTCCGTAGG-Dabcyl-3′ |
| K-ras MB2 | K-ras codon 12 GGT to GTT mutation | 5′-Texas-red-CCTACGCCAACAGCTCCGTAGG-Dabcyl-3′ |
| Survivin MB1 | Survivin cDNA from 27 to 43 nucleotide | 5′-Cy3-CTGAGAAAGGGCTGCCAGTCTCAG-Dabcyl-3′ |
| Survivin MB2 | Survivin cDNA from 33 to 50 nucleotide | 5′-FITC-TGGTCCTTGAGAAAGGGCGACCA-Dabcyl-3′ |
| GAPDH MB | GAPDH cDNA from 504 to 521 nucleotide | 5′-Cy3-GAGTCCTTCCACGATACCGACTC-Dabcyl-3′ |
|
| ||
| DNA Targets | Synthesized Oligonucleotide sequences | |
| K-ras WT (GGT) | 5′-GTA GTT GGA GCT GGT GGC GTA GGC AAG AGTGCCTTGACGATACAGCTAATT CAG-3′ | |
| K-ras Mut 1 (GAT) | 5′-GTA GTT GGA GCT GAT GGC GTA GGC AAG AGTGCCTTGACGATACAGCTAATT CAG-3′ | |
| K-ras Mut 2 (GTT) | 5′-GTA GTT GGA GCT GTT GGC GTA GGC AAGAGTGCCTTGACGATACAGCTAATT CAG-3′ | |
| Survivin | 5′-CCTGCCTGGCAGCCCTTTCTCAAGGACCACCGCATCTCTACATTCAAGAAC-3′ | |
Determination of specificity of the MBs in solution
The oligonucleotide targets for each MB, shown in Table 1, were synthesized at Integrated DNA Technologies, Inc. (Coralville, IA). Two methods were used to determine the specificity of the MBs:
Examination of binding of the MBs to specific DNA targets using a fluorescence microplate reader
200 nM of MBs were mixed with 1 μM of oligonucleotide targets in 100 μl of Opti-MEM medium (Invitrogen) in 96-well plates. Opti-MEM medium was used because our cellular study was performed using this medium, which gave a higher signal to noise ratio than other buffers in the cells. After incubating at 37°C (survivin MB) or 50°C (K-ras MB) for 60 minutes, fluorescence intensity in each well was measured by a fluorescence microplate reader (Bioteck FL600 Fluorometer, Winooski, VT).
Detection of thermal profile of the MB/target hybridizations using a thermal cycler
The thermal profiles of K-ras MBs hybridizing to their oligonucleotide targets or nonspecific targets were examined using a Bio-Rad iCycler (BioRad Laboratories, Hercules, CA). 200 nM of K-ras MBs were mixed with 1 μM of various DNA targets in 50 μl of Opti-MEM medium. The relative fluorescence unit was then measured at temperatures ranging from 15 to 80°C. Specifically, the temperature was first held at 95°C for 3 minutes. After the first cycle of 80°C for 10 minutes, the fluorescence intensity was recorded at the end of each cycle. The temperature was decreased by 5°C increments to 15°C with each step lasting for 10 minutes. Cy 3 or Texas-red fluorescent dye was used as an internal control to correct for intrinsic changes of fluorescence with temperature.
Next, we examined whether K-ras MBs could detect the presence of mutant mRNA in total RNA samples isolated from pancreatic cancer cell lines. Total RNAs from pancreatic cancer cell lines were isolated using RNA Bee kit (Tel-test, Friendswood, TX). 100 nM of K-ras MB1 or K-ras MB2 was added to 100 μl of Opti-MEM medium containing 1 μg of total RNA. The mixtures were incubated at 50°C for 60 minutes and the fluorescence units in each samples were then examined using a fluorescence microplate reader.
Specific detection of mutant K-ras and survivin gene expression in pancreatic cancer cells using the MBs
For the detection of gene expression in fixed cells, human pancreatic cancer cell lines, PANC-1, Capan-2, MIA PaCa-2 and BXPC-3, and the control normal cell line HDF were plated on chamber slides and then fixed with ice-cold acetone for 8 minutes. After air drying, the slides were stained immediately or stored in a −80°C freezer until use. A mixture of 200 nM of survivin MB and 50 nM of K-ras MB1 or K-ras MB2 diluted in Opti-MEM medium was incubated with the fixed cells at 50°C for 60 minutes. The slides were washed briefly with PBS and examined under a confocal microscope (LSM 510 Meta, Carl Zeiss Microimaging, Inc., Thornwood, NY). The fluorescent images were taken using the same instrument setting for each color.
For detection of the cancer cells expressing tumor marker genes in human cancer tissues, 5 μm frozen sections of pancreatic normal and cancer tissues, fixed with ice-cold acetone for 8 minutes, were incubated with 100 or 200 nM of K-ras or survivin MB for 60 minutes and then counterstained with 10 μg/ml Hoechst 33342 (Molecular Probes, Eugene, OR). The slides were observed under a Nikon fluorescence microscope (Nikon Eclipse E800, Nikon Instrument Inc. Melville, NY). Fluorescence images were taken using Optronics Magnafire digital imaging system (Meyer Instrument, Houston, TX).
Real-time RT PCR
Two micrograms of RNA samples from pancreatic cancer cell lines were amplified with an Omniscript RT kit using an oligo dT primer (QIAGEN Inc, Chatsworth, CA) to generate 20 μl of cDNAs. One to 2 microliters of cDNA was then quantified by Real-Time PCR with primer pairs for K-ras exon 1 or β-actin using QuantiTect SYBR Green PCR kit (QIAGEN Inc) and detected by Bio-Rad iCycler (BioRad Laboratories). The primer pairs for detection of K-ras gene expression were E1 Forward 5′-ATAAACTTGTGGTAGTTGGAGCT-3′ and E2 Reverse, 5′-CACAAA-GAAAGCCCTCCCCA-3′. Amplification of the β-actin gene was used as an internal control for Real-time RT-PCR. The primer pairs for the β-actin gene were β-actin forward, 5′-AAAGACCTGTACGCCAACACAGTGCT-GTCTGG-3′ and β-actin reverse, 5′-CGTCATACTCCTGCTTGCTGAT-CCACATCTGC-3′. The starting quantity of PCR products from amplification of the K-ras gene was standardized with the starting quantity of β-actin products for each sample to obtain the relative level of gene expression.
Immunofluorescence staining
Frozen sections of normal and cancer tissues from pancreatic cancer patients were incubated with a goat anti-human survivin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or a mouse monoclonal anti-cytokeratin 8 antibody (Sigma-Aldrich, St. Louis, MO) for 1 hour. After washing with PBS, the slides were incubated with either biotin-conjugated donkey anti-goat or anti-mouse secondary antibody for 1 hour and then streptavidin-FITC for 30 minutes (Pierce, Rockford, IL). The slides were then examined under a Nikon fluorescence microscope.
RESULTS
K-ras and survivin MBs bound specifically to their oligonucleotide targets in solution
Analysis of K-ras mutation types of pancreatic cancer patients from five previous studies showed that K-ras codon 12 GGT to GAT and GGT to GTT mutations were the most common types of K-ras mutations. Over 75% of the cancer patients have one of these mutations (Table 2).16,26–29 To determine the feasibility of detection of mutant K-ras mRNA in pancreatic cancer cells, we designed and synthesized K-ras MBs targeting those mutations including GGT to GAT (K-ras MB1) or GGT to GTT mutation (K-ras MB2).
Table 2.
K-ras point mutations in pancreatic cancer cell lines and tissues.
| Cell lines and cancer tissues | K-ras mutationb | Alteration | Predicated products | Percentage of the mutation in pancreatic cancer tissues among 188 pancreatic cancer casesa |
|---|---|---|---|---|
| PANC-1 | Yes, codon 12 | GGT to GAT | GLY to ASP | 42.6 % (80/188) |
| Capan-2 | Yes, codon 12 | GGT to GTT | GLY to VAL | 32.4% (61/188) |
| MIA PaCa-2 | Yes, codon 12 | GGT to TGT | GLY to CYS | 1.1% (2/188) |
| BXPC-3 | NO | NO | GLY | N/A |
| HDF | NO | NO | GLY | N/A |
| Pancreatic cancer patient #1c | Yes, codon 12 | GGT to GAT | GLY to ASP | N/A |
| Pancreatic cancer patient #2c | Yes, codon 12 (two mutations) |
GGT to GAT GGT to TGT |
GLY to ASP GLY to CYS |
N/A |
| Pancreatic cancer patient #5c | Yes, codon 12 | GGT to GTT | GLY to VAL | N/A |
Percentage of each K-ras codon 12 mutation in pancreatic cancer tissues was summarized from five studies.16, 26–29
Types of K-ras point mutation in PANC-1, Capan-2 and BXPC-3 cell lines were confirmed in our laboratory by sequencing genomic DNA, which was consistent with the previous reports.26,27
Presence of K-ras mutations in pancreatic cancer tissue samples collected in our laboratory was determined by mutant-enriched PCR as described in the Methods. N/A:Not applicable.
To determine the specificity of the MBs, we examined the binding of the MBs to their DNA or RNA targets in vitro and evaluated the specificity of the binding by measuring changes in fluorescent signal after incubating the MBs with corresponding and control targets. After mixing K-ras MB1 or MB2 with K-ras wild type (WT), Mut 1 (GGT to GAT) or Mut 2 (GGT to GTT) target, the fluorescence intensity in each group was measured using a fluorescence microplate reader. We found that a stronger fluorescent signal was produced (2–15 fold) when the K-ras MBs were mixed with their specific K-ras mutant targets, compared to non-specific K-ras targets (Fig. 1A). Further, the K-ras MBs were highly specific to K-ras DNA target. The relative fluorescence units in K-ras MB1 or MB2 mixed with their specific mutant K-ras targets were 33 to 51 fold higher than K-ras MBs mixed with survivin DNA targets (Fig. 1A). We also detected about 7-fold higher fluorescence intensity in survivin MB mixed with its oligonucleotide target as compared with that mixed with K-ras target (Fig. 1A). The results from this study demonstrated that K-ras and survivin MBs bound to specific oligonucleotide sequences and produced strong fluorescent signals in vitro. In addition, K-ras MBs differentially bind to K-ras targets with a single base mutation.
Figure 1.
Examination of specific binding of MBs to their DNA targets in solution. (A) Selective binding of K-ras MBs to specific mutant K-ras targets and specific hybridization of survivin MB to its DNA target. K-ras MB1, K-ras MB2 or survivin MB were mixed with various oligonucleotide targets for 1 hour. Relative fluorescence units (RT-U) were measured by a fluorescence microplate reader. The number in the bar figure is the mean fluorescence unit of four repeat samples. As shown, 2–3 fold higher fluorescence units were detected in K-ras MB1 mixing with Mut 1 target than mixing with WT or Mut 2 target. The differences between fluorescence units of K-ras MB2 mixed with K-ras Mut 2, and K-ras MB2 mixed with WT or K-ras Mut 1 were 4 and 15 fold. For both K-ras MB1 and MB2, there was a significant difference between the relative fluorescence obtained when the MBs were mixed with specific mutant K-ras targets and those with nonspecific mutants or WT K-ras target (Student’s t-test: P < 0.001 for all groups). On the other hand, the fluorescence intensity was 33 to 50 fold higher when K-ras MBs were mixed with its corresponding K-ras targets than that mixed with a survivin target. Survivin MB also showed a high specificity for survivin target. (B) Thermal profile of K-ras MBs hybridizing to DNA targets with wild type, specific and nonspecific K-ras mutations. K-ras MBs were mixed with oligonucleotide targets for K-ras wild type (WT), Mut 1 and Mut 2 in 96-well PCR plates. The plates were placed immediately in a BioRad iCycler and relative fluorescence units were measured by the end of each temperature cycle using filters for Cy3 and Texas-red. Relative fluorescence units from each group were normalized with relative fluorescence units of Cy3 or Texas red dye at each temperature to correct the intrinsic changes of fluorescence signal due to temperature change. Significant differences in the fluorescence intensity were detected between K-ras MBs mixed with specific K-ras mutant target and those with WT K-ras or nonspecific K-ras mutant target (Curve statistic analysis by Student’s t-test: K-ras MB1: P< 1×10−9; K-ras MB2: P< 1×10−13). (C) Detection of the mutant K-ras mRNAs in total RNA isolated from pancreatic cancer cell lines. 1μg of total RNA was mixed with 100 nM of either K-ras MB1 or K-ras MB2 for one hour and RFU was measured using a fluorescence microplate reader. The number in the bar figure is the mean fluorescence unit of three repeat samples
To determine optimal hybridization temperatures for K-ras MBs, we examined the effect of temperature on specific binding of the MBs to DNA targets. Using a thermal cycle, we monitored changes in the fluorescence signal at temperatures ranging from 15 to 80°C after mixing the K-ras MBs with different DNA targets. We found that in solution, K-ras MB1 selectively hybridized to K-ras Mut 1 while K-ras MB2 preferentially bound to K-ras Mut 2 DNA target at temperatures ranging from 15 to 55°C (Fig. 1B). Both K-ras MBs showed a low binding affinity with K-ras WT or nonspecific mutant K-ras target. However, when the temperature was over 55°C, K-ras MBs began to dissociate from their targets and the stem of the MBs started to break apart, resulting in increased fluorescent signal in the mixtures of K-ras MBs and nonspecific DNA targets (Fig. 1B).
We further examined whether K-ras MBs bind to specific mRNA targets in solution. Although previous publications showed types of K-ras mutations in pancreatic cancer cell lines, we examined K-ras mutations in the cancer cell lines in our laboratory to confirm the presence of specific K-ras mutations in the cells.26,30 Consistent with previous reports, we found that the PANC-1 cell line has a K-ras codon 12 GGT to GAT mutation. The Capan-2 cell line has a GGT to GTT mutation. The BXPC-3 cell line contains a wild type K-ras gene (Table 2). The K-ras mutation in the MIA PaCa-2 cell line was referenced from previous studies.26,30 We isolated total RNA from those cell lines and mixed total RNA with either K-ras MB1 or K-ras MB2. As shown in Figure 1C, we found that K-ras MB1 produced a higher fluorescence signal in PANC-1 RNA sample while K-ras MB2 generated a stronger fluorescence in the group with Capan-2 RNA. Therefore, K-ras MBs are able to recognize a single base change in both DNA and mRNA targets. It is feasible to use MBs for detecting expression of mutant K-ras and survivin genes in pancreatic cancer cells.
Delivery of K-ras and survivin MBs produced fluorescent signals in pancreatic cancer cells expressing mutant K-ras and survivin genes
Although the thermal profile of the K-ras MBs in solution showed that the MBs selectively bound to specific K-ras mutant targets at temperature ranging from 15 to 55°C, our initial study on the pancreatic cancer cell lines suggested that incubation of the fixed cells with K-ras MBs at lower temperature, such as below 37°C, produced a high background fluorescence in the cells expressing a wild type or nonspecific mutant K-ras mRNA (data not shown). The background was significantly reduced when the incubation temperature was raised to 42 to 50°C. Therefore, we used an incubation temperature of 50°C for our study.
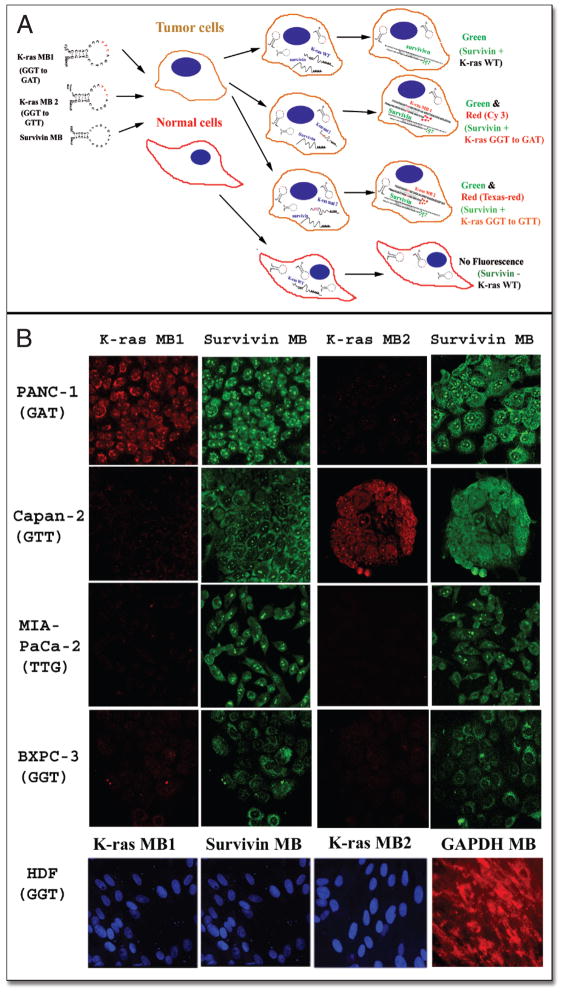
Since pancreatic cancer cells consist of heterogeneous cell populations that express different tumor markers, it is important to develop a method that can detect the expression of several tumor marker genes simultaneously in single cells, which would increase sensitivity and specificity for detection. We examined the feasibility of detection of both mutant K-ras and survivin mRNA in single cells. The mechanism of the action for the detection of pancreatic cancer cells using K-ras and survivin MBs is illustrated in Figure 2A. We incubated acetone-fixed pancreatic cancer and normal cells with a mixture of 50 nM of either K-ras MB1 (Cy3) or MB2 (Texas-red) and 200 nM of survivin MB (FITC) at 50°C for 60 minutes. We used a low concentration of K-ras MBs than survivin MB since our pilot study showed that although high K-ras MB concentrations produced strong fluorescence signal in pancreatic cancer cells with a specific K-ras mutation, nonspecific fluorescence was also increased in the cells containing a wild type or a different mutant K-ras gene. After a brief wash, the slides were observed under a confocal microscope. We found that K-ras MBs preferentially produced strong fluorescent signals in the pancreatic cancer cells with specific K-ras mutations. As shown in Figure 2B, after incubation of K-ras MB1 with PANC-1 and Capan-2 cells, a stronger fluorescent signal was produced in PANC-1 cells that contained a GGT to GAT mutation compared to Capan-2 cells that expressed a GGT to GTT mutant K-ras mRNA. On the other hand, brighter fluorescence was observed in Capan-2 cells after incubation with K-ras MB2, which detects a GGT to GTT mutation (Fig. 2B) compared to PANC-1 cells. We further examined the specificity of the K-ras MBs on pancreatic cancer cell lines that express a different type of mutant K-ras gene or contain a wild type K-ras mRNA. We found that incubation of either K-ras MB1 or MB2 with MIA PaCa-2 cell line, which expresses a GGT to TGT mutant K-ras gene, did not generate a strong fluorescence signal in the cells (Fig. 2B). Moreover, BXPC-3 cells express a wild type K-ras gene and both K-ras MBs failed to generate a strong fluorescent signal in the cells. However, regardless of fluorescence intensities generated by K-ras MBs, simultaneous delivery of survivin MB into the pancreatic cancer cells produced intermediate to strong green fluorescence in all tumor cell lines. In PANC-1 and Capan-2 cells, which have both survivin and mutant K-ras gene expression, we observed both green and red fluorescence signals, suggesting that MBs were able to examine the expression of multiple genes simultaneously in single cells. On the other hand, delivery of the K-ras MBs and survivin MB into normal cell line HDF failed to produce fluorescence signals (Fig. 2B). Incubation of HDF cells with a GAPDH MB produced a strong fluorescence signal, suggesting that mRNAs were preserved well in HDF cells (Fig. 2B).
Figure 2.
Detection of pancreatic cancer cells using MBs targeting tumor marker genes. (A) Schematic illustration of detection of tumor marker gene-expressing cells using MB probes. MBs targeting different gene transcripts are labeled with different fluorophores and the expression of several genes can be detected simultaneously in single cells. In this study, MBs designed to target tumor marker mRNAs such as mutant K-ras and survivin were delivered into fixed cells. Although single cells received all MBs, only the cells expressing the specific genes produced fluorescent signals. For example, in normal cells which lack survivin gene expression and have a wild type K-ras gene, delivery of the MBs doesn’t generate fluorescent signals. However, pancreatic cancer cells express survivin and/or mutant K-ras genes and delivery of MBs into the cancer cells produced green (survivin only) or green and red fluorescence (survivin and mutant K-ras genes). (B) MB-imaging of pancreatic cancer cells expressing specific mutant K-ras and tumor marker survivin gene. Pancreatic cancer and normal cell lines were cultured in chamber slides and fixed with ice-cold acetone. The cells were then incubated with a mixture of MBs containing either K-ras MB1-Cy3 (GGT to GAT) or K-ras MB2-Texas red (GGT to GTT), and survivin MB-FITC. Fluorescent images were taken under a confocal microscope using a 40x lens. The same exposure time was used to take all images for each color. The cells with red fluorescence were pancreatic cancer cells expressing specific mutant K-ras as detected either by K-ras MB1 or K-ras MB2. The cells expressing survivin gene showed green fluorescence. In addition to K-ras and survivin MBs, normal cell line HDF was also incubated with GAPDH MB-cy3 as a control (Red). SYTOX blue (Molecular Probes) was used as counterstaining for nuclei (Blue). Images of K-ras MB1 and survivin MB for HDF cells were taken from the same field of confocal microscope. Fluorescence image of GAPDH MB was taken from a different area of HDF cells.
We further quantified the fluorescence intensity from the images taken from the confocal microscope. Our results confirmed the microscopic observation. After incubating with K-ras MB1, the mean fluorescence intensity in PANC-1 cells was 3.5, 5.2, 7.3 or 11 fold higher than that detected in BXPC-3, Capan-2, MIA PaCa-2 or HDF cells respectively. On the other hand, delivery of K-ras MB2 produced a high level of fluorescence in Capan-2 cells, which was 3, 9, 11, or 20 fold higher than that detected in PANC-1, BXPC-3, HDF or MIA PaCa-2 cells respectively (Fig. 3A).
Figure 3.
Analysis of fluorescent signal produced in pancreatic cancer cells after delivery of K-ras MBs targeting mutant K-ras mRNAs in the cells. (A) Quantitative analysis of fluorescence intensity in K-ras MB-labeled cells. Fluorescence images were obtained from K-ras MB-labeled cells under a confocal microscope using the same instrument setting for either Cy3 or Texas red. The numbers in the bar figure are the mean fluorescence units from three to four images with three areas measured in each image. Significant differences were detected in the relative fluorescence unit of the cells with a specific K-ras mutation after delivery of K-ras MB1 or K-ras MB2 compared to the cells with a wild type or nonspecific K-ras gene (Student’s t-test: P < 1×10−5 for all groups). Similar results were obtained from three independent experiments. (B) Quantification of the level of K-ras mRNA in pancreatic cancer cell lines by Real-Time RT PCR. cDNAs were amplified with K-ras primer pairs using a Bio-Rad iCycler. Amplification of β-actin gene from the same cDNA samples was used as an internal control. The numbers in the figure were the average numbers of two to three repeat samples calculated from ratios of the starting quantity of survivin gene and the starting quantity of β-actin gene. The levels of K-ras mRNA may represent both mutant and wild type K-ras gene expression since pancreatic cancer cell lines have heterozygous K-ras mutations.
To determine that differential fluorescent signals in pancreatic cancer cells produced by the K-ras MBs were indeed due to the specific binding of the MBs to mutant K-ras transcripts but not to differences in the levels of K-ras gene expression, we examined K-ras gene expression in pancreatic cancer cell lines by Real-Time RT PCR. We detected high levels of K-ras mRNA in PANC-l, Capan-2 and MIA PaCa-2 cell lines (Fig. 3B). However, the cell lines expressing a wild type K-ras, such as BXPC-3 and HDF, showed a low level of K-ras gene expression. Since we detected a high level of K-ras mRNA in MIA PaCa-2 cells that had a low level of fluorescence signal after incubation with both K-ras MBl and MB2, it is unlikely that the difference in fluorescence intensity detected in those cell lines was entirely the result of levels of gene expression. In addition, the detection of strong fluorescent signals in Capan-2 cells by K-ras MB2 but not K-ras MB1 or in PANC-1 cells by K-ras MB1 but not K-ras MB2 further supported our conclusion. The results of this study demonstrate that K-ras MBs bound selectively to mutant K-ras mRNA and generated a strong fluorescent signal in pancreatic cancer cells.
K-ras and survivin MBs were able to detect cancer cells in frozen tissue sections
At present, the pathological diagnosis of pancreatic cancer mainly depends on morphological classification and immunohistochemical staining with tissue or tumor markers. Development of novel and simple approaches for the detection of cancer cells by examining the expression of multiple tumor marker genes on the same tissue section may increase the sensitivity and specificity. To address this issue, we examined the feasibility of detecting cancer cells on frozen tissue sections using the MBs. Frozen tissue sections of paired pancreatic normal and cancer tissues were incubated with 100 nM of either K-ras MB1 or MB2 for 60 minutes. The slides were observed under a fluorescence microscope. Results from examination of tissue samples from five pancreatic cancer patients showed that K-ras MBs were able to detect cancer cells expressing specific mutant K-ras mRNAs on frozen tissue sections. For example, we detected cells with red fluorescence on frozen sections of cancer tissues from patient # 1 and #2, after incubation with K-ras MBl (GGT to GAT) but not K-ras MB2 (Fig. 4A). We also detected the cells with a bright fluorescence signal on frozen sections of patients #4 and #5 after incubating with K-ras MB2 but not K-ras MB1 (Fig. 4A). To confirm the presence and types of K-ras mutations, we used a mutant-enriched PCR procedure to examine the genomic DNA isolated from pancreatic cancer tissues of the patients. Consistent with the MB-detection, results of DNA sequencing demonstrated that patient #1 had a GGT to GAT mutation and patient # 5 had a GGT to GTT mutation (Fig. 4A). Interestedly, we found two types of K-ras mutations in the tissue sample of the patient #2, including a GGT to GAT and a GGT to TGT mutation. Since we used frozen tissue blocks rather than microdissection to obtain the total DNA, it is still unclear whether two K-ras mutations were from the same tumor cell population or from heterogeneous tumor cell populations. Examination of frozen tissue sections of paired normal pancreatic tissue from patient #1 and #2 after incubating the K-ras MBl or patient # 4 and #5 following delivery of K-ras MB2 failed to detect any cells with strong fluorescent signals (Fig. 4A, representative images from normal pancreatic tissues of Patient # 1 and #5).
Figure 4.
Specific imaging of pancreatic cancer cells expressing mutant K-ras and survivin mRNAs on frozen tissue sections of pancreatic cancer tissues. Frozen tissue sections were incubated with K-ras MB1 or K-ras MB2 and counterstained with Hoechst 33342. All fluorescent images were taken by Nikon Eclipse E800 fluorescence microscope under a 40 × lens using an Optronics Magnafire digital imaging system. (A) Detection of expression of specific mutant K-ras genes in pancreatic cancer cells on frozen sections using K-ras MBs. K-ras MB1 detected the cancer cells expressing a GGT to GAT mutant K-ras gene on frozen sections of pancreatic cancer tissues from patients #1 and #2. However, bright red fluorescent cells were found on frozen sections of pancreatic cancer tissues from patient #5, which had a K-ras GGT to GTT mutation, only after incubation with K-ras MB2. The frozen sections from normal pancreatic tissues (patient #1 and #5) did not show bright red fluorescence signals following incubation with either K-ras MB1 (patient #1) or K-ras MB2 (patient #5). (B) Frozen sections from patient #2 were incubated with either K-ras MB1 or K-ras MB2 and the fluorescent images were taken under a fluorescence microscope. The sections were then stained with H&E and observed under a bright field of the microscope. The same areas were identified and compared with previous fluorescent images. Yellow arrows indicate pancreatic cancer cells expressing a GGT to GAT mutant K-ras gene. White arrows show the absence of fluorescence in normal stromal cells in K-ras MB 1 labeled section or in pancreatic cancer cells in K-ras MB2 labeled section. (C) Bright fluorescence was detected in normal pancreatic tissues after incubation with GAPDH MB-Cy3, suggesting that the absence of fluorescence in K-ras MB labeled normal pancreatic tissues was not a result of degradation of mRNAs in the frozen sections.
To determine whether the K-ras MB positive cells detected on the frozen tissue sections are indeed cancer cells, we incubated the frozen tissue sections from patient #2 with either K-ras MB1 or K-ras MB2 and then stained with H&E for the morphology of the MB positive cells. Our result indicated that K-ras MB1 positive cells on the frozen section have characteristics of pancreatic cancer cells (Fig. 4B).
We further showed that lack of a strong fluorescence in normal pancreatic tissues after incubating with K-ras MBs was not due to the degradation of mRNAs by RNAase, which is present in normal pancreatic tissues. We observed a strong fluorescence signal in the tissue sections after incubation with a control GAPDH MB (Fig. 4C).
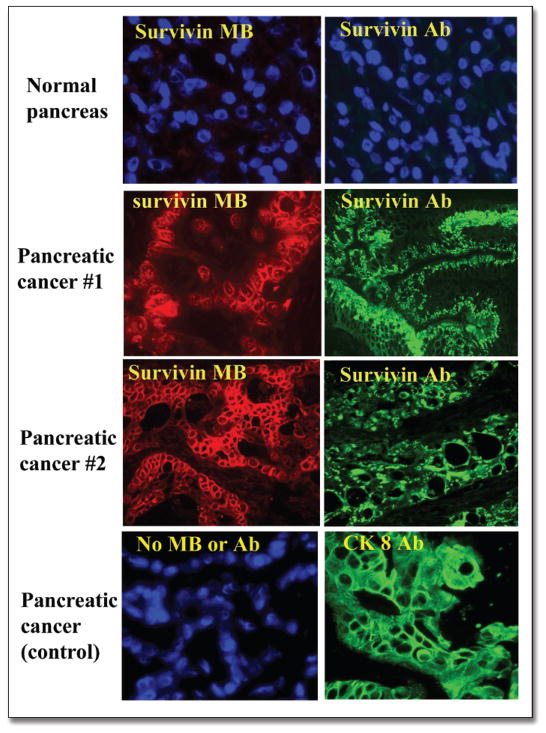
We also examined the feasibility of detection of survivin gene expression in pancreatic cancer tissues by survivin MB. At first, expression of survivin in pancreatic cancer tissues was demonstrated by immunofluorescence labeling with an anti-survivin antibody. We showed that survivin protein was highly expressed in pancreatic cancer tissues but was undetectable in normal pancreas (Fig. 5). Next, the frozen tissue sections from cancer and normal tissues were incubated with survivin MB-Cy3 for 60 minutes and observed under a fluorescence microscope. A high level of fluorescence signal was detected in the cancer cells on the frozen sections of pancreatic cancer but not in normal tissues (Fig. 5). Immuofluorescence labeling with an anti-cytokeratin 8 antibody showed that survivin-expressing cells on frozen sections were epithelial-type cancer cells in pancreas (Fig. 5).
Figure 5.
Detection of pancreatic cancer cells using survivin MB and a survivin antibody. Frozen sections from normal and pancreatic cancer tissues were incubated with either survivin MB or survivin antibody. The slides were examined under a fluorescence microscope after labeling with a secondary detection antibody and FITC-streptavidin. As shown, survivin MB detected a high level of survivin mRNA in pancreatic cancer cells but not in normal pancreatic tissues. Immunofluorescence labeling with survivin antibody further confirmed the presence of survivin protein in the pancreatic cancer cells. An anti-cytokeratin 8 antibody was used as a positive control for pancreatic cancer cells.
Our results with in situ detection of tumor marker gene expression in pancreatic cancer tissues indicate that MB-imaging of tumor marker genes may provide a simple and specific means for pathologists to identify cancer cells on frozen tissue sections or other sources of clinical samples, such as fine needle aspirates and cellular fraction of body fluids.
DISCUSSION
Increasing evidence has indicated that molecular beacons have a great advantage in detecting specific oligonucleotide sequences compared to linear probes.4,31 The design of MBs allows specific binding of the MBs to their target nucleotide sequences and reports the hybridization by generating a fluorescent signal without the separation of unbound probes from the MB-target complex since free MBs do not fluoresce. Therefore, MBs should be an excellent tool for detecting specific nucleotide sequences, such as mRNA and DNA, with a high signal to noise ratio in intact cells as well as in solution. The ability of the MB probes to detect specific target molecules without the washing-away or separation of unbound probes also provided us with an opportunity to detect intracellular mRNA molecules. It has been shown that the detection limit of preformed MB/β-actin mRNA duplexes microinjected into the cells was 10 mRNA molecules, suggesting that MB technology is a very sensitive method for detecting mRNAs in cells.12 Although previous studies have shown that it is feasible to detect mRNAs in cells using MBs,12–14,32 the significance of using this technology to address issues in cancer research and clinical applications has not been explored. Methods of applying MB technology for clinical research have yet to be developed.
In this study, we developed a simple MB approach for the detection of tumor marker gene expression in pancreatic cancer cells. We designed the MBs targeting tumor marker mRNAs that are found in pancreatic cancer, such as mutant K-ras and survivin. From the study of pancreatic cancer cell lines and tissues, we demonstrated that the K-ras MBs selectively bind to mutant K-ras mRNAs in fixed cancer cells, resulting in strong fluorescence signals in the cells. Previous studies on mutation detection by MBs were carried out in solution. We showed that properly designed MBs can detect single base mutations in intact cells. By optimizing hybridization conditions such as temperature, buffer condition, MB concentration and method and time of fixation, we could achieve a high fluorescent signal in cancer cells expressing specific mutant K-ras mRNA and reduce the background or nonspecific fluorescence in cells expressing a different type of mutant K-ras mRNA or containing a wild type K-ras gene. Although we only detected 2 to 15 fold higher fluorescence intensity in solution when K-ras MBs mixed with specific mutant K-ras DNA target than that mixed with nonspecific K-ras target, 3 to 20 fold higher levels of differences were observed when K-ras MBs were delivered into the cells expressing specific mutant K-ras mRNA compared to the cells expressing a wild type or non-specific K-ras gene. It is possible that a higher level of fluorescent signal observed in the cells expressing specific K-ras mRNA is due to different hybridization dynamics between a K-ras MB with a short DNA target and a K-ras MB with an mRNA molecule inside a cell. In addition, results of Real-Time RT PCR showed that the levels of K-ras gene expression in pancreatic cancer cells with K-ras mutations were higher than the cells expressing a wild type K-ras gene. Although the level of K-ras mRNAs detected by Real-Time RT PCR may include both mutant and wild type K-ras mRNAs because most K-ras mutations in cancer cells are heterozygous, it is also possible that the difference in fluorescence intensity was further enhanced in the cells with a high level of mutant K-ras gene expression.
During last decade, extensive studies have been carried out to detect K-ras mutations in blood, pancreatic juice and cancer tissue samples from pancreatic cancer patients using PCR or mutant-enriched PCR.16,33 Utilization of MBs to detect K-ras mutations specifically in PCR products of DNA samples isolated from lung cancers has been reported.34 Although identification of K-ras mutations by PCR is a fairly sensitive molecular approach, the procedures for PCR and subsequent assays for the identification of mutations are very time-consuming, making them difficult as a routine clinical procedure. A major advantage of using a MB-based approach as compared to PCR and immunohistochemistry is that a mixture of MBs targeting multiple tumor-specific mRNAs can be delivered into single cells at the same time, and the expression of all these markers can be observed in a single assay using a fluorescence microscope.
In addition, K-ras mutations were also found in 30–40% of patients with pancreatic duct cysts or chronic pancreatitis.35,36 A better way to identify a cancer cell is to detect expression of mutant K-ras gene together with several other tumor marker genes. We examined the expression of another tumor marker gene, survivin, in pancreatic cancer cell lines using a MB targeting survivin mRNA. Since survivin is not found in normal pancreatic tissues but is highly expressed in over 70% of pancreatic cancer tissues including early stage carcinomas,23 a combination of the detection of survivin with mutant K-ras mRNAs may further enhance specificity of the detection. We demonstrated that delivery of survivin MBs into cells produced a strong fluorescent signal in cancer cells but not in normal cells. A mixture of survivin and K-ras MBs generated different fluorescent signals in pancreatic cancer cell lines expressing survivin and/or specific mutant K-ras mRNA. To detect pancreatic cancer cells in clinical samples, it is important to identify a few abnormal cells that are mixed with large amounts of normal cells. MB-imaging individual cells expressing tumor marker genes may allow the detection of those cancer cells.
In this study, we showed that it is feasible to identify pancreatic cancer cells through detection of both mutant K-ras and survivin mRNAs using the MB-technology. This technology offers an opportunity to detect specific expression of several tumor marker genes in single cells using a simple procedure. Our results demonstrated that the MBs could be used for determination of the presence of mutations in tumor suppressor genes or oncogenes as well as for the examination of expression of tumor marker genes in the cancer cells. In order to detect point mutations, it is necessary to synthesize the MBs that are specific for each mutation in the gene. Thus, a limitation of using MBs is that it is best for genes with a few hot-spot mutations, such as K-ras codon 12 mutations in pancreatic cancers. However, the ability of MBs to detect the expression of genes that are highly expressed in tumor cells but lacking or low in normal cells, such as the survivin gene, should allow us to develop approaches for detecting cancer cells in many common types of human cancers.
Traditional methods for classifying the cancer cells on tissue sections or aspirates of fine needle biopsy involve thorough examination of the morphology of the cells after H&E staining or immunostaining with antibodies for cellular and tumor-related protein markers. Although in situ hybridization method has been used to detect gene expression in tissue sections, it is very time-consuming and usually involves a high background since unbound probes also produce a fluorescent signal. In this study, we found that MBs could be used to detect the expression of tumor marker genes and to identify specific mutations on frozen tissue sections. The procedure for the detection of mRNA by MBs is very simple and the results can be observed within one hour after a brief washing without extensive washing steps and days of procedures as required by in situ hybridization protocol. Development of this MB approach for detection of gene expression on tissue section should provide us with a new method to identify and classify the types of cancer cells on tissue sections, aspirates from fine needle biopsy, blood and exfoliated cells in body fluids.
In summary, we have developed a simple MB approach to examine the expression of tumor marker genes in human cancer cells. We demonstrated that MBs can be used to identify cancer cells expressing oncogenes with point mutations as well as overexpressing tumor marker genes. The results of our study indicate that molecular imaging of cancer cells with MBs has great potential for the development of a simple and specific procedure for cancer detection.
Acknowledgments
We would like to thank Dr. Prasanthi Karna for quantitative analysis of K-ras MB labeled fluorescence images. We would also like to thank Dr. Mark Behlke for suggestions on the design of K-ras MBs and survivin MB-Cy3 and for providing MBs and DNA targets during our pilot study for this research project. We appreciate helpful discussions and collaborations with Drs. Wei Zhou, Andrew Tsourkas, and Gang Bao. This research was supported by the funds from the Wallace H. Coulter Foundation, NIH CA80017 and NIH CA095643.
ABBREVIATIONS
- Dabcyl
4-({4′-(dimethylamino)phenyl]azo) benzoic acid
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IAP
inhibitor of apoptosis protein
- MB
Molecular beacon
- Mut
mutation
- WT
Wild type
References
- 2.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–9. [PubMed] [Google Scholar]
- 3.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 5.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci USA. 1999;96:6171–6. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frutos AG, Pal S, Quesada M, Lahiri J. Method for detection of single-base mismatches using bimolecular beacons. J Am Chem Soc. 2002;124:2396–7. doi: 10.1021/ja012374d. [DOI] [PubMed] [Google Scholar]
- 8.Broude NE. Stem-loop oligonucleotides: A robust tool for molecular biology and biotech-nology. Trends Biotechnol. 2002;20:249–56. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]
- 9.Fang X, Li JJ, Tan W. Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal Chem. 2000;72:3280–5. doi: 10.1021/ac991434j. [DOI] [PubMed] [Google Scholar]
- 10.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat Biotechnol. 2002;20:171–6. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, Galizia G, Lieto E, Goodman SN, Romans KE, Kinzler KW, Vogelstein B, Choti MA, Montgomery EA. Counting alleles reveals a connection between chromosome 18q loss and vascular invasion. Nat Biotechnol. 2001;19:78–81. doi: 10.1038/83572. [DOI] [PubMed] [Google Scholar]
- 12.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA. RNA hybridization in living cells. Proc Natl Acad Sci USA. 1998;95:11538–43. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlette J, Tan W. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal Chem. 2001;73:5544–50. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 14.Fang X, Mi Y, Li JJ, Beck T, Schuster S, Tan W. Molecular beacons: Fluorogenic probes for living cell study. Cell Biochem Biophys. 2002;37:71–81. doi: 10.1385/CBB:37:2:071. [DOI] [PubMed] [Google Scholar]
- 15.Shah R, El-Deiry WS. p53-Dependent activation of a molecular beacon in tumor cells following exposure to doxorubicin chemotherapy. Cancer Biol Ther. 2004:3. doi: 10.4161/cbt.3.9.1053. [DOI] [PubMed] [Google Scholar]
- 16.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–54. [PMC free article] [PubMed] [Google Scholar]
- 17.Minamoto T, Mai M, Ronai Z. K-ras mutation: Early detection in molecular diagnosis and risk assessment of colorectal, pancreas, and lung cancers—a review. Cancer Detect Prev. 2000;24:1–12. [PubMed] [Google Scholar]
- 18.Futakawa N, Kimura W, Yamagata S, Zhao B, Ilsoo H, Inoue T, Sata N, Kawaguchi Y, Kubota Y, Muto T. Significance of K-ras mutation and CEA level in pancreatic juice in the diagnosis of pancreatic cancer. J Hepatobiliary Pancreat Surg. 2000;7:63–71. doi: 10.1007/s005340050156. [DOI] [PubMed] [Google Scholar]
- 19.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. Embo J. 1998;17:2215–23. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17:3247–59. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- 21.Altieri DC. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol Med. 2001;7:542–7. doi: 10.1016/s1471-4914(01)02243-2. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127–34. [PubMed] [Google Scholar]
- 23.Satoh K, Kaneko K, Hirota M, Masamune A, Satoh A, Shimosegawa T. Expression of survivin is correlated with cancer cell apoptosis and is involved in the development of human pancreatic duct cell tumors. Cancer. 2001;92:271–8. doi: 10.1002/1097-0142(20010715)92:2<271::aid-cncr1319>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Sarela AI, Verbeke CS, Ramsdale J, Davies CL, Markham AF, Guillou PJ. Expression of survivin, a novel inhibitor of apoptosis and cell cycle regulatory protein, in pancreatic adenocarcinoma. Br J Cancer. 2002;86:886–92. doi: 10.1038/sj.bjc.6600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apple SK, Hecht JR, Novak JM, Nieberg RK, Rosenthal DL, Grody WW. Polymerase chain reaction-based K-ras mutation detection of pancreatic adenocarcinoma in routine cytology smears. Am J Clin Pathol. 1996;105:321–6. doi: 10.1093/ajcp/105.3.321. [DOI] [PubMed] [Google Scholar]
- 26.Berrozpe G, Schaeffer J, Peinado MA, Real FX, Perucho M. Comparative analysis of mutations in the p53 and K-ras genes in pancreatic cancer. Int J Cancer. 1994;58:185–91. doi: 10.1002/ijc.2910580207. [DOI] [PubMed] [Google Scholar]
- 27.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ, Kern SE. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–4. [PubMed] [Google Scholar]
- 28.Nomoto S, Nakao A, Ando N, Takeda S, Kasai Y, Inoue S, Kaneko T, Takagi H. Clinical application of K-ras oncogene mutations in pancreatic carcinoma: Detection of micrometastases. Semin Surg Oncol. 1998;15:40–6. doi: 10.1002/(sici)1098-2388(199807/08)15:1<40::aid-ssu7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 29.Hruban RH, Sturm PD, Slebos RJ, Wilentz RE, Musler AR, Yeo CJ, Sohn TA, van Velthuysen ML, Offerhaus GJ. Can K-ras codon 12 mutations be used to distinguish benign bile duct proliferations from metastases in the liver? A molecular analysis of 101 liver lesions from 93 patients. Am J Pathol. 1997;151:943–9. [PMC free article] [PubMed] [Google Scholar]
- 30.Moore PS, Sipos B, Orlandini S, Sorio C, Real FX, Lemoine NR, Gress T, Bassi C, Kloppel G, Kalthoff H, Ungefroren H, Lohr M, Scarpa A. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 31.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci USA. 1998;95:8602–6. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dirks RW, Molenaar C, Tanke HJ. Methods for visualizing RNA processing and transport pathways in living cells. Histochem Cell Biol. 2001;115:3–11. doi: 10.1007/s004180000214. [DOI] [PubMed] [Google Scholar]
- 33.Shibata K, Mori M, Kitano S, Akiyoshi T. Detection of ras gene mutations in peripheral blood of carcinoma patients using CD45 immunomagnetic separation and nested mutant allele specific amplification. Int J Oncol. 1998;12:1333–8. doi: 10.3892/ijo.12.6.1333. [DOI] [PubMed] [Google Scholar]
- 34.Clayton SJ, Scott FM, Walker J, Callaghan K, Haque K, Liloglou T, Xinarianos G, Shawcross S, Ceuppens P, Field JK, Fox JC. K-ras point mutation detection in lung cancer: Comparison of two approaches to somatic mutation detection using ARMS allele-specific amplification. Clin Chem. 2000;46:1929–38. [PubMed] [Google Scholar]
- 35.Queneau PE, Adessi GL, Thibault P, Cleau D, Heyd B, Mantion G, Carayon P. Early detection of pancreatic cancer in patients with chronic pancreatitis: Diagnostic utility of a K-ras point mutation in the pancreatic juice. Am J Gastroenterol. 2001;96:700–4. doi: 10.1111/j.1572-0241.2001.03608.x. [DOI] [PubMed] [Google Scholar]