Abstract
Purpose
The molecular drivers that determine histology in lung cancer are largely unknown. We investigated whether microRNA (miR) expression profiles can differentiate histological subtypes and predict survival for non-small cell lung cancer.
Experimental design
We analyzed miR expression in 165 adenocarcinoma (AD) and 125 squamous cell carcinoma (SQ) tissue samples from the Environmental And Genetics in Lung cancer Etiology (EAGLE) study using a custom oligo array with 440 human mature antisense miRs. We compared miR expression profiles using t-tests and F-tests and accounted for multiple testing using global permutation tests. We assessed the association of miR expression with tobacco smoking using Spearman correlation coefficients and linear regression models, and with clinical outcome using log-rank tests, Cox proportional hazards and survival risk prediction models, accounting for demographic and tumor characteristics.
Results
MiR expression profiles strongly differed between AD and SQ (global p<0.0001), particularly in the early stages, and included miRs located on chromosome loci most often altered in lung cancer (e.g., 3p21-22). Most miRs, including all members of the let-7 family, were down-regulated in SQ. Major findings were confirmed by QRT-PCR in EAGLE samples and in an independent set of lung cancer cases. In SQ, low expression of miRs down-regulated in the histology comparison was associated with 1.2 to 3.6-fold increased mortality risk. A 5-miR signature significantly predicted survival for SQ.
Conclusions
We identified a miR expression profile that strongly differentiated AD from SQ and had prognostic implications. These findings may lead to histology-based therapeutic approaches.
Introduction
The two most common histologic subtypes of non small cell lung cancer (NSCLC) are squamous cell carcinomas (SQ) and adenocarcinomas (AD), mainly derived from epithelial cells that line the larger airways and the peripheral small airways, respectively (1). Demographic and etiological characteristics of AD and SQ have been described (2;3), and inherited predisposition to distinct histological types has been identified (4;5). Mutations in the RAS proto-oncogene (6) and epidermal growth factor receptor (EGFR) (7) have been mostly found in lung adenocarcinoma, and are typically mutually exclusive (8). While these and other differences are documented, the precise molecular features that characterize and distinguish the two histologies are largely unknown. Consequently, treatment across NSCLC lesions is similar and shows comparable poor efficacy. A refined understanding of the underlying histological distinctions may help elucidate histology-specific patterns that can be exploited therapeutically and improve selection of molecular markers for early detection. We sought to explore microRNA (miR) expression differences by histology and tobacco smoking and their significance in terms of prognosis and treatment of lung cancer.
MiRs are a class of short, highly conserved non-coding RNAs involved in numerous developmental processes. MiRs regulate gene expression through incomplete basepairing to a complementary sequence in the 3′ untranslated region (UTR) of a target mRNA, resulting in translational repression and, to a lesser extent, accelerated turnover of the target transcript (9). Deregulation of miRs has been linked to cancer initiation and progression, indicating that miRs may act as tumor suppressor genes or oncogenes and can target apoptosis (10).
MiR expression studies in lung cancer cell lines or animal models have mostly focused on the let-7 family of miRs, which act as tumor suppressors by repressing cell proliferation and regulating the RAS oncogene (11). Let-7 may also target cell cycle effectors (10) and in turn be down-regulated by the MYC oncogene (12). A few studies in humans identified different miR expression profiles associated with lung cancer survival (13–15). Overall, these studies were relatively small and had limited power to account for other potential risk factors for lung cancer.
We conducted a large miR expression study in 290 subjects, including 165 AD and 125 SQ patients, within the Environment And Genetics in Lung cancer Etiology (EAGLE) study (16). Using a custom-made miR microarray, detailed epidemiological and clinical data, and lung tissue blocks, we investigated whether miR expression profiles can distinguish AD from SQ or predict survival. We also explored the effect of tobacco smoking on miR expression. The large study size allowed adjustment or stratification for potential confounders (e.g., age, sex, smoking, and tumor stage, grade, size, lymph node involvement, presence of metastasis and histology) when appropriate. We confirmed our major findings by QRT-PCR in 49 samples from EAGLE and 76 samples from an independent set of lung cancer cases.
Methods
EAGLE Samples
The EAGLE study design and subjects’ characteristics have been previously described (16). Briefly, EAGLE is a population-based case-control study of lung cancer conducted in Italy between 2003 and 2005, which includes 2100 primary lung cancer cases and 2120 age-, sex- and residence-matched population controls. The study was approved by the institutional review boards of the enrolling hospitals and the National Cancer Institute. All participating subjects signed an informed consent form. Lung cancer histology and presence of malignant cells in the formalin-fixed, paraffin-embedded (FFPE) tissue blocks were ascertained by the EAGLE local pathologists and reviewed by a pathologist from the National Cancer Institute. For the present study, we excluded subjects who received chemotherapy and/or radiation therapy prior to the study in order to avoid potential confounding effects on miR expression due to treatment. Moreover, we excluded tissue blocks with mixed histologies or insufficient tumor material. All cases meeting inclusion criteria with sufficient tissue available were used. The analytic sample included 290 cases (Table 1).
Table 1.
Characteristics of EAGLE patients whose samples were used in miR array analysis
| Subjects characteristics | Adenocarcinoma | Squamous cell carcinoma | |
|---|---|---|---|
| N. = 165
|
N.= 125
|
||
| Agea | >=67 yrs | 66 (46%) | 77 (54%) |
| <67 yrs | 99 (67%) | 48 (33%) | |
| Sex | Males | 89 (42%) | 122 (58%) |
| Females | 76 (96%) | 3 (4%) | |
| Smoking | Current | 70 (52%) | 64 (48%) |
| Formerb | 61 (50%) | 60 (50%) | |
| Never | 34 (97%) | 1 (3%) | |
| Cig. per dayc | >=20 | 71 (43%) | 94 (57%) |
| <20 | 60 (67%) | 30 (33%) | |
| Stage | IA | 32 (56%) | 25 (44%) |
| IB | 33 (55%) | 27 (45%) | |
| IIA | 10 (71%) | 4 (29%) | |
| IIB | 33 (46%) | 38 (54%) | |
| IIIA | 32 (65%) | 17 (35%) | |
| IIIB | 14 (52%) | 13 (48%) | |
| IV | 11 (92%) | 1 (8%) | |
| Life status | Alive | 88 (59%) | 61 (41%) |
| Deceased for lung cancer | 67 (54%) | 56 (46%) | |
| Deceased for other causes | 10 (56%) | 8 (44%) | |
Age range for the entire group of patients was 39.08 to 79.74 (median = 66.87).
For adenocarcinoma, age range was 39.08 to 79.45 (median = 65.04), and for squamous cell carcinoma, age range was 41.95 to 79.74 (median = 69.56).
Former smokers were subjects who quit smoking six months or more prior to the study
Total number of subjects varies because of missing data for this variable.
Chip array analysis and quality control procedures
We used a custom-made two-channel oligo array including 713 human, mammalian and viral mature antisense miRs plus 2 internal controls with 7 serial dilutions, using one EBV cell line as reference sample (17). The in-house miR array was tested for hybridization efficiency (intensity range log10 2–16), reproducibility and concordance with Ambion oligo set arrays (data not shown). Real time PCR validations of the array were performed on selected miRs (18). For the present study, we focused on the human miRs (n=440). Details on the oligo array, RNA extraction, normalization and quality control procedures are reported in the Supplemental Material (SM) #1.
Statistical analysis
Class comparisons
Tests for expression differences between histologies were conducted for individual miRs using 2-sided t-tests or F-tests, considering p-values<0.05 as significant. Adjustment or stratification for age, sex, histology, tumor characteristics, and smoking was performed when appropriate (details in SM1).
The Benjamini and Hochberg method was used to estimate the false discovery rate (FDR) (19). To account for multiple testing, global permutation tests with 10,000 permutations were conducted for each class comparison to confirm overall significance of the expression profile differences (details on permutation tests in SM1).
Association between miR expression and tobacco smoking
When relating miR expression to a specified continuous outcome (e.g., for the analysis of cigarettes per day), Spearman correlation coefficients were calculated as a measure of correlation and to compute parametric p-values. In addition, linear regression models were used to examine the association between cigarettes per day (CPD) and miR expression data using the lm function in R statistical package1.
Survival analysis
For survival analyses, patients who were alive at the date of last follow-up (n=149) or who died due to causes other than lung cancer (n=18) were censored. Univariate Cox proportional hazards models were fit to test individual miR expression levels for association with survival. Regression coefficients from these models were tested using a 2-sided Wald test, considering p-values<0.05 as significant. First, we examined the association between expression of individual miRs and survival in unadjusted and adjusted analyses. For SQ, we excluded the 3 female patients and 1 never smoker and adjusted for age and stage. For AD, we adjusted for age, stage, sex and smoking. We also conducted the same analyses restricted to subjects with resectable tumors (stages IA, IB, IIA, IIB, and IIIA) to identify miRs with prognostic potential for surgical candidates. Kaplan-Meier survival curves were plotted for individuals with miR expression levels above and below the median using the R statistical package.
To test combinations of miRs that could predict risk of dying from lung cancer, we used the supervised principal component method of Bair and Tibshirani (20) in BRB-ArrayTools (21). We calculated the first three principal component linear combinations of expression levels from miRs that were univariately correlated with survival at p<0.01 (using Cox regression), producing three “super-miRs”. A Cox proportional hazards model was fit to relate survival time to these three “super-miRs”. This method provides a regression coefficient (weight) for each “super-miR” for calculation of a prognostic index, or weighted combination of the “super-miRs”. To avoid overfitting due to the initial supervised selection of miRs to use in defining the prognostic index, we used ten-fold cross-validation. A high prognostic index value corresponded to a high predicted hazard of death due to lung cancer with correspondingly poor predicted survival. Kaplan-Meier survival curves were plotted for cases predicted to have above average risk (prognostic index above the median in the cross-validated model) and for cases predicted to have below average risk (prognostic index below the median in the cross-validated model). Details on the cross-validation procedure and permutation test are in SM1.
Correlation between miRs and mRNA expression
We estimated the predicted targets of the top miRs differentiating the two histology groups using TargetScan (22). We measured mRNA expression of the putative target genes using Affymetrix U133A microarray chips (23) in 47 snap-frozen tissue samples from adenocarcinoma patients for whom we also had miR expression data. We estimated the correlation (based on a linear model) between miR expression and gene expression for each miR-probe pair, and computed a global permutation p-value (using 1000 permutations) based on the number of miR-probe correlations that were significant (p-value<0.01) at the individual level.
Molecular function classification of genes targeted by miRs in the histology comparison
We used Gene Ontology (GO) (24),to assign the targeted genes whose expression was significantly correlated with the top miRs differentiating the histology groups to functional categories and rank-ordered the GO categories using GoMiner (25).
Confirmation and replication of miR expression
We confirmed array results by QRT-PCR in 49 samples from EAGLE that had sufficient tumor microRNA remaining after the array analysis. We also replicated our analysis in 76 fresh frozen lung cancer tissue samples from an independent Caucasian population obtained from the Cooperative Human Tissue Network (CHTN). The 49 EAGLE and 76 CHTN samples are described in SM2. We selected 5 miRs to confirm the array results by QRT-PCR using TaqMan microRNA assays (Applied Biosystems) (procedure details in SM1).
Results
Analysis of adenocarcinoma (AD) vs squamous cell carcinoma (SQ)
In the overall unadjusted analysis, miR expression profiles strongly differentiated AD from SQ (class comparison global permutation test p<0.0001), with 127 miRs differentially expressed at p<0.05 and with 86 of the 127 miRs differentially expressed at p<0.001 (list in SM3). To verify whether these results were affected by gender, age or smoking, we repeated the same analyses in male smokers only (84 AD, 121 SQ), adjusting for age. In this adjusted analysis, 34 miRs were confirmed at p<0.0001 (Figure 1 and Table 2), of which 32 (94%) were up-regulated in AD vs. SQ.
Figure 1.
Hierarchical clustering of normalized miR expression levels for the 34 miRs that displayed the strongest statistical significance for differential expression between adenocarcinoma and squamous cell carcinoma samples after adjustment for age in male smoker patients. Average linkage and 1-Pearson correlation as distance metric were used for the clustering.
Table 2.
34 miRs that significantly (at p<0.0001) differentiate adenocarcinoma from squamous cell carcinoma samples in male smoker patients (n. subjects=205)
| microRNAs | p-valuea | FDRb | GM in ADc | GM in SQd | Fold-change AD/SQe | Unique ID |
|---|---|---|---|---|---|---|
| hsa-let-7g | < 1e-07 | < 1e-07 | 0.19 | 0.09 | 2.18 | MIMAT0000414 |
| hsa-let-7b | 1e-07 | 6.6e-06 | 0.49 | 0.27 | 1.83 | MIMAT0000063 |
| hsa-let-7c | 1e-07 | 6.6e-06 | 0.35 | 0.18 | 1.95 | MIMAT0000064 |
| hsa-miR-29a | 2e-07 | 7.9e-06 | 0.35 | 0.18 | 1.98 | MIMAT0000086 |
| hsa-let-7f | 2e-07 | 7.9e-06 | 0.18 | 0.09 | 2.14 | MIMAT0000067 |
| hsa-miR-453# | 4e-07 | 1.13e-05 | 1.25 | 1.46 | 0.86 | MIMAT0001630 |
| hsa-let-7d | 4e-07 | 1.13e-05 | 0.46 | 0.30 | 1.54 | MIMAT0000065 |
| hsa-miR-98 | 6e-07 | 1.32e-05 | 0.25 | 0.12 | 2.06 | MIMAT0000096 |
| hsa-let-7i | 6e-07 | 1.32e-05 | 0.25 | 0.13 | 1.84 | MIMAT0000415 |
| hsa-miR-26a | 8e-07 | 1.58e-05 | 0.63 | 0.28 | 2.27 | MIMAT0000082 |
| hsa-miR-509-3p | 1e-06 | 1.8e-05 | 0.69 | 0.89 | 0.78 | MIMAT0002881 |
| hsa-miR-30b | 1.4e-06 | 2.31e-05 | 0.21 | 0.11 | 1.85 | MIMAT0000420 |
| hsa-miR-146b-5p | 2.1e-06 | 2.97e-05 | 0.05 | 0.03 | 1.71 | MIMAT0002809 |
| hsa-miR-106b | 2.1e-06 | 2.97e-05 | 0.11 | 0.07 | 1.52 | MIMAT0000680 |
| hsa-let-7a | 2.3e-06 | 3.04e-05 | 0.27 | 0.16 | 1.73 | MIMAT0000062 |
| hsa-mir-663 | 3.5e-06 | 4.31e-05 | 1.32 | 0.94 | 1.40 | MIMAT0003326 |
| hsa-miR-30d | 3.7e-06 | 4.31e-05 | 0.63 | 0.39 | 1.62 | MIMAT0000245 |
| hsa-miR-17 | 4.7e-06 | 5.17e-05 | 0.09 | 0.05 | 1.60 | MIMAT0000070 |
| hsa-miR-498& | 5.3e-06 | 5.52e-05 | 1.31 | 0.87 | 1.51 | MIMAT0002824 |
| hsa-miR-26b | 6.4e-06 | 6.34e-05 | 0.40 | 0.23 | 1.75 | MIMAT0000083 |
| hsa-let-7e | 1.11e-06 | 1.e-04 | 0.49 | 0.37 | 1.32 | MIMAT0000066 |
| hsa-mir-654-5p# | 1.28e-06 | 1.15e-04 | 0.53 | 0.43 | 1.24 | MIMAT0003330 |
| hsa-miR-181a | 1.65e-06 | 1.42e-04 | 0.57 | 0.38 | 1.52 | MIMAT0000256 |
| hsa-miR-103 | 2.21e-05 | 1.78e-04 | 0.38 | 0.25 | 1.51 | MIMAT0000101 |
| hsa-miR-195 | 2.25e-05 | 1.78e-04 | 0.21 | 0.12 | 1.66 | MIMAT0000461 |
| hsa-miR-191 | 2.38e-05 | 1.81e-04 | 0.40 | 0.29 | 1.41 | MIMAT0000440 |
| hsa-miR-20a | 2.8e-05 | 1.99e-04 | 0.05 | 0.03 | 1.61 | MIMAT0000075 |
| hsa-miR-106a | 2.81e-05 | 2.32e-04 | 0.07 | 0.05 | 1.56 | MIMAT0000103 |
| hsa-miR-29c | 3.46e-05 | 2.32e-04 | 0.43 | 0.26 | 1.66 | MIMAT0000681 |
| hsa-miR-29b | 3.51e-05 | 2.32e-04 | 0.46 | 0.26 | 1.79 | MIMAT0000100 |
| hsa-miR-491-5p | 5.51e-05 | 3.52e-04 | 1.11 | 0.84 | 1.33 | MIMAT0002807 |
| hsa-miR-19b | 5.76e-05 | 3.56e-04 | 0.07 | 0.04 | 1.59 | MIMAT0000074 |
| hsa-miR-107 | 7.63e-05 | 4.58e-04 | 0.37 | 0.25 | 1.45 | MIMAT0000104 |
| hsa-miR-16 | 9.43e-05 | 5.49e-04 | 0.16 | 0.10 | 1.56 | MIMAT0000069 |
MiRs are sorted by the parametric p-value from the univariate test. Model adjusted by age.
Parametric p-value;
FDR= False Discovery Rate calculated by the method of Benjamini and Hochberg (17);
Geometric mean of miR expression in adenocarcinoma (AD) samples compared to the EBV reference sample;
Geometric mean of miR expression in squamous cell carcinoma (SQ) samples compared to the EBV reference sample;
Ratio of geometric mean ratios of miR expression in adenocarcinoma/squamous cell carcinoma.
MiRs located on chromosome 14q32.2;
MiRs located on chromosome 19q13.42
We explored the correlation of the top 34 miRs differentiating the two histology groups with the mRNA expression of the TargetScan-predicted targets of these miRs. As expected, each miR is predicted to target hundreds of genes (Table 3). Expression of 5 of the 34 top miRs (14.7%) was found to be overall significantly associated with the expression of their putative target genes, while only about 2 were expected by chance. Results summaries are reported in Table 3, and the corresponding list of probes, genes, correlation coefficients and p-values are shown in SM The GO (24) functional categories indicate that these genes are mainly involved in chemical/cellular homeostasis, peroxisomal transport and G-protein signaling (SM5)
Table 3.
Correlation between miRs differentiating adenocarcinoma from squamous cell carcinoma and mRNA expression of predicted target genes
| MicroRNAs | N. genesa | N. probesa | N. correlations | Global p-value |
|---|---|---|---|---|
| hsa-miR-181a | 617 | 708 | 59 | <0.001 |
| hsa-miR-191 | 384 | 450 | 40 | 0.001 |
| hsa-miR-107 | 374 | 434 | 21 | 0.008 |
| hsa-miR-103 | 394 | 497 | 22 | 0.017 |
| hsa-let-7b | 731 | 754 | 25 | 0.029 |
The corresponding genes and probes are listed in SM4
Four miRs among the 34 exhibited >2.0 fold-difference: miR-26a, Let-7g, let-7f, and miR-98 (p<0.0001, Table 2). Interestingly, miR-26a and let-7g are located on chromosome 3p21-22 (26), a common area of chromosomal alterations in lung cancer. MiR-98 is localized to chromosome X within the same cluster of let-7f-2 and is considered part of the let-7 family. Notably, all members of the human let-7 family on our chip (let-7a, b, c, d, e, f, g, i, miR-98 and miR-202) (27) differentiated the two histology groups (p<0.0001) and were up-regulated in AD vs. SQ.
In comparing histologies (p<0.05), we also identified two groups of miRs clustered <10kb apart from each other in two genomic regions that are often gained or lost in lung cancer (28;29): chromosome 19q13.42 with 7 miRs (i.e., miR-498, miR-520b, miR-517*, miR-373*, miR-526b, miR-518c*, miR-520d-3p, plus let-7e, adjacent on chromosome 19q13.41) and chromosome 14q32.2 also with 7 miRs (miR-453, miR-654-5p, miR-299-3p, miR-381, miR-432, miR-342-3p, miR-370).
We verified whether the differences in miR expression between AD and SQ vary by stage. We found that miR expression differed dramatically by histology in stages I and II (SM6-7), but not in the advanced stages, even when samples from stages IIIB and IV were pooled (n=57 AD and 31 SQ, global p-value=0.195, SM8). Specifically, in comparing 65 AD vs. 52 SQ stage I cases and 43 AD vs. 42 SQ stage II cases, there were 99 and 76 miRs, respectively, whose expression strongly differentiated the two histology groups (global p-value<0.0001 for each of stage I and II). Among the 76 miRs differentiating the histologies in stage II, 55 (71%) also differentiated the histologies in stage I and included all members of the let-7 family. The largest difference in miR expression (3.4-fold overall and 3.5-fold in male smokers only) was observed for miR-21 in stage II samples (p-value<10−5). Notably, expression of this miR was not significantly different by histology in stage I cancers. We observed comparable results in the analysis restricted to the male smokers only, with expression of 89 and 90 miRs differentiating histologies in stages I and II, respectively, and only 2 miRs differentially expressed in stages IIIB+IV (data not shown).
Effect of cigarette smoking on miR expression
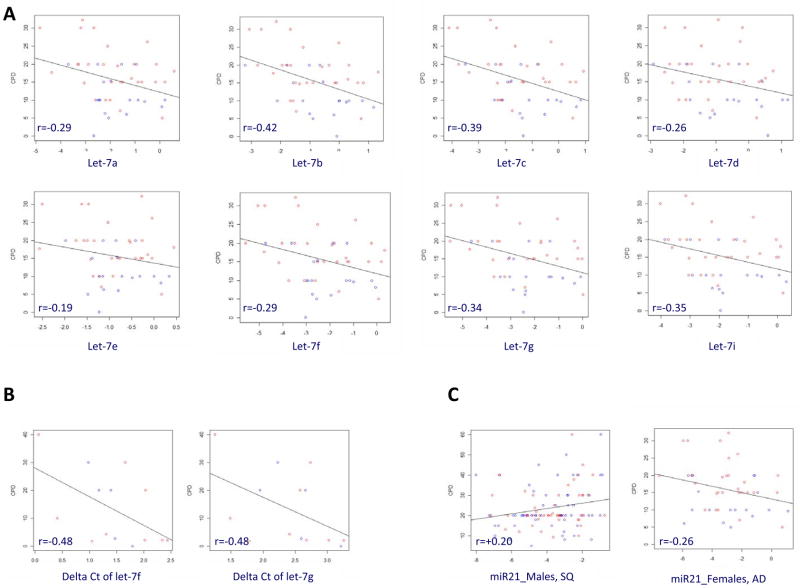
We assessed whether tobacco smoking affected miR expression by comparing miR expression in smokers vs. non smokers. Given the differences in miR expression by histology, we focused this analysis on adenocarcinoma only, the histological group with a wide range of smoking histories, including never smokers (Table 1). No overall significant results were observed by smoking among the AD cases, even when subjects were stratified by sex and adjusted by age (data not shown), although expression of most miRs, including all members of the let-7 family, appeared to be generally down-regulated in the smokers. To further explore the role of smoking, we analyzed the correlation between smoking intensity (number of cigarettes per day) and expression of miRs that were downregulated in SQ compared to AD (let-7 family) and one miR that was upregulated in the same comparison (miR21) in smokers. We found an inverse correlation for the let-7 family in female adenocarcinoma patients, independent from age (Figure 2A). We confirmed this inverse association in CHTN tissue samples from adenocarcinoma female cases (Figure 2B). We could not analyze the effect of smoking in squamous cell carcinoma because we had only 3 females in this group. In males, there was no association between smoking intensity and miR expression in either SQ or AD in both the EAGLE and CHTN samples (data not shown). No major differences were observed by current vs. former smoking status, years of smoking, age at smoking initiation or exposure to environmental (passive) smoking (data not shown). The miR21 analysis showed a significant increase of miR expression by cigarettes per day in males with SQ (p=0.02) (Figure 2C), but no significant association in adenocarcinoma either in males (data not shown) or females, although there was a suggestive down-regulation of the expression in females (Figure 2C).
Figure 2.
On the top (A), correlation of cigarettes per day (CPD) with expression of members of the let-7 family in adenocarcinoma (AD) female patients based on array analysis in EAGLE; on the bottom left (B), correlation of CPD with expression of members of the let-7 family in AD female patients based on QRTPCR analysis in CHTN samples; on the bottom right (C), correlation of CPD with expression of miR21 in SQ male patients and AD females patients. Regression lines from linear regression models and Spearman correlation coefficients are reported. All p-values for EAGLE samples are significant at level 0.05 except let-7e (p=0.20), let-7d (p=0.09) and miR21 in females (p=0.09). P-values for the CHTN samples based on 14 subjects are p=0.08 and p=0.09 for let-7f and let-7g, respectively. Colors in the figures represent smoking status (red=current smokers; blue=former smokers).
Survival analysis
Both the unadjusted survival analysis and the analysis adjusted by age, sex, histology, stage, and smoking status (n=290 for all stages, n=251 for early stages) produced no more significant miRs than would have been expected by chance for SQ and AD combined. Due to the observed biological differences in the miR expression comparisons and the disparate covariate distributions (SQ patients were nearly all male ever smokers), survival analyses were conducted separately for AD and SQ patients.
Adenocarcinoma
Although 12 miRs (parametric p-value<0.05) were identified in the unadjusted univariate survival analyses of the AD sample and 15 were identified in the analyses restricted to the early stage, the global tests for the associations were not significant (permutation p-value of global test=0.27 and 0.20, respectively). Similarly, 11 miRs were associated with survival in the analyses adjusted by age, sex, smoking and stage, and 10 were expected by chance (SM9). A risk prediction analysis for combined miR expression did not show significant associations with survival (data not shown).
Squamous cell carcinoma
Both the unadjusted and adjusted analyses showed significant results for the association of miR expression with survival from SQ. Because there were only 3 females and 1 non-smoker, we focused on male smokers. Results based on stages I, II and IIIA (n=107) are discussed below. Results including all stages (n=121, with 25 miRs with p<0.05) are reported in SM10.
Using a cut-off of p<0.05, there were 33 miRs individually associated with mortality risk in the unadjusted analysis (global permutation p=0.026) and 36 in the age- and stage-adjusted analysis (SM11). Many of these miRs also differentiated the histology groups. Expression of most miRs was inversely related to mortality, ranging from 1.2 to 3.6-fold increased risks for subjects with low expression. Given the strong individual miR associations, we fit a risk prediction model for the combined effect of miRs on survival. Since age (p=0.96) and stage (p=0.46, including only stages I, II, and IIIA) were not associated with survival in this group, and subjects only included male smokers, we conducted an unadjusted analysis. Among the 107 SQ patients included in this analysis, 51 were Stage I, 39 stage II, and 17 stage IIIA, and the number of deaths due to lung cancer were 22, 17, and 9, respectively.
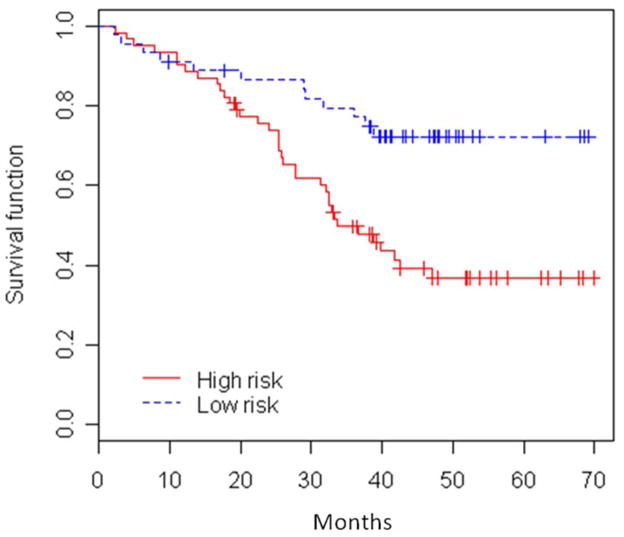
We found 5 miRs (miR-25, miR-34c-5p, miR-191, let-7e, and miR-34a) whose expression strongly predicted SQ survival for the 107 male smokers with early stage SQ tumors. Using the cross-validated supervised principal component method, 62 patients (36 lung cancer deaths) were predicted to have high mortality risk (prognostic index above median) and 45 patients (12 lung cancer deaths) were predicted to have low mortality risk (prognostic index below median). Figure 3 displays the Kaplan-Meier curves comparing the two risk groups (p-value for the permutation test was 0.017 based on 10,000 permutations); the corresponding results from the individual miR analysis of the 5 miRs are shown in Table 4.
Figure 3.
Kaplan-Meier survival curves for male smoker squamous cell carcinoma patients based on cross-validated multi-miR models developed using supervised principal component analysis (PCA) method (18) (p-value=0.017, permutation test based on 10,000 permutations). Among the 107 squamous cell carcinoma patients included in this analysis, 51 were in Stage I, 39 in stage II, and 17 in stage IIIA, and the number of deaths for lung cancer were 22, 17, and 9, respectively.
Table 4.
Association of miR expression with survival of squamous cell carcinoma for the 5 miRs included in the risk prediction model
| MicroRNAs | Hazard Ratio | 95% CI | p-value |
|---|---|---|---|
|
Unadjusted analysis
| |||
| hsa-miR-25 | 0.46 | 0.28–0.75 | 0.002 |
| hsa-miR-191 | 0.52 | 0.33–0.80 | 0.003 |
| hsa-let-7e | 0.46 | 0.27–0.80 | 0.006 |
| hsa-miR-34c-5p | 0.31 | 0.13–0.74 | 0.008 |
| hsa-miR-34a | 0.47 | 0.26–0.84 | 0.011 |
|
| |||
|
Adjusted analysisa
| |||
| hsa-miR-25 | 0.44 | 0.27–0.72 | 0.001 |
| hsa-miR-191 | 0.51 | 0.33–0.89 | 0.003 |
| hsa-let-7e | 0.45 | 0.26–0.79 | 0.006 |
| hsa-miR-34c-5p | 0.32 | 0.13–0.77 | 0.011 |
| hsa-miR-34a | 0.48 | 0.27–0.85 | 0.011 |
Analysis adjusted for stage and age
Confirmation and replication of array results by QRT-PCR
We analyzed 5 miRs by QRT-PCR in 49 samples remaining from the EAGLE study and 76 CHTN samples. Specifically, let-7g, miR-26a, and let-7f were chosen to confirm the histology comparison (fold change >2 in the array comparison), let-7g and let-7f were used to confirm the smoking comparison, and miR-638 and miR-107 were chosen to confirm the survival results. High expression of miR-638 was a risk factor and high expression of miR-107 was a protective factor for mortality from SQ in the array data. The three miRs chosen for the histology comparison showed a similar pattern in the QRT-PCR-based results in both the EAGLE samples and the CHTN samples (SM12). Similarly, the QRT-PCR results for the two miRs selected for the smoking comparison (Figure 2B) and the two miRs selected for the association with SQ survival (SM13) were consistent with the array data. Of note, the array results were based on FFPE samples while CHTN samples were fresh frozen. Our findings confirm (30) that both samples types elicit comparable miR results.
Discussion
Recent clinical trials show that different histological subtypes of NSCLC have different responses to therapy (31–34). Yet, current treatment is similar across histologies because histology-specific molecular targets that could be therapeutically exploited are lacking. In the largest study of miR expression in lung cancer to date, we found a miR profile that strongly and consistently differentiated AD from SQ. We confirmed the largest differences in an independent set of lung cancer cases and identified a set of putative target genes whose expression was correlated with the expression of miRs differentiating the two histology groups, providing preliminary evidence of miR-target correlations for further experimental validation. Expression differences by histology were highly significant in the early stage tumors but not significant in the advanced stages, suggesting that in advanced, less differentiated tumors, miR expression loses histology specificity. This finding implies that efforts to exploit these differences for mechanistic insight or therapeutic benefit should focus on early stage tumors. Given the different miR profiles by histology, we conducted survival analyses separately in the two histology groups, with a rigorous statistical approach. Many of the miRs down-regulated in early stage SQ vs. AD were associated with increased risk of mortality from SQ, suggesting a role for these miRs in the repression of genes involved in lung cancer progression. This finding may also provide insight into why some early stage, apparently surgically cured, patients recur with a fatal outcome. Among these miRs, we identified a five-miR signature that strongly predicted survival from SQ.
All members of the let-7 family highly differentiated AD from SQ and increased mortality risk in SQ, suggesting that this group exerts its influence most profoundly within the early stage SQ histology. Pioneering studies described let-7 miR as a negative regulator of the oncogenic family of RAS, and repressor of cell proliferation pathways and NSCLC growth (11;27;35). As it has been observed for other pathways (e.g., the EGFR pathway in non small cell lung cancer (36)) or other tumors (e.g., the NRAS/BRAF mutations in melanoma (37)), it is possible that let-7 expression alterations and KRAS mutations are mutually exclusive in lung carcinogenesis, with let-7 having a more predominant effect in SQ and KRAS mutations in AD. If further confirmed by mutational analyses, this finding may improve our understanding of the carcinogenetic pathways leading to non small cell lung cancer. Among the let-7 miRs, let-7g showed the largest fold-change together with miR 26a, an hypoxia-induced miR known to decrease pro-apoptotic signaling (38). Interestingly, both let-7g and miR-26a are localized to chromosome 3p21-22. Allele loss and genetic alteration at this locus are the most frequent and earliest genomic abnormalities in NSCLC (39) and are more common in SQ than AD (40;41). We also found histological differences for miRs clustering at chromosome 14q32 and 19q13, both loci involved in lung cancer development and progression (28;29). All these loci have been extensively investigated for potential tumor suppressor genes without clear success. However, if the chromosomal alterations involve miRs that target distant genes, the potentially relevant lung cancer genes likely extend beyond these loci.
Other miRs strongly differentiating the two histology groups included miR-29a, which affects apoptosis (42) and epigenetic normalization of NSCLC (43), and miR-21, which acts as an oncogene or “oncomiR” in many tumor types and plays an important role in tumor metastasis (44;45). Interestingly, in our study miR-21 strongly differentiated the histology groups with high levels in AD in stage II but not in stage I tumors, suggesting that miR-21 may be a marker of tumor progression in AD, identifying tumors on the verge of acquiring metastatic potential.
The large majority of miRs were down-regulated in SQ vs. AD in our study. This pattern suggests that miRs that function as tumor suppressors, like let-7 and miR-29a, may be more relevant for SQ tumorigenesis because they exhibited poor expression in this tumor type. In contrast, miRs with oncogenic potentials in many tissue types, like miR-21 and -26a, may be crucial for AD development since they were over-expressed in AD compared to SQ. If functionally confirmed, these miRs may identify histology-specific therapeutic targets, especially for surgically resectable lesions.
The only previous study showing miR expression differences among lung cancer histologies found 5 mature miRs and 1 miR precursor differentiating 65 AD from 39 SQ (14) in an unadjusted analysis of Caucasians and African Americans, with different smoking patterns than Southern Europeans. Dissimilarity in sample sizes, analytical approaches, assays and ethnic groups may have contributed to the discrepancies between studies. Moreover, differential expression and variable ratio of mature vs. immature miRs in specific subpopulations of malignant tissues (45;46) may account for some differences.
We examined the impact of tobacco smoking on miR expression and obtained interesting preliminary findings. The comparison between smokers and never smokers did not elicit significant results, possibly because of the small number of never smokers (only 5 in males). In smokers, we found that the expression of each member of the let-7 family, which has tumor suppression potential, was inversely associated with number of cigarettes per day in females but not in males. We confirmed this finding also in the independent samples from the CHTN. Down-regulation of let-7 by tobacco smoking was also observed in rats (47). The gender discrepancy might be related to hormonal effects, since estrogen levels may modify miR expression (48). Sex and smoking status may have differential impact on miR expression as has been observed for mutational patterns of EGFR in non small cell lung cancer (49). Interestingly, expression of miR21, which acts as an oncogene, increased with increasing cigarettes per day in SQ but not AD, suggesting a histology-specific response to tobacco carcinogens. However, small numbers or other unknown factors may have contributed to these results, and larger studies and functional tests are necessary to confirm our findings. If confirmed, they may provide an important piece in the mechanistic puzzle relating tobacco smoking to lung cancer development.
In the survival analysis, we found that low expression of several miRs was associated with up to 4-fold excess mortality from SQ overall and in the subgroup including only male smokers with stage I to IIIA. Interestingly, many of these miRs were down-regulated in SQ vs. AD even in stage I, suggesting an early role for these miRs in regulation of tumor progression. We confirmed the findings by QRT-PCR for selected miRs.
Lower expression of let-7e, miR-34a, miR-34c-5p, miR-25, and miR-191 constituted a “poor survival” signature. Reduced expression of the let-7 family has been previously correlated with poor post-operative survival in lung cancer (13). MiR-34a and, to a lesser extent, miR-34c are targets of p53 and are involved in p53-dependent apoptosis, cell cycle arrest, senescence, and DNA damage response (50). MiR34a has been recently found to be associated with poor survival of NSCLC in a study investigating the role of the miR34 family (51) (Table 5). Recently, Mascaux et al. found that miR-34c expression progressively decreased from normal bronchial tissues of nonsmokers to SQ (52), while Liu et al., (53) identified miR34c as a growth-suppressive miR in murine lung cancer, suggesting that this miR is involved in bronchial carcinogenesis from the very early steps of this process. MiR-25, which targets important cell cycle regulators, like AURKA (p<1e-05, miRBase database), may contribute to the change from bronchioalveolar stem cell to lung cancer stem cells (54) with metastatic potential. Finally, expression of miR-191 has been found associated with survival from SQ (55) and acute myeloid leukemia (56). Although risk prediction models should be interpreted cautiously (57) and need confirmation in multiple studies, taken together these findings suggest that targeting these miRs in the treatment of SQ may prove fruitful.
Table 5.
MiRs associated with worse survival from lung cancer across studies
| MicroRNAs | Direction | N. cases (replication) | Histology | Stages | Ethnicity | Assay (n. miRs) | Tissue type | Author |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-25 | Down | 107 male smokers | SQ | I, II, IIIA | Caucasians | Custom-made microarray chip (440) | FFPE | Landi et al., 2009 (this study) |
| hsa-miR-34c-5pa | Down | |||||||
| hsa-miR-191 | Down | |||||||
| hsa-let-7e | Down | |||||||
| hsa-miR-34a | Down | |||||||
|
| ||||||||
| Has-miR-34a | Down | 70 | NSCLC | I, II, III | Not reported | RT-PCR (3) | FFPE | Gallardo et al., 2009 (51) |
|
| ||||||||
| hsa-miR-146b | Up | 54 | SQ | I, II, III | Not reported | MirVana miRNA Bioarray (328) | Snap-frozen | Raponi et al., 2009 (55) |
| hsa-miR-191 | Up | |||||||
| hsa-miR-206 | Down | |||||||
| hsa-miR-299-3p | Down | |||||||
| hsa-miR-155 | Up | |||||||
| hsa-miR-15a | Up | |||||||
| hsa-miR-122a | Down | |||||||
| hsa-miR-513b | Down | |||||||
| hsa-miR-184 | Down | |||||||
| hsa-miR-511 | Up | |||||||
| hsa-miR-100 | Up | |||||||
| hsa-miR-10a | Up | |||||||
| hsa-miR-453 | Down | |||||||
| hsa-miR-379 | Down | |||||||
| hsa-miR-202 | Down | |||||||
| hsa-miR-21 | Up | |||||||
| hsa-miR-126 | Up | |||||||
| hsa-miR-494 | Down | |||||||
| hsa-miR-432 | Down | |||||||
| hsa-miR-370 | Down | |||||||
|
| ||||||||
| hsa-let-7a | Down | 112 (65) | NSCLC | I, II, III | Asians | RT-PCR Human Panel-Early Access Kit (157) | Snap-frozen | Yu et al., 2008 (15) |
| hsa-miR-221 | Down | |||||||
| hsa-miR-137 | Up | |||||||
| hsa-miR-372 | Up | |||||||
| hsa-miR-182* | Up | |||||||
|
| ||||||||
| hsa-miR-21 | Up | 48 | NSCLC | Not reported | Not reported | RT-PCR (2) | Snap-frozen | Markou et al., 2008 (58) |
|
| ||||||||
| hsa-miR-155 | Up | 65 (32) | AD | I, II, III, IV | Caucasians + African Americans | MirVana miRNA Detection Kit (352) | Not reported | Yanaihara et al., 2006 (14) |
| hsa-let-7a2 | Up | |||||||
|
| ||||||||
| hsa-let-7 | Down | 143 | NSCLC | I, II, III | Asians | RT-PCR (1) | Not reported | Takamizawa et al., 2004 (13) |
In bold, miRs confirmed in more than one study
Identified as a growth-suppressive miR in murine lung cancer (53)
Although hsa-miR-513 was not included in the top 5 miRs in our study, it was borderline significant in our univariate analysis (permutation p-value=0.0468)
Previously, few studies explored the association of miR expression with survival from lung cancer and found different sets of miRs (Table 5). Four of the 5 miRs identified by our risk prediction model were also found to be associated with lung cancer survival in other studies, while we could not confirm associations with some of the previously identified miRs. As Table 5 shows, there is large heterogeneity across studies with regard to subjects’ and tumors’ characteristics, sample size, assays, analytical approaches, and tissue types, which likely explain the discrepant results. Results may also be affected by the different numbers of miRs tested and different cut-offs used to exclude miRs with low intensity across multiple samples or arrays. As an example, Yu et al (15) found a group of 5 mature miRs that together predicted survival in 57 SQ and 60 AD stage I-III Asian patients. We could not verify this finding since three of the five miRs identified in this study were excluded from our analyses because they had more than 50% missing data across subjects. However, of the remaining two miRs, one (let-7a) was associated with survival in our study.
In conclusion, in the largest study of miR expression in lung cancer to date, miR expression profiles strongly differed between AD and SQ, suggesting that different sets of miRs contribute to the pathogenesis of different NSCLC histologies and may become targets of histology-specific treatment in the future. Among miRs whose expression was reduced in SQ from the early stages, we identified a profile that predicted survival for SQ. These miRs may have important implications for prognosis and treatment of this histology subgroup of lung cancer.
Supplementary Material
Translational relevance.
Recent clinical trials for non small cell lung cancer showed that response to therapy may vary by histology. We identified a microRNA signature that strongly differentiated histology subtypes, had prognostic implications and predicted survival from squamous cell carcinoma. These findings may suggest targets for histology-specific treatments of non small cell lung cancer in the future.
Acknowledgments
This work was supported by the Intramural Research Program of National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics, and a 2006 National Cancer Institute Director’s Innovation Award to M.T.L. We are extremely grateful to the EAGLE participants and the large number of collaborators2 that made the EAGLE study possible. We also thank the Cooperative Human Tissue Network that provided samples and data for the replication study, and Drs. Giuseppe Giaccone and Adi Gazdar for their insightful comments.
Footnotes
Reference List
- 1.World Health Organization Classification of Tumors. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. [Google Scholar]
- 2.Gabrielson E. Worldwide trends in lung cancer pathology. Respirology. 2006;11:533–8. doi: 10.1111/j.1440-1843.2006.00909.x. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 4.Gao Y, Goldstein AM, Consonni D, et al. Family history of cancer and nonmalignant lung diseases as risk factors for lung cancer. Int J Cancer. 2009;125:146–52. doi: 10.1002/ijc.24283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landi M, Chatterjee N, Yu K, et al. A Genome-wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma. The American Journal of Human Genetics. 2009 doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52(9 Suppl):2665s–9s. [PubMed] [Google Scholar]
- 7.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–65. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 9.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang TC, Yu D, Lee YS, Wentzel EA, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 14.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Landi MT, Consonni D, Rotunno M, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin P, Wang E, Ren J, et al. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J Transl Med. 2008;6:39. doi: 10.1186/1479-5876-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 20.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Grocock RJ, van DS, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Choi JS, Zheng LT, Ha E, et al. Comparative genomic hybridization array analysis and real-time PCR reveals genomic copy number alteration for lung adenocarcinomas. Lung. 2006;184:355–62. doi: 10.1007/s00408-006-0009-0. [DOI] [PubMed] [Google Scholar]
- 29.Choi YW, Choi JS, Zheng LT, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung Cancer. 2007;55:43–51. doi: 10.1016/j.lungcan.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scagliotti GV, Parikh P, von PJ, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch FR, Spreafico A, Novello S, Wood MD, Simms L, Papotti M. The prognostic and predictive role of histology in advanced non-small cell lung cancer: a literature review. J Thorac Oncol. 2008;3:1468–81. doi: 10.1097/JTO.0b013e318189f551. [DOI] [PubMed] [Google Scholar]
- 33.Chang GC, Tsai CM, Chen KC, et al. Predictive factors of gefitinib antitumor activity in East Asian advanced non-small cell lung cancer patients. J Thorac Oncol. 2006;1:520–5. [PubMed] [Google Scholar]
- 34.Ardizzoni A, Boni L, Tiseo M, et al. Cisplatin- versus carboplatin-based chemotherapy in first-line treatment of advanced non-small-cell lung cancer: an individual patient data meta-analysis. J Natl Cancer Inst. 2007;99:847–57. doi: 10.1093/jnci/djk196. [DOI] [PubMed] [Google Scholar]
- 35.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–62. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 37.Platz A, Egyhazi S, Ringborg U, Hansson J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008;1:395–405. doi: 10.1016/j.molonc.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. The International Lung Cancer Chromosome 3p21. 3 Tumor Suppressor Gene Consortium. Cancer Res. 2000;60:6116–33. [PubMed] [Google Scholar]
- 40.Mitsudomi T, Oyama T, Nishida K, et al. Loss of heterozygosity at 3p in non-small cell lung cancer and its prognostic implication. Clin Cancer Res. 1996;2:1185–9. [PubMed] [Google Scholar]
- 41.Braga E, Senchenko V, Bazov I, et al. Critical tumor-suppressor gene regions on chromosome 3P in major human epithelial malignancies: allelotyping and quantitative real-time PCR. Int J Cancer. 2002;100:534–41. doi: 10.1002/ijc.10511. [DOI] [PubMed] [Google Scholar]
- 42.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 43.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 45.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–2. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 46.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 47.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De FS. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2008 Oct 24;23:806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 49.Toyooka S, Matsuo K, Shigematsu H, et al. The impact of sex and smoking status on the mutational spectrum of epidermal growth factor receptor gene in non small cell lung cancer. Clin Cancer Res. 2007;13:5763–8. doi: 10.1158/1078-0432.CCR-07-0216. [DOI] [PubMed] [Google Scholar]
- 50.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 51.Gallardo E, Navarro A, Vinolas N, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009 Sep 7; doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 52.Mascaux C, Laes JF, Anthoine G, et al. Evolution of microRNA expression during human bronchial squamous carcinogenesis. Eur Respir J. 2009;33:352–9. doi: 10.1183/09031936.00084108. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Sempere LF, Galimberti F, et al. Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer Res. 2009;15:1177–83. doi: 10.1158/1078-0432.CCR-08-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian S, Ding JY, Xie R, et al. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun. 2008;377:668–73. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 55.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 56.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–9. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106:2824–8. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696–704. doi: 10.1373/clinchem.2007.101741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.