Abstract
Rationale
Ox-PAPC accumulates in atherosclerotic lesions, is pro-atherogenic, and influences the expression of over 1000 genes in endothelial cells.
Objective
To elucidate the major pathways involved in Ox-PAPC action, we conducted a systems analysis of endothelial cell gene expression after exposure to Ox-PAPC.
Methods and Results
We used the variable responses of primary endothelial cells from 149 individuals exposed to Ox-PAPC to construct a network consisting of 11 groups of genes or modules. Modules were enriched for a broad range of GO pathways, some of which have not been previously identified as major Ox-PAPC targets. Further validating our method of network construction, modules were consistent with relationships established by cell biology studies of Ox-PAPC effects on endothelial cells. This network provides novel hypotheses about molecular interactions, as well as candidate molecular regulators of inflammation and atherosclerosis. We validate several hypotheses based on network connections and genomic association. Our network analysis predicted that the hub gene CHAC1 was regulated by the ATF4 arm of the unfolded protein response pathway and here we showed that ATF4 directly activates an element in the CHAC1 promoter. We show that variation in basal levels of HMOX1 contribute to the response to Ox-PAPC, consistent with its position as a hub in our network. We also identified GPR39 as a regulator of HMOX1 levels and showed that it modulates the promoter activity of HMOX1. We further showed that OKL38/OSGN1, the hub gene in the blue module, is a key regulator of both inflammatory and anti-inflammatory molecules.
Conclusions
Our systems genetics approach has provided a broad view of the pathways involved in the response of endothelial cells to Ox-PAPC and also identified novel regulatory mechanisms.
Keywords: Endothelial Cells, Oxidized Phospholipids, Gene Expression, Heme Oxygenase, Network, Systems Genetics, Genome-Wide Association Studies
Introduction
Elevated levels of circulating low density lipoprotein (LDL) result in the retention of LDL in the vessel wall, where the LDL undergoes aggregation and oxidative modification1. Oxidized LDL has inflammatory activities that are strongly regulated by oxidized phospholipids2, 3 that have been shown to be present in human and mouse atherosclerotic lesions where inflammation is increased4 and in apoptotic cells, a component of atherosclerotic lesions3. Plasma levels of specific phospholipid oxidation products of palmitoyl-arachidonyl-phosphatidylcholine (Ox-PAPC), reacting with the antibody EO6, have been shown to be prognostic for the development of atherosclerosis3.
Global gene expression studies of the responses of ECs to Ox-PAPC have shown that hundreds of genes are affected and that the affected genes show little overlap with those of classic innate immunity responses such as NFκB activation5 or other characterized inflammatory pathways. PEIPC and POVPC components of Ox-PAPC have been shown to mimic most of the effects of Ox-PAPC. Several inflammatory functions regulated by Ox-PAPC have been described including induction of a number of pro-inflammatory cytokines, inhibition of LPS action, induction of heme oxygenase 1 (HMOX1)2, 3, the activation of the sterol biosynthetic pathway, procoagulant pathways and the unfolded protein response pathway. Some of the functions of Ox-PAPC are anti-inflammatory while others are pro-inflammatory. Although aspects of the receptor regulation and signaling mechanisms of Ox-PAPC have been described, it is important to obtain an integrated view of the Ox-PAPC network in order to effectively target the induced pathways.
We reported previously that ECs from different strains of mice6 or from different heart transplant donors7 showed striking differences in the responses to Ox-PAPC in part due to genetic variation. In this study, to globally define the relationships among the molecules that respond to Ox-PAPC treatment in ECs, a systems genetics approach was employed. Systems-based approaches attempt to examine biologic processes more broadly than traditional reductionist approaches, which focus on individual genes or pathways. They often employ high-throughput technologies such as expression arrays that generate genome-wide data. Data are then examined using statistical and modeling methods, such as biologic networks in which the components of the systems are represented as `nodes' and their interactions as `edges'. In `systems genetics', natural variations are used to perturb this system to allow modeling of the interactions. A fundamental concept of systems biology is that the whole is greater than the sum of the parts, and that by examining the system as a whole, intrinsic novel properties may be identified that could not be derived linearly from the additive effects of the individual components.
For our studies, we took advantage of naturally-occurring gene expression variation in the human population. Global gene expression levels in early passage EC cultures from 149 different heart transplant donors were examined with and without treatment with Ox-PAPC for 4 hours. An unbiased co-expression algorithm that models a scale-free gene network of the most responsive genes identified 11 modules of highly correlated genes that were enriched for a diverse array of novel functions. Analysis of this network revealed previously unknown effects of Ox-PAPC, most prominently an inhibition of cell division. Genes not previously known to be major regulators of Ox-PAPC action were identified as hub genes in modules and new aspects of their regulation are reported. Follow-up validation studies focused on HMOX1, a known regulator of inflammation and a highly connected gene in one of the modules. Greater basal HMOX1 expression was associated with reduced responses of inflammatory genes to Ox-PAPC. A combination of network and GWAS studies revealed a regulatory role of G-protein-coupled receptor 39 (GPR39) in HMOX1 expression, which was confirmed by knock-down studies. Using promoter-reporter constructs for HMOX1, we further showed that GPR39 controls the transcription of HMOX1 through a particular promoter region. We also examined other network-based hypotheses. For example, we showed that ATF4 directly regulates the expression of CHAC1, as predicted by the network model, and identified a specific regulatory element. Based on the network analysis, we identified OKL38/OSGIN1 as the major hub gene in the blue module and in subsequent studies determined that this gene regulates both basal and Ox-PAPC induced levels of important inflammatory and anti-inflammatory genes. The elucidation of the Ox-PAPC network and the discovery of novel pathways induced by Ox-PAPC in ECs by this network will be useful for formulating hypotheses for investigating the role of oxidized phospholipids in atherosclerosis.
Methods
Cell Culture and Treatments
158 HAEC cultures were isolated from aortic explants of 149 unique heart transplant donors in the UCLA transplant program and grown to confluence in 100mm dishes as previously described8. 9 of these cultures were duplicates (Supplemental Methods). We have previously validated our method of endothelial cell isolation on multiple cultures. We demonstrated that greater than 95% of the cells were Factor 8 positive and took up dil-acetyl LDL8. In more recent studies we have demonstrated in multiple EC preparations that greater than 95% of the cells were PECAM positive (data not shown). At 90% – 100% confluence, cells were treated in duplicate with either M199 media (Mediatech) containing 1% FBS (Hyclone), or additionally with 40 μg/ml Ox-PAPC. For PEIPC experiments, treatment media contained either 5 μg/ml PEIPC or 50 μg/ml Ox-PAPC. Production of Ox-PAPC and synthesis of PEIPC have been described9, 10. We previously determined that the level of Ox-PAPC in the vessel wall of hypercholesterolemic rabbits is approximately 10× the concentration used in these studies11. mRNA was harvested after 4 hours of treatment.
Gene Expression Profiling
Cytoplasmic RNA was hybridized to Affymetrix HT-HU133A microarrays as described previously12. 629 arrays were used to quantify gene expression, corresponding to 158 cultures and 149 unique donors (Supplemental Methods). Intensity values were normalized with robust multi-array average (RMA) normalization method as previously described12. For the PEIPC experiment, Illumina HumanRef-8v2 microarrays were used according to manufacturers protocol and data was normalized in Bead Studio (Illumina) with the `rank invariant' method. Array data is available in GEO accessions GSE20060 and GSE30169.
Network Construction and Visualization
Duplicate expression measurements were averaged per condition and per donor. The 2000 most Ox-PAPC regulated genes were identified by comparing the untreated and Ox-PAPC treated values across the population with a paired t-test. The 2000 most Ox-PAPC regulated genes corresponded to those with t-test p-values < 1.0e-28. 2000 transcripts were used so that the network would be inclusive of the most significant genes that were differentially expressed in response to Ox-PAPC, yet small enough to be visualized. Average transcript measurements for the 2000 transcripts in both conditions, untreated and treated with Ox-PAPC, were used in network construction. The pair-wise adjacency matrix between genes was used to determine `topological overlap' between gene pairs13, 14. The topological overlap matrix was raised to the power of 12 to emphasize the difference between transcripts with many connections and transcripts with few connections. The topological overlap matrix was used to identify `modules' of highly co-expressed genes and the clustering dendogram of the topological overlap matrix was cut with the `dynamic hybrid' method to define modules15. 11 modules were identified and arbitrarily named by colors. The `grey module' contained the genes with dissimilar expression patterns and was not considered a true module. All of these analyses were performed in R using the Weighted Gene Co-expression Network Analysis (WGCNA) package15. Network visualization was performed in Cytoscape v2.6.316. For details of the network visualizations refer to Supplemental Methods.
Genotyping and Association Analysis
Genomic DNA was isolated from HAECs using the DNeasy kit with RNase treatment (Qiagen). For HMOX1 microsatellite genotyping, a FAM-labeled PCR product was generated using the primers 5' GAG CCT GCA GCT TCT CAG AT 3' and 5' ACA GCT GAT GCC CAC TTT CT 3'. Microsatellite alleles were then classified as `short' (<29 repeats) or `long' (≥29 repeats). SNP Genotypes were acquired as previously described12 (Supplemental Methods). The 147 unique donors with complete gene expression and genotyping data were used in association analysis. Genotype-gene expression associations (ie. eQTL) were tested using linear regression with the `−linear' option in PLINK 1.417.
RT-qPCR, Western Blotting, and siRNA Knock-down
qRT-PCR was performed using the Roche LightCycler 480 (Roche Diagnostics, Indianapolis, IN). siRNA experiments were performed as previously described18. siRNAs used in this study were HMOX1 siRNA#1 (Qiagen cat# SI00033089), HMOX1 siRNA#2 (Qiagen cat# SI00033096), GPR39 siRNA#1 (Qiagen cat# SI00430416), GPR39 siRNA#2 (Qiagen cat# SI00430423), GRID1 siRNA#1 (Qiagen cat# SI00430836), and GRID1 siRNA#2 (Qiagen cat# SI00430843). OKL38 siRNAs were (Qiagen SI 00665182, SI66500196).
Heme Oxygenase Activity Assay and Protein Quantification
Heme oxygenase activity was measured by bilirubin generation as previously described19 (Supplemental Methods).
Luciferase Constructs and Assays
The CHAC1 promoter and deletion constructs were generated with PCR from a fosmid (Bacpac clone# G248P8704A5) and cloned into pGL3Basic (Promega). The HMOX1 luciferase constructs were described previously (−4.5 kb = pHOGL3/4.520 and −9.4 kb = pHOGL3/9.421).
Results
Network Modeling Identifies Modules of Tightly Co-expressed Genes
We sought to obtain a systems-level view of the responses of endothelial cells to Ox-PAPC in order to provide a framework for mechanistic studies. To achieve this, primary cultured human aortic endothelial cells (HAECs, or the `system') were exposed for 4 hours to either control medium or medium containing Ox-PAPC (40 μg/ml) and transcript levels of the Ox-PAPC responsive genes (the `elements' of the system) were quantified using global expression arrays. This time period was chosen based on previous time-course experiments to permit detection of a robust yet early response. The HAEC cultures obtained in this study were not patients but heart transplant donors whose hearts were deemed healthy enough for transplant by the transplant surgeons prior to tissue collection. All clinical information, including age and sex, remained anonymous in this study (sex was independently determined from the X chromosome genotypes). However, the average age of the donors in the transplant program is known to be about 25 years. Clinical phenotypes such as use of medications and hormonal status were not known. Since most cells were used at passages 4–7, most acute drug and hormonal effects would likely be lost by this passage. Of about 19,000 genes analyzed, the 2,000 transcripts most affected by Ox-PAPC (paired t-test values <1e-28) across the population (see below) were used for network analysis.
Since Ox-PAPC is a mixture of several oxidation products of PAPC, we tested whether the Ox-PAPC response in HAECs was similar to that observed after treatment with the most potent single phospholipid component of Ox-PAPC: 1-palmitoyl-2-(5,6)-epoxyisoprostane E2-sn-glycero-3-phosphocholine (PEIPC)9. HAECs from a single donor were treated with 5 μg/ml PEIPC or 50 μg/ml Ox-PAPC. Gene expression arrays quantified the gene expression levels at baseline and after treatment, and the fold change values were calculated for each transcript. A highly conserved response was observed between Ox-PAPC and PEIPC (p-value < 2e-16). Furthermore, the average fold change for the 2000 genes from 149 donors used to construct the network was highly correlated with the fold change in these same genes by PEIPC in the donor tested (p-value < 2e-16, Online Figure I). These data demonstrate that PEIPC has a very similar effect to Ox-PAPC.
To model an unbiased co-expression network, we took advantage of naturally-occurring variations in the human population that perturbed individual gene expression patterns with and without Ox-PAPC treatment. We have previously shown that the transcriptomic responses to Ox-PAPC are determined largely by genetic factors12. A co-expression network algorithm, WGNCA15, based on `topological overlap' of gene expression both with and without Ox-PAPC treatment was used to construct the network. Topological overlap is a metric of similarity reflecting the relationship strength between gene pairs. It depends on the correlation between genes, as well as shared correlations with other genes. The network was visualized using Cytoscape software16 (Figure 1A). In concordance with other biologic networks, our co-expression network was modeled to exhibit a scale-free structure so that few genes had many connections and most genes had few connections22, 23.
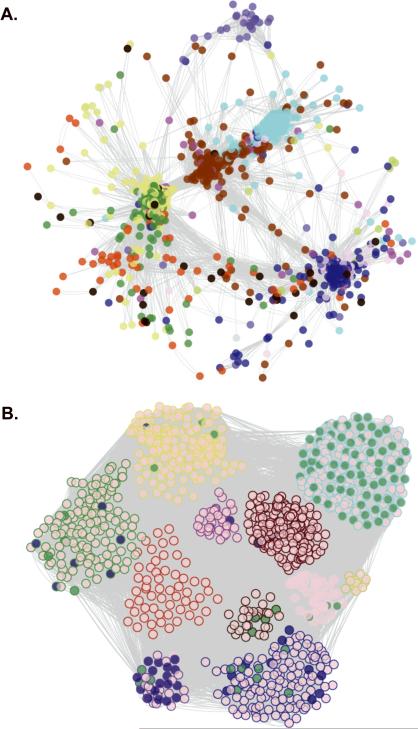
Figure 1. Visualization and Functional Enrichment of the HAEC Gene Co-Expression Network.
In the whole network, 2000 transcripts were organized into 11 modules. The number of module transcripts were: black (93), blue (299), brown (292), green (205), greenyellow (40), magenta (78), pink (82), purple (52), red (158), turquoise (373), yellow (270). The grey module had 58 genes but was not considered a real module. For ease of visualization, the 1090 most connected network transcripts (topological overlap scores > 0.9035) are shown. Circles (nodes) represent genes and lines (edges) represent topological overlap (similar to correlation) between transcript pairs. A. Genes are arranged by topological overlap, so that small distances indicate high correlation and node colors indicate module membership. B. Module membership is indicated by the node outline color and functional enrichment is illustrated by the node fill color. Transcripts belonging to the cell cycle pathway are shaded in green, while ER stress genes (translational initiation, response to unfolded protein, response to ER stress, and protein folding) are shaded in blue. Nodes were arranged manually in B according to module membership.
Genes were grouped into color-coded modules based on similarity in patterns of expression across the sample population. The network of endothelial genes most significantly induced by Ox-PAPC consisted of 11 modules of tightly connected genes (Figure 1 and Online Figures II–XII). A full list of the transcripts included in the network analysis, arranged by modules in order of highest to lowest connectivity, are included in Online Table I. Multiple probe set IDs for the same gene on the HT-HG133A array were treated as independent transcripts, which is why more than one node may appear in a module with the same gene symbol. This approach was taken so that alternative transcripts would be maintained in the network.
Co-Expressed Modules Are Enriched For Functional Pathways
Co-expression is evidence for co-regulation and suggests involvement in similar cellular functions. To examine whether genes in a given module were in the same pathway we tested for enrichment of each module based on Gene Ontology categories. Several modules exhibited enrichment for pathway ontologies (Table 1). These included novel as well as previously reported pathways that are affected by Ox-PAPC treatment. A particularly striking, novel observation was seen in the turquoise module that was highly enriched for the cell cycle genes that were down-regulated after 4 hours of Ox-PAPC treatment (Figure 1B and Online Figure III). Another enriched pathway in the turquoise module not previously implicated in the response of ECs to Ox-PAPC treatment was nucleobase, nucleoside, nucleotide and nucleic acid metabolism. Other modules/pathways not previously observed included endocytosis enriched in the black module (Online Figure X) and golgi organization in the brown module (Online Figure VII).
Table 1.
Gene Ontology Enrichments in Modules of the HAEC Gene Co-Expression Network
| Module | GO ID | Enrichment p-value | GO Term |
|---|---|---|---|
| black | GO:0006897 | 7.8e-04 | endocytosis |
| blue | GO:0000096 | 1.4e-08 | sulfur amino acid metabolic process |
| blue | GO:0009064 | 6.2e-06 | glutamine family amino acid metabolic process |
| blue | GO:0045444 | 6.3e-05 | fat cell differentiation |
| blue | GO:0032273 | 8.6e-05 | positive regulation of protein polymerization |
| brown | GO:0044419 | 3.4e-11 | interspecies interaction between organisms |
| brown | GO:0007030 | 9.8e-06 | golgi organization |
| green | GO:0006509 | 3.3e-06 | membrane protein ectodomain proteolysis |
| green | GO:0030833 | 6.0e-05 | regulation of actin filament polymerization |
| greenyellow | GO:0016477 | 1.5e-03 | cell migration |
| magenta | GO:0045736 | 2.0e-04 | negative regulation of cyclin-dependent protein kinase activity |
| pink | GO:0048598 | 1.9e-04 | embryonic morphogenesis |
| purple | GO:0006457 | 4.3e-12 | protein folding |
| purple | GO:0006986 | 1.9e-08 | response to unfolded protein |
| red | GO:0007204 | 2.9e-08 | elevation of cytosolic calcium ion concentration |
| turquoise | GO:0006l39 | 1.5e-23 | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| turquoise | GO:0010467 | 9.5e-23 | gene expression |
| turquoise | GO:0042254 | 1.1e-12 | ribosome biogenesis |
| turquoise | GO:003l577 | 7.0e-10 | spindle checkpoint |
| turquoise | GO:005l436 | 3.6e-06 | negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle |
| turquoise | GO:0000077 | 2.3e-05 | DNA damage checkpoint |
| turquoise | GO:0006268 | 2.7e-05 | DNA unwinding during replication |
| turquoise | GO:0007093 | 3.4e-05 | mitotic cell cycle checkpoint |
| turquoise | GO:0006281 | 4.5e-05 | DNA repair |
| turquoise | GO:0006261 | 8.3e-05 | DNA-dependent DNA replication |
| turquoise | GO:0009262 | 8.5e-05 | deoxyribonucleotide metabolic process |
| turquoise | GO:0006284 | 9.2e-05 | base-excision repair |
| turquoise | GO:0006298 | 9.2e-05 | mismatch repair |
| yellow | GO:0007l65 | 3.2e-10 | signal transduction |
| yellow | GO:0016070 | 3.2e-07 | RNA metabolic process |
Molecular interactions determined by the network revealed novel relationships in previously reported Ox-PAPC-responsive pathways such as the UPR. The genes in the UPR pathway segregated into two modules, the purple and blue, which corresponded to the X-box binding protein 1 (XBP1) (Online Figure IV) and activating transcription factor 4 ATF4 (Figure 2) arms of the UPR, respectively. Clustering of the first principal components of the gene expression in each module showed that the blue and purple modules were most similar to each other relative to all other modules in the network, suggesting that these two modules share functional characteristics and capture biological regulatory mechanisms (Online Figure XIII).
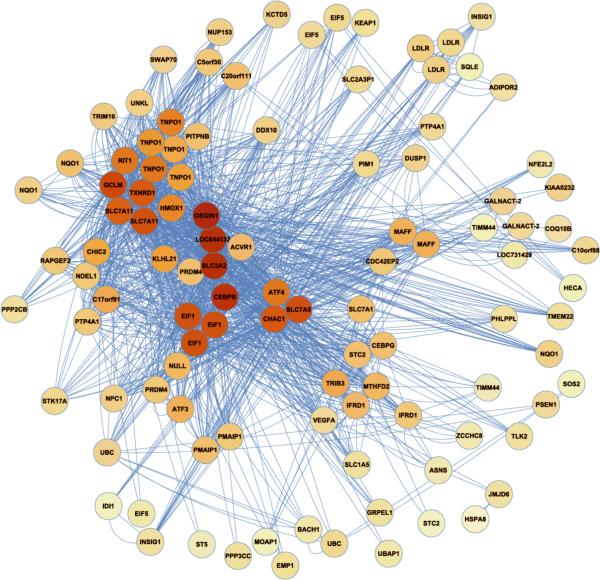
Figure 2. The Blue Module.
Transcripts (nodes) are shaded by the degree of intra-modular connectivity, where red is the most connected (i.e. `hubs'). Lines connecting the transcripts (`edges') signify co-expression. This is a simplified view of the blue module. Refer to Online Figure II for a more thorough representation.
Enrichment for sulfur amino acid metabolic processes and glutamine family amino acid metabolic processes in the blue module (Table 1) is consistent with the evidence that treatment of endothelial cells with Ox-PAPC increases oxidative stress24. In particular, enrichment for sulfhydryl proteins in the blue module supports the novel mechanistic hypothesis that covalent interaction of certain Ox-PAPC compounds with free sulfhydryl groups trigger the Ox-PAPC response. PEIPC has been show to covalently bind to proteins25. Our studies suggest that the allylic epoxide group on PEIPC, which readily binds sulfhydryls, is most likely the major active site on this molecule9. Furthermore, we identified modules representing pathways previously reported as regulated by Ox-PAPC including regulation of actin filament polymerization in the green module (Online Figure VI), and elevation of cytosolic calcium ion concentration in the red module (Online Figure VIII) indicating the ability of co-expression network modeling to provide novel as well as established relationships in the response of ECs to Ox-PAPC (Table 1, Figure 1B). Knowledge of the components and connectivity of these genes will be useful in identifying signal transduction pathways regulating these processes.
Hub Genes Are Likely To Be Key Regulators
In addition to large-scale functional enrichments, our network analysis identifies hub genes that are potentially important regulators of major pathways activated in response to Ox-PAPC. One such gene is OKL38/OSGIN1 that was the most connected gene in the blue module (Figure 2). This gene was previously identified by us as a redox gene regulated by Ox-PAPC but its pivotal role was not recognized26. Supporting the hypothesis that OKL38 plays a key role, we have identified a number of inflammatory and anti-inflammatory genes regulated by OKL38 in knock-down studies (Online Figure XIV). Two OKL38 siRNAs were used that achieved ~50% of OKL38 transcript levels (data not shown). We demonstrated that knock-down of OKL38 increased both basal and Ox-PAPC induced levels of IL-8, ATF4 and KLF4, all molecules previously associated with the endothelial cell inflammatory responses3, 7. This is the expected result if the expression levels of these genes are controlled by increased oxidative stress and OKL38 protects against oxidative stress. In addition we observed that knock-down of OKL38 dramatically increased both basal and Ox-PAPC induced levels of HMOX1 suggesting a close relationship between the levels of these two molecules. Two other members of the blue module showed no effect of OKL38 knock-down (data not shown).
Genes at GWAS Loci for Coronary Artery Disease Are Regulated By Ox-PAPC, Exhibit Regulation of Gene Expression, and Are In The HAEC Network
Next we performed systems genetics analyses with the goal of elucidating the disease genes and pathways of atherosclerosis. To this end, we integrated results from a recent comprehensive GWAS analysis of coronary artery disease (CAD) with our endothelial systems data in two ways. First, we queried genes residing under GWAS peaks and their local neighborhoods for regulation by Ox-PAPC in our HAEC co-expression network. Secondly, we identified genes whose expression exhibited regulation by the peak GWAS SNPs.
We employed CAD GWAS loci identified by H. Schunkert and colleagues27 using 22,233 cases and 64,762 controls with additional genotyping for peak loci in 56,682 additional individuals. For our analysis, we began with 31 genes under 25 association peaks that were measured in our HAEC population (Online Table II). 5 genes (LDLR, NT5C2, SH2B3, UBE2Z, and COL4A1) were regulated by Ox-PAPC by at least an average of 1.1-fold across the HAEC population (p-values < 1e-21). 4 of these genes were included in the HAEC network and resided in the blue (LDLR and UBE2Z), green (NT5C2), and brown (SH2B3) modules (Online Table II). To explore the network neighborhoods we identified the transcripts most connected to these genes in our network and visualized these as `GWAS gene-centered sub-networks' of the overall network. An example is shown for NT5C2 in Online Figure XV. Taken together, these data provide evidence that LDLR, UBE2Z, NT5C2 and possibly COL4A1 are not only positional candidates for CAD, but that these genes may play a role in disease based on regulation by Ox-PAPC and their connectivities to other Ox-PAPC regulated genes. LDLR is already known to play a major role in CAD, and serves as further evidence that regulation by Ox-PAPC is a valid approach for defining CAD genes.
To achieve our second systems genetics aim, we identified transcripts whose expression at baseline, after Ox-PAPC treatment, or whose Ox-PAPC fold induction exhibited regulation, and thus expression Quantitative Trait Loci (eQTL), at peak GWAS SNPs. In other words, we searched for genes whose expression may be the causal intermediate between genetic variation and CAD. Of the 16 GWAS SNPs genotyped in the HAEC data, 4 harbored an eQTL (p-value < 1e-6) (Online Table II). 3 of these loci regulated expression traits that were distal/trans to the GWAS locus. However, rs11556924 on chromosome 7q32.2 significantly (p-value = 7.8e-9) regulated baseline expression of KLHDC10 (kelch domain containing 10) that is located within the locus. These data prioritize KLHDC10 for further investigation as a causal gene at the 7q32.2 locus for CAD.
Validation of Causal Interactions: CHAC1 is Directly Regulated by ATF4
We previously identified the CHAC1 gene as a target of the unfolded protein response (UPR) based on its position as a hub gene in a module enriched for UPR genes7, 28. CHAC1 is induced in response to UPR activators such as thapsigargin and siRNA knock-down of several UPR genes, including ATF4, ATF3, and CHOP reduced CHAC1 induction28. CHAC1 is highly conserved, from bacteria to mammals but, prior to these studies, its function had not been established. Subsequently, we have shown that it has a key role in apoptosis (data not shown); therefore, the precise mechanism of its regulation is of considerable interest.
Our network modeling revealed that ATF4 and CHAC1 were connected by a particularly strong edge and that they had many shared neighbors (genes with shared edges) (Figure 2). This suggested that ATF4 might directly regulate the expression of the CHAC1 gene. To address this question, we constructed a series of promoter constructs coupled to luciferase and tested them for activity following transfection of 293 cells (Figure 3). Co-transfection of ATF4 robustly induced the expression of the CHAC1 promoter, and this required an ATF/CREB element at −267 of the promoter (Figure 3). Additionally, ATF4 co-transfection was sufficient to activate CHAC1 transcription in sequential internal promoter-reporter deletion constructs, where the −267 ATF/CREB site remained intact (Figure 3), and scrambling the −267 ATF/CREB site was sufficient to block the ATF4 effect on CHAC1 promoter induction (data not shown). Importantly, plasmid over-expression of ATF3, spliced XBP1 or CHOP (data not shown) did not affect CHAC1 promoter activity. These data highlight the ATF4 interaction with CHAC1 via an ATF/CREB site as the major determinant of CHAC1 regulation. Additionally, these data indicate that ATF3 and CHOP do not directly activate the CHAC1 promoter. This demonstrates the power of the network modeling to highlight functional gene interactions and provide important information on complex signaling networks that can be further refined using directed strategies.
Figure 3. ATF4 directs CHAC1 promoter activity.
A. Luciferase expression from sequential CHAC1 promoter-reporter constructs is shown, as normalized to CMV-renilla, and co-transfected with plasmids expressing βGal, ATF4, ATF3 or sXBP1. B. Shown are luciferase expression results from CHAC1 promoter-reporter constructs containing a major CREB/ATF site and sequential internal deletions that were co -transfected with bGal, ATF4, ATF3 and sXBP1 expression plasmids.
In addition to wide-scale functional enrichments, the network recovers several known interactions. For example, in the blue module the UPR transcription factor ATF4 was tightly coupled to its known targets, including ASNS29, TRIB330 and VEGFA31 (Figure 2). In addition NRF2/NFE2L2 clustered in the blue module with several of its known target genes, including NQO132, GCLM32, and TXRD133 (Figure 2).
Heme Oxygenase 1 (HMOX1), A Gene Previously Implicated in Protection Against Atherosclerosis, Affects Many Genes In the Network and Suppresses the Response of Inflammatory Genes to Ox-PAPC
Among the many hypotheses of gene-gene interactions generated by the network, we focused on the role of HMOX134 and its regulation to illustrate the applications of the network model. We focused on this gene based upon the importance of HMOX1 in animal35–39 and human40–42 studies. HMOX1 itself is a `hub' located in the blue module (Figure 2) along with a number of genes previously identified as responsive to Ox-PAPC, including NAD(P)H dehydrogenase quinone 1 (NQO1)24, nuclear factor erythroid-derived 2-like 2 (NRF2/NFE2L2)32, ATF418, vascular endothelial growth factor A (VEGFA)43, and low density lipoprotein receptor (LDLR)44.
There was marked baseline variation among 149 EC donors as a 9-fold difference was observed between extremes of HMOX1 basal expression (Figure 4A). In contrast, only 2-fold difference among donors was observed in HMOX1 transcript levels in cells treated with Ox-PAPC. Taken together, these data suggest that there is a ceiling to the expression of HMOX1 in EC in this model system. We verified that HMOX1 mRNA induction was closely reflected at the levels of protein and heme oxygenase enzymatic activity in HAECs and in HeLa cells (Online Figure XVI). These data confirm that HMOX1 mRNA expression was a suitable means for evaluating HMOX1 activity, and support the hypothesis that variation in its basal activity significantly impacts the Ox-PAPC response.
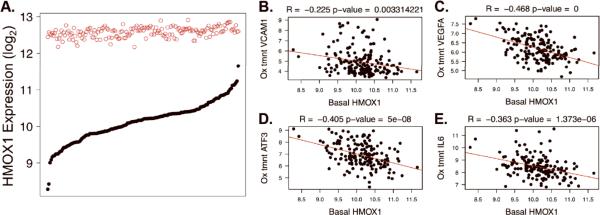
Figure 4. HMOX1 Variation and Correlation with Pro-Inflammatory Gene Expression.
A. Gene expression intensity (y-axis, log2 scale) is shown for the HMOX1 transcript at baseline (solid black circles) and after treatment with Ox-PAPC (open red circles) in the HAEC population. 149 donors are ranked along the x-axis according to baseline expression. B–E. Basal HMOX1 levels are plotted on the x-axes against the Ox-PAPC treated levels of VCAM1 (B), VEGFA (C), ATF3 (D), and IL6 (E) on the y-axes. R and p-values result from Pearson Correlation. The data shown in 4A is an expansion of data from 96 donors published previously in the same manner (ref 12).
These findings led us to hypothesize that basal levels of the enzyme would protect against the effects of Ox-PAPC. To model a causal relationship whereby basal HMOX1 levels would influence the induction of genes by Ox-PAPC we calculated the correlations between HMOX1 basal levels and the levels of genes in various pathways after treatment with Ox-PAPC. Many of the key molecules that coordinate the aforementioned endothelial responses were strongly negatively correlated to baseline HMOX1 expression, including the adhesion molecule VCAM1 (p=3.0e-3), genes regulated by the UPR including VEGFA (p < 1.0e-10), ATF3 (p=5.0e-8), and the inflammatory cytokine IL6 (p=1.4e-6) (Figure 4B–E). When we examined the distribution of HMOX1 correlated genes in the overall network we found that HMOX1 correlated genes were dispersed among all of the modules in the network (Figure 5A). These data support the hypothesis that basal HMOX1 modulates the Ox-PAPC response of genes from a diverse set of pathways. Whereas most genes were negatively correlated to basal HMOX1 (meaning greater basal HMOX1 corresponded to less Ox-PAPC treated levels of the other transcript), exceptions were predominantly found in the red module (Figure 5A, Online Table III). Such positively correlated genes included VTI1B (R = 0.55, p = 7.6e-15), ZMAT3 (R = 0.52, p = 5.6e-13), SOX4 (R = 0.50, p = 8.2e-12), CERK (R = 0.48, p = 3.3e-11), and SESN1 (R = 0.48, p = 6.0e-11). Unlike other genes in the red module, these molecules are not implicated in calcium signaling and have not been grouped together in a Gene Ontology category.
Figure 5. Regulation by HMOX1 in the Network.
A. Node border color signifies module membership and the fill indicates whether the Ox-PAPC treated expression level of a gene was correlated with HMOX1 basal levels (red is negative and green is positive correlation). siRNA-mediated knock-down of HMOX1 is shown for two siRNAs with the scrambled control in B. The fold changes of IL1b, IL6, MCP1, LDLR and INSIG1 were normalized to the fold changes measured in the scrambled control in C. mRNA was measured by RT-qPCR and averages ± s.d are shown. Asterisks indicate p-values less than 0.05.
Based upon the importance of HMOX1 in animal35–39, 45–47 and human studies48–50 and the observation that basal HMOX1 levels were more variable than post Ox-PAPC-treatment, we hypothesized that greater baseline HMOX1 levels would be protective against exposure to Ox-PAPC. To test this we correlated individual HMOX1 basal levels with the degree to which each individual responded to Ox-PAPC. The number of genes regulated above a given threshold per individual was defined as responsiveness to Ox-PAPC. Baseline HMOX1 expression was highly correlated with individual responsiveness at thresholds above 2-fold, and most significantly at the 7.5-fold threshold (R=−0.57) Taken together, these data strongly support the hypothesis that HMOX1 plays a critical role in mitigating multiple cellular responses to Ox-PAPC.
To directly test the role of HMOX1 levels on target gene expression, the HMOX1 transcript was silenced using two different siRNAs to 10–15% baseline values (Figure 5B). The expression of the pro-inflammatory cytokines IL1β, IL6, and MCP1 was found to be more highly induced after HMOX1 knock-down compared to the control (Figure 5C). Further, the cholesterol regulatory genes LDLR and INSIG1 were also more highly induced in response to Ox-PAPC as a result of HMOX1 knock-down, suggesting that sterol regulation may be dependent on HMOX1 (Figure 5C).
Regulation of HMOX1 Expression by Nuclear Factor (Erythroid-Derived 2)-Like 2 (NRF2)
In an effort to discover how basal HMOX1 transcript was regulated, we examined the relationship between HMOX1 and NRF2 that was also in the blue module. NRF2 has been shown to activate HMOX1 transcription upon translocation to the nucleus and binding to ARE elements in the HMOX1 promoter51. In the absence of Ox-PAPC treatment, NRF2 and HMOX1 were negatively correlated across the 149 donors (R = −0.3, p = 7.2e-5), meaning that greater NRF2 expression corresponded to less HMOX1 expression (Online Figure XVIIA). Surprisingly, correlations between NRF2 and HMOX1 were not significant after exposure to Ox-PAPC and no significant correlation existed between untreated NRF2 levels and treated HMOX1 levels (Online Figure XVIIB-C). These data were consistent with the studies demonstrating a role of other transcription factors regulating HMOX1 induction and the cell type dependent regulation by NRF252. However, NRF2 levels were highly correlated, at baseline and after Ox-PAPC treatment, with other known target antioxidant molecules including NAD(P)H dehydrogenase quinone 1 (NQO1), CCAAT/enhancer-binding protein beta (CEBPB), glutamate-cysteine ligase modifier subunit (GCLM), thioredoxin reductase 1 (TXNRD1), as well as glutathione- and glucuronyl- transferases (GSTK1 and B3GAT3) (Online Figure XVIII). These observations confirmed that expression data from our sample population were able to recover known molecular relationships, and confirmed the role of NRF2 in oxidant gene regulation. Nonetheless, these data demonstrate that NRF2 levels could explain only a small portion of HMOX1 basal variation. These results suggest that mechanisms other than NRF2 modulate basal HMOX1 levels.
HMOX1 Expression Does Not Exhibit Robust Local Genetic Regulation
We investigated the possibility of basal HMOX1 transcript regulation by a HMOX1 promoter microsatellite polymorphism. This dinucleotide GT expansion has been associated with various inflammatory diseases, including CAD, in several populations (reviewed in41). In these studies, longer alleles of the (GT) repeat were associated with disease. The mechanism by which longer alleles convey risk has been suggested to occur through the altered expression of HMOX140. Most convincing was the recent report by Taha et.al. who showed that short microsatellite alleles produced more HMOX1 than long alleles, and that short alleles were protective against oxidative stress in human umbilical vein endothelial cells (HUVEC)42. To test for differences in HMOX1 expression as a function of promoter alleles in HAECs, we genotyped the (GT)n promoter microsatellite and associated the dichotomized genotypes (short alleles < 29 (GT) repeats and long alleles ≥ 29 (GT) repeats in concordance with previous studies41) to HMOX1 expression levels. HMOX1 expression at baseline was associated to the microsatellite polymorphism in the whole population (n = 149, p = 1.9e-3) and in males (n = 113, p = 1.9e-3) but not in females (n=36, p = 0.533) (Online Figure XIX, top row). Where significant, HMOX1 expression was greater in individuals with more (GT) repeats. The fold change of HMOX1 induction was significant in the whole population (p = 2.4e-3) and in males (p = 2.0e-3) (Online Figure XIX, bottom row). In the case of HMOX1 fold change upon treatment with Ox-PAPC, individuals with longer alleles displayed lesser fold change values. These results contradict the previous findings, which suggested that short alleles would produce more HMOX142.
Next, we examined SNPs located in or near HMOX1 to test for additional local-regulatory variation. 62 SNPs were genotyped with minor allele frequency (MAF) greater than 5% that were within 100kb of the HMOX1 transcriptional start site on chromosome 22. Of these SNPs, the most significant association with the basal levels of HMOX1 was for rs5995385 (p = 9.0e-3, MAF = 0.39), downstream of HMOX1. After correcting for the multiple SNPs tested in the local region, this association became insignificant, suggesting that no local- SNPs significantly regulated HMOX1 basal expression.
Since we did not observe robust regulation of HMOX1 mRNA expression by local- genetic variation we hypothesized that variation at the HMOX1 locus may perturb HMOX1 function by other means. In particular, we focused on rs2071747, which is a missense mutation in the seventh amino acid of HMOX1 and converts an aspartic acid residue to histidine. Although this SNP was not genotyped in our population, it was in perfect linkage disequilibrium (R2 = 1, D' = 1) in the HapMap CEPH population with the HMOX1 intronic SNP rs5995097 that was genotyped in our population. We did not detect a significant effect of this polymorphism on HMOX1 protein localization, as determined by immunohistochemistry, or on HMOX1 enzymatic activity (data not shown). Taken together, these data suggest that HMOX1 was not strongly regulated by local- variants.
Distal Regulation of HMOX1 by GPR39
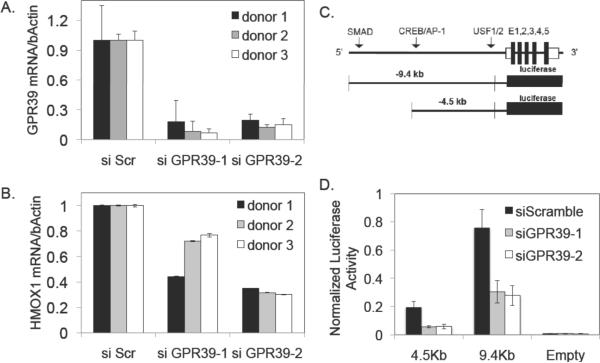
Using genome-wide association, we next tested whether distal- DNA variants associated to HMOX1 expression. Of the 718,374 SNPs that were polymorphic with MAF greater than 5%, 3 showed association to HMOX1 with a p-value < 1e-5 (Online Table IV). Each of these distally- associated SNPs were located within gene introns and suggested that the distally-located genes regulated the expression levels of HMOX1. The three genes were G-protein-coupled receptor 39 (GPR39), hypothetical protein LOC283521, and glutamate receptor, ionotropic, delta 1 (GRID1). We tested whether GPR39 and GRID1 exhibited regulation of HMOX1 basal levels using siRNA-mediated gene knock-down by two independent siRNA molecules (Figure 6A). GRID1 exhibited significant silencing, but no effect was observed in HMOX1 expression. However, GPR39, which was silenced to 10–20% of control levels, demonstrated consistent regulation of HMOX1. As predicted from the association data, GPR39 reduced the baseline expression of HMOX1 in all three donors tested by about 50% (Figure 6B). We performed linear regression for HMOX1 basal expression levels after correcting for GPR39 genotypes (rs12618338) to explore whether additional loci perturbed HMOX1 independently of GPR39. After multiple test correction no SNPs exhibited significant association to HMOX1 expression.
Figure 6. GPR39 Regulates Basal and Ox-PAPC treated Levels of HMOX1.
Two siRNAs were used against GPR39 and compared to the scrambled control in A. The baseline values of HMOX1 expression after GPR39 knock-down are shown in B. Results from luciferase HMOX1 promoter constructs (C) are shown in D for control (scrambled) and two GPR39 siRNA knock-downs. Luciferase activity was normalized to CMV-renilla. Plotted are averages ± s.d.
To further examine the mechanism by which GPR39 regulates HMOX1, we utilized two previously studied HMOX1 promoter-reporter constructs20, 21 (kindly provided by Dr. Anupam Agarwal, University of Alabama, Birmingham) (Figure 6C). Attempts to study HMOX1 regulation with these constructs directly in EC were unsuccessful primarily due to low transfection efficiencies and, therefore, we utilized HeLa cells that also express GPR39. Luciferase activity decreased upon GPR39 knock-down for both the −4.5kb (~75% reduction) and −9.4kb (~50% reduction) HMOX1 luciferase constructs. The fact that suppression was observed for both constructs, but was more pronounced for the −4.5 kb construct, suggests that GPR39 positively regulates HMOX1 expression by modulating transcription factor binding in the −4.5kb promoter region of the HMOX1 promoter (Figure 6D).
Discussion
It is clear that oxidized phospholipids significantly influence the expression of well over 1,000 genes in HAECs7, 12. While biochemical studies have revealed a number of important mechanisms underlying this response, the overall pathways involved are poorly understood. We have employed a systems genetics approach to model a biologic network, in which common, naturally-occurring variations in the human population perturb responses to oxidized phospholipids. The approach integrates aspects of systems biology, in particular, network modeling, and genome-wide association. This resulting systems level view emphasizes the diverse effects of oxidized phospholipids and it provides the basis for many hypotheses relating to molecular interactions and genetic regulatory mechanisms.
The co-expression network we generated is a simplified model of the molecular interactions involved in responses to Ox-PAPC at the transcript level. In this model, genes are represented as nodes and the transcript correlations across genetic perturbations as edges. We assume that the connections identified result from shared regulatory mechanisms, implying related functions. Of course, the model is incomplete and should be considered hypothesis generating. Our efforts in the current study were focused on the 2000 most regulated genes by Ox-PAPC to 1) define relationships and generate hypotheses likely to be most important to Ox-PAPC action, 2) to simplify the network for visualization purposes, and 3) reduce computational time required for analyses. One caveat of our network is that, by focusing on only 2000 transcripts, this network would not be able to capture relationships among all transcripts measured in the cell. Likewise, our network would not capture post-transcriptional relationships. To demonstrate the predictive power of the model, and to elucidate molecular mechanisms of clinical interest, we explored several hypotheses using experimental perturbation and genetic association. In addition to the several known pathways reflected in this network, we observed novel interactions between GPR39 and HMOX1, between ATF4 and CHAC1, and between OKL38 and redox genes. These are discussed below.
The gene co-expression network consisted of the genes most affected by Ox-PAPC in 149 donors and defines the relationship between genes and pathways. The network, consisting of 11 modules of highly connected genes (Figure 1, Online Figures II–XII), was constructed from transcripts quantified with and without Ox-PAPC treatment. The modules were enriched for pathways previously described for the action of Ox-PAPC, and others that have not been identified previously. The fact that our network was constructed with a large number of individuals, each likely contributing thousands of genetic variations that perturbed the responses to Ox-PAPC, enabled modeling of fine-grained gene relationships within and between modules. Since the precise connections are sometimes difficult to visualize in the figures, we have included the top 10 most connected genes, their connection strength, and the corresponding modules for all 2000 genes input for network construction. This file is in Cytoscape network format16 so that the relationships may be easily visualized for genes of interest (Online Table V).
We previously reported a network analysis of Ox-PAPC treated EC using a small number of donors7. The present network, consisting of a much larger number of individuals, enabled modeling of significantly improved relationships. For example, known relationships among UPR genes were more accurately capitulated in the present study. In addition, important candidate genes, not previously identified as UPR genes, are suggested by their strong connectivity in this module. The striking enrichment for Gene Ontology categories observed in the network supports the hypothesis that co-expressed genes share functional significance (Table 1). Furthermore, known biological relationships were identified in modules of our network.
The co-expression network approach allows the formulation of three different kinds of hypotheses. First, it is possible to infer likely functions of genes whose functions are unknown if they are highly connected to genes with known functions. We have previously used this approach to show that CHAC1, a highly conserved protein whose function was previously unknown, plays a role in the UPR28. We now identify a specific interaction (ATF4-CHAC1) based on our network analysis. Similarly, it can be hypothesized that genes in the turquoise module are likely to have a role in cell cycling, that genes in the purple module are likely to be members of the UPR, and so on. Second, the network modeling can provide information about novel mechanistic interactions between the genes. For example, the result that certain anti-oxidant genes, such as NRF2 and HMOX1, are tightly connected with numerous UPR genes in the blue module, provides evidence of interactions between these molecules, and recent studies support this possibility58,59. Similarly, we have now implicated OKL38 in redox functions in the blue module. Importantly, we have identified OKL38 as an important regulator of HMOX1. In this study we have shown that the basal levels of HMOX1 varied considerably in our group of 149 donors; however, HMOX1 levels induced by Ox-PAPC were very similar for all donors (Figure 4A). We have now demonstrated that knockdown of OKL38 increased the Ox-PAPC induced levels of HMOX1 by 3-fold (Online Figure XIV). This suggests that OKL38 and HMOX1 have an important combined relationship in redox regulation. Third, network modeling, when performed as a function of genetic variation, allows the formulation of hypotheses concerning genetic regulatory mechanisms. An example of this is the regulation of HMOX1 by GPR39, discussed below.
In this report, we have focused on hypotheses relating to HMOX1. Our laboratories as well as those of others have implicated HMOX1 in protection against atherosclerosis35, 36, 38, 53, 54, ischemia/reperfusion injury39, 55, and restenosis46, 56, 57. In humans, a variable (GT)n dinucleotide repeat in the HMOX1 promoter has been associated with EC function42, as well as clinical diseases including pulmonary disease, cardiovascular disease, renal transplantation, and idiopathic miscarriage (refer to Table 1 in ref41 for specific citations). Our data did not replicate previous reports that protective microsatellite alleles produced more HMOX1 mRNA. This discrepancy could be a cell type difference between HUVECs and HAECs. Since HMOX1 basal levels varied ~9-fold among EC donors, whereas treated levels varied ~2-fold (Figure 4A), we hypothesized that variable responses to Ox-PAPC for highly induced gene expression traits would predominantly be determined by baseline levels.
When we analyzed how baseline HMOX1 variation affected the network we found that the expression of many pro-inflammatory genes in several modules was inversely correlated with HMOX1 expression (Figures 4B–E, 5A), with the notable exception of genes in the red module, which were positively correlated with basal HMOX1 levels (Figure 5A, Table 2). Furthermore, baseline expression differences in HMOX1 significantly predicted individual responsiveness to Ox-PAPC (R= −0.57), such that greater HMOX1 protected against induction of endothelial transcripts by Ox-PAPC. To experimentally test the role of HMOX1 in our network we silenced HMOX1 with siRNA and observed that pro-inflammatory molecules were affected (Figure 5B,C). A more detailed study of donors exhibiting either extremely high or low basal HMOX1 levels (as demonstrated in Figure 4A) would be an attractive strategy for identifying molecular regulators of HMOX1 but such a study would require additional samples to be collected. In addition, failure to identify HMOX1 local-/cis-regulatory elements may be a power issue. However, undetected variants would likely have small effect sizes. This is supported by our recent identification of over 1000 cis-regulatory elements of high significance in our previous study of only 96 donors12. It should also be noted that since the HAECs in this study were grown to confluence using culture conditions in vitro it is likely that the data collected in this study would not fully recapitulate the in vivo gene expression profiles of HAECs.
Table 2.
Greater HMOX1 Baseline Values Prevented The Induction Of Key Molecules In Ox-PAPC Induced Pathways Of Interest
| Gene | R | p | Pathway |
|---|---|---|---|
| ATF3 | −0.40 | 5.01e-08 | UPR/ER homeostasis |
| CEBPB | −0.41 | 2.74e-08 | UPR/ER homeostasis |
| CHAC1 | −0.37 | 6.90e-07 | UPR/ER homeostasis |
| SLC7A11 | −0.36 | 1.23e-06 | UPR/ER homeostasis |
| TRIB3 | −0.343 | 7.28e-06 | UPR/ER homeostasis |
| SLC2A3 | −0.26 | 5.62e-04 | UPR/ER homeostasis |
| VEGFA | −0.47 | 1.58e-10 | angiogensis/UPR |
| IL6 | −0.36 | 1.37e-06 | proinflammatory cytokine |
| IL8 | −0.28 | 2.41e-04 | proinflammatory cytokine |
| LDLR | −0.25 | 1.05e-03 | sterol homeostasis |
| E2F1 | −0.33 | 1.63e-05 | cell cycle |
| HMOX1 | 0.39 | 2.38e-07 | redox |
| TXN | −0.34 | 6.01e-06 | redox |
| VCAM1 | −0.22 | 3.31e-03 | adhesion molecule |
To test for genetic trans-regulation of HMOX1 basal levels, we carried out GWAS and identified three loci with suggestive p-values. The peak SNP at one of the loci occurred within the GPR39 locus on human chromosome 2. To test the possible relationship of GPR39 with HMOX1 expression, we used siRNA knock-down with two separate GPR39 siRNAs in HAECs from three different individuals. In all cases, GPR39 knock-down resulted in a substantial (approximately 40%) decrease in HMOX1 expression as compared to scrambled controls (Figure 6). In contrast to GPR39, siRNA experiments for GRID1 at the chromosome 10 locus were negative (data not shown). To further examine the mechanism by which GPR39 regulates HMOX1, we performed promoter-reporter studies. These indicated that GPR39 regulates HMOX1 expression by interaction with elements in the proximal 4.5 kb upstream region of HMOX1, although the precise site of interaction is unclear.
GPR39 is a G-Protein coupled receptor related to the Ghrelin/neurotensin receptor sub- family. Knockout mice are viable and have been characterized by increased body weight and altered intestinal function. This receptor is constitutively active in a number of cell types and its activity has been shown to be increased by zinc in some cell types. The constitutive activity is mediated by Gαq which stimulates CRE mediated transcription, and Gα12/13 which stimulates SRE mediated transcription (reviewed in58). We hypothesize that GPR39 regulates the HMOX1 promoter due to the activation of CRE, which most likely lies within the 4.5KB promoter region. In neurons GPR39 overexpression protected against cell death, oxidative stress and ER stress59. The latter results are consistent with a role of GPR39 in activating expression of HMOX1.
Although it has been clear for two decades that atherosclerosis is an inflammatory disease, the underlying mechanisms responsible for the inflammation remain poorly understood. GWAS studies performed for atherosclerosis have been successful in identifying only a very small fraction of the genetic component27;60. Analysis of functional genetic variations identified through systems genetics approaches, such as that affecting GPR39, should complement the larger population studies that directly examine links to clinical disease. The Ox-PAPC network serves as an additional resource for integration with conventional GWAS studies to elucidate gene interactions and regulatory relationships that can be tested in mechanistic studies.
Supplementary Material
Novelty and Significance.
What is known?
Atherosclerosis is a chronic inflammatory disease. The mechanisms underlying the initiation and propagation of inflammation remain poorly understood.
Accumulating evidence suggests that oxidized phospholipids derived from lipoproteins entrapped in the vessel wall activate endothelial cells and are responsible in part for the initiation of inflammation.
In vitro studies with cultured endothelial cells from different donors have shown that oxidized phospholipids influence the expression of hundreds of genes that regulate inflammation in a donor-specific manner.
What new information does this article contribute?
We used a systems biology approach utilizing endothelial gene expression from 150 donors and identified 11 groups of functionally related genes that played an important role in the response to oxidized phospholipids. We also identified key regulators of each group.
Heme oxygenase 1 (HMOX1), which showed 9 fold variation in transcript levels among donors, was identified as a key regulator of one of these groups of inflammatory genes.
A nucleotide polymorphism in G-protein coupled receptor 39 (GPR39) was shown to associate with the levels of HMOX1 and inflammatory genes; this was confirmed by biochemical studies.
Summary.
We have previously demonstrated that hundreds of endothelial genes are affected by oxidized phospholipids and that this response differs among humans. This may explain the variation in susceptibility to atherosclerosis among individuals. In this study, we have utilized network modeling to obtain a global understanding of the processes underlying the inflammatory responses in endothelial cell cultures from 150 human donors. We grouped the genes that exhibited similar changes, in response to oxidized phospholipids, into 11 modules that showed dramatic enrichment for known biological pathways. An important aspect of such network modeling is that it allows one to see “the forest for the trees.” Within each module, likely interactions between individual genes were predicted. This allowed us to identify critical genes, such as OKL38, as novel regulators of the endothelial response to oxidized phospholipids. Also, HMOX1 was shown to be highly connected to genes in the inflammatory and unfolded protein response groups. In a complementary genetics approach, we mapped the expression levels of selected genes to genomic loci, providing novel regulatory information. This systems-based approach to inflammatory responses has particular significance for the understanding of the complex genetic and environmental interactions contributing to common forms of atherosclerosis.
Acknowledgments
We thank the Atherosclerosis Research Unit (ARU) at UCLA for help collecting the HAECs and for valuable discussions. We thank Dr. Anupam Agarwal from the University of Alabama, Birmingham for the HMOX1 luciferase constructs.
Sources of Funding This research was funded by NIH grant PO1-HL030568 (AJL, JAB), NIH Grant HL-064731 (JAB), NIH training grant HL069766 (CER), NIH Grant ES016959 (JAA) an American Heart Association pre-doctoral fellowship (CER) and an American Heart Association post-doctoral fellowship (SDL, MC).
Non-standard Abbreviations and Acronyms
- EC
Endothelial Cell
- OxPL
Oxidized Phospholipid
- Ox-PAPC
Oxidized Palmitoyl-Arachidonyl-Phosphatidylcholine
- PEIPC
1-Palmitoyl-2-(5,6)-Epoxyisoprostane E2-sn-glycero-3-Phosphocholine
- LDL
Low Density Lipoprotein
- eQTL
Expression Quantitative Trait Locus
Footnotes
Disclosures Wen-Pin Yang Ph.D. and Paul Kayne Ph.D. are employees and shareholders of Bristol-Myers Squibb.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levitan I, Volkov S, Subbaiah PV. Oxidized ldl: Diversity, patterns of recognition, and pathophysiology. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2009.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berliner JA, Leitinger N, Tsimikas S. The role of oxidized phospholipids in atherosclerosis. J Lipid Res. 2009;50(Suppl):S207–212. doi: 10.1194/jlr.R800074-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witztum JL, Steinberg D. The oxidative modification hypothesis of atherosclerosis: Does it hold for humans? Trends Cardiovasc Med. 2001;11:93–102. doi: 10.1016/s1050-1738(01)00111-6. [DOI] [PubMed] [Google Scholar]
- 5.Berliner JA, Watson AD. A role for oxidized phospholipids in atherosclerosis. The New England journal of medicine. 2005;353:9–11. doi: 10.1056/NEJMp058118. [DOI] [PubMed] [Google Scholar]
- 6.Shi W, Haberland ME, Jien ML, Shih DM, Lusis AJ. Endothelial responses to oxidized lipoproteins determine genetic susceptibility to atherosclerosis in mice. Circulation. 2000;102:75–81. doi: 10.1161/01.cir.102.1.75. [DOI] [PubMed] [Google Scholar]
- 7.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, Nelson SF, Horvath S, Berliner JA, Kirchgessner TG, Lusis AJ. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci U S A. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navab M, Hough GP, Stevenson LW, Drinkwater DC, Laks H, Fogelman AM. Monocyte migration into the subendothelial space of a coculture of adult human aortic endothelial and smooth muscle cells. The Journal of clinical investigation. 1988;82:1853–1863. doi: 10.1172/JCI113802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung ME, Berliner JA, Koroniak L, Gugiu BG, Watson AD. Improved synthesis of the epoxy isoprostane phospholipid peipc and its reactivity with amines. Org Lett. 2008;10:4207–4209. doi: 10.1021/ol8014804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson A, Subbanagounder G, Welsbie D, Faull K, Navab M, Jung M, Fogelman A, Berliner J. Structural identification of a novel pro-inflammatory epoxyisoprostane phospholipid in mildly oxidized low density lipoprotein. Journal of Biological Chemistry. 1999;274:24787–24798. doi: 10.1074/jbc.274.35.24787. [DOI] [PubMed] [Google Scholar]
- 11.Subbanagounder G, Leitinger N, Schwenke DC, Wong JW, Lee H, Rizza C, Watson AD, Faull KF, Fogelman AM, Berliner JA. Determinants of bioactivity of oxidized phospholipids: Specific oxidized fatty acyl groups at the sn-2 position. Arterioscler Thromb Vasc Biol. 2000;20:2248–2254. doi: 10.1161/01.atv.20.10.2248. [DOI] [PubMed] [Google Scholar]
- 12.Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, Yordanova R, Tilford C, Guan B, He A, Gargalovic PS, Kirchgessner TG, Berliner JA, Lusis AJ. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86:399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 14.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P, Horvath S. Wgcna: An r package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gargalovic PS, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Baruch-Oren T, Berliner JA, Kirchgessner TG, Lusis AJ. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:2490–2496. doi: 10.1161/01.ATV.0000242903.41158.a1. [DOI] [PubMed] [Google Scholar]
- 19.Haider A, Olszanecki R, Gryglewski R, Schwartzman ML, Lianos E, Kappas A, Nasjletti A, Abraham NG. Regulation of cyclooxygenase by the heme-heme oxygenase system in microvessel endothelial cells. Journal of Pharmacology and Experimental Therapeutics. 2002;300:188–194. doi: 10.1124/jpet.300.1.188. [DOI] [PubMed] [Google Scholar]
- 20.Deshane J, Kim J, Bolisetty S, Hock TD, Hill-Kapturczak N, Agarwal A. Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. Journal of Biological Chemistry. 2010;285:16476–16486. doi: 10.1074/jbc.M109.058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traylor A, Hock T, Hill-Kapturczak N. Specificity protein 1 and smad-dependent regulation of human heme oxygenase-1 gene by transforming growth factor-œ≤1 in renal epithelial cells. American Journal of Physiology - Renal Physiology. 2007;293:F885–F894. doi: 10.1152/ajprenal.00519.2006. [DOI] [PubMed] [Google Scholar]
- 22.Barabasi AL. Scale-free networks: A decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 23.Barabasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 24.Lee S, Gharavi NM, Honda H, Chang I, Kim B, Jen N, Li R, Zimman A, Berliner JA. A role for nadph oxidase 4 in the activation of vascular endothelial cells by oxidized phospholipids. Free Radic Biol Med. 2009;47:145–151. doi: 10.1016/j.freeradbiomed.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gugiu BG, Mouillesseaux K, Duong V, Herzog T, Hekimian A, Koroniak L, Vondriska TM, Watson AD. Protein targets of oxidized phospholipids in endothelial cells. J Lipid Res. 2008;49:510–520. doi: 10.1194/jlr.M700264-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Li R, Chen W, Yanes R, Lee S, Berliner JA. Okl38 is an oxidative stress response gene stimulated by oxidized phospholipids. Journal of Lipid Research. 2007;48:709–715. doi: 10.1194/jlr.M600501-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NEE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJP, Khaw K-T, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WHW, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JCM, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. Chac1/mgc4504 is a novel proapoptotic component of the unfolded protein response, downstream of the atf4-atf3-chop cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. Atf4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. Journal of Biological Chemistry. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 30.Shang Y-Y, Zhong M, Zhang L-P, Guo Z-X, Wang Z-H, Zhang Y, Deng J-T, Zhang W. Tribble 3, a novel oxidized low-density lipoprotein-inducible gene, is induced via the activating transcription factor 4–c/ebp homologous protein pathway. Clinical and Experimental Pharmacology and Physiology. 2010;37:51–55. doi: 10.1111/j.1440-1681.2009.05229.x. [DOI] [PubMed] [Google Scholar]
- 31.Roybal CN, Hunsaker LA, Barbash O, Jagt DLV, Abcouwer SF. The oxidative stressor arsenite activates vascular endothelial growth factor mrna transcription by an atf4-dependent mechanism. Journal of Biological Chemistry. 2005;280:20331–20339. doi: 10.1074/jbc.M411275200. [DOI] [PubMed] [Google Scholar]
- 32.Jyrkkanen HK, Kansanen E, Inkala M, Kivela AM, Hurttila H, Heinonen SE, Goldsteins G, Jauhiainen S, Tiainen S, Makkonen H, Oskolkova O, Afonyushkin T, Koistinaho J, Yamamoto M, Bochkov VN, Yla-Herttuala S, Levonen AL. Nrf2 regulates antioxidant gene expression evoked by oxidized phospholipids in endothelial cells and murine arteries in vivo. Circ Res. 2008;103:e1–9. doi: 10.1161/CIRCRESAHA.108.176883. [DOI] [PubMed] [Google Scholar]
- 33.Reichard JF, Motz GT, Puga A. Heme oxygenase-1 induction by nrf2 requires inactivation of the transcriptional repressor bach1. Nucleic acids research. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S, Li R, Kim B, Palvolgyi R, Ho T, Yang QZ, Xu J, Szeto WL, Honda H, Berliner JA. Ox-papc activation of pmet system increases expression of heme oxygenase-1 in human aortic endothelial cell. J Lipid Res. 2009;50:265–274. doi: 10.1194/jlr.M800317-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikawa K, Sugawara D, Goto J, Watanabe Y, Kawamura K, Shiomi M, Itabe H, Maruyama Y. Heme oxygenase-1 inhibits atherogenesis in watanabe heritable hyperlipidemic rabbits. Circulation. 2001;104:1831–1836. doi: 10.1161/hc3901.095897. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa K, Sugawara D, Wang X, Suzuki K, Itabe H, Maruyama Y, Lusis AJ. Heme oxygenase-1 inhibits atherosclerotic lesion formation in ldl-receptor knockout mice. Circ Res. 2001;88:506–512. doi: 10.1161/01.res.88.5.506. [DOI] [PubMed] [Google Scholar]
- 37.Tulis DA, Durante W, Peyton KJ, Evans AJ, Schafer AI. Heme oxygenase-1 attenuates vascular remodeling following balloon injury in rat carotid arteries. Atherosclerosis. 2001;155:113–122. doi: 10.1016/s0021-9150(00)00552-9. [DOI] [PubMed] [Google Scholar]
- 38.Yet SF, Layne MD, Liu X, Chen YH, Ith B, Sibinga NE, Perrella MA. Absence of heme oxygenase-1 exacerbates atherosclerotic lesion formation and vascular remodeling. FASEB J. 2003;17:1759–1761. doi: 10.1096/fj.03-0187fje. [DOI] [PubMed] [Google Scholar]
- 39.Yet SF, Tian R, Layne MD, Wang ZY, Maemura K, Solovyeva M, Ith B, Melo LG, Zhang L, Ingwall JS, Dzau VJ, Lee ME, Perrella MA. Cardiac-specific expression of heme oxygenase-1 protects against ischemia and reperfusion injury in transgenic mice. Circ Res. 2001;89:168–173. doi: 10.1161/hh1401.093314. [DOI] [PubMed] [Google Scholar]
- 40.Chen YH, Lin SJ, Lin MW, Tsai HL, Kuo SS, Chen JW, Charng MJ, Wu TC, Chen LC, Ding YA, Pan WH, Jou YS, Chau LY. Microsatellite polymorphism in promoter of heme oxygenase-1 gene is associated with susceptibility to coronary artery disease in type 2 diabetic patients. Hum Genet. 2002;111:1–8. doi: 10.1007/s00439-002-0769-4. [DOI] [PubMed] [Google Scholar]
- 41.Exner M, Minar E, Wagner O, Schillinger M. The role of heme oxygenase-1 promoter polymorphisms in human disease. Free Radic Biol Med. 2004;37:1097–1104. doi: 10.1016/j.freeradbiomed.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Taha H, Skrzypek K, Guevara I, Nigisch A, Mustafa S, Grochot-Przeczek A, Ferdek P, Was H, Kotlinowski J, Kozakowska M, Balcerczyk A, Muchova L, Vitek L, Weigel G, Dulak J, Jozkowicz A. Role of heme oxygenase-1 in human endothelial cells: Lesson from the promoter allelic variants. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30:1634–1641. doi: 10.1161/ATVBAHA.110.207316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimman A, Mouillesseaux KP, Le T, Gharavi NM, Ryvkin A, Graeber TG, Chen TT, Watson AD, Berliner JA. Vascular endothelial growth factor receptor 2 plays a role in the activation of aortic endothelial cells by oxidized phospholipids. Arterioscler Thromb Vasc Biol. 2007;27:332–338. doi: 10.1161/01.ATV.0000252842.57585.df. [DOI] [PubMed] [Google Scholar]
- 44.Yeh M, Cole AL, Choi J, Liu Y, Tulchinsky D, Qiao JH, Fishbein MC, Dooley AN, Hovnanian T, Mouilleseaux K, Vora DK, Yang WP, Gargalovic P, Kirchgessner T, Shyy JY, Berliner JA. Role for sterol regulatory element-binding protein in activation of endothelial cells by phospholipid oxidation products. Circ Res. 2004;95:780–788. doi: 10.1161/01.RES.0000146030.53089.18. [DOI] [PubMed] [Google Scholar]
- 45.Wang LJ, Lee TS, Lee FY, Pai RC, Chau LY. Expression of heme oxygenase-1 in atherosclerotic lesions. Am J Pathol. 1998;152:711–720. [PMC free article] [PubMed] [Google Scholar]
- 46.Tulis DA, Durante W, Liu X, Evans AJ, Peyton KJ, Schafer AI. Adenovirus-mediated heme oxygenase-1 gene delivery inhibits injury-induced vascular neointima formation. Circulation. 2001;104:2710–2715. doi: 10.1161/hc4701.099585. [DOI] [PubMed] [Google Scholar]
- 47.Kronke G, Bochkov VN, Huber J, Gruber F, Bluml S, Furnkranz A, Kadl A, Binder BR, Leitinger N. Oxidized phospholipids induce expression of human heme oxygenase-1 involving activation of camp-responsive element-binding protein. J Biol Chem. 2003;278:51006–51014. doi: 10.1074/jbc.M304103200. [DOI] [PubMed] [Google Scholar]
- 48.Rueda B, Oliver J, Robledo G, Lopez-Nevot MA, Balsa A, Pascual-Salcedo D, Gonzalez-Gay MA, Gonzalez-Escribano MF, Martin J. Ho-1 promoter polymorphism associated with rheumatoid arthritis. Arthritis Rheum. 2007;56:3953–3958. doi: 10.1002/art.23048. [DOI] [PubMed] [Google Scholar]
- 49.Sheu CC, Zhai R, Wang Z, Gong MN, Tejera P, Chen F, Su L, Thompson BT, Christiani DC. Heme oxygenase-1 microsatellite polymorphism and haplotypes are associated with the development of acute respiratory distress syndrome. Intensive Care Med. 2009;35:1343–1351. doi: 10.1007/s00134-009-1504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song F, Li X, Zhang M, Yao P, Yang N, Sun X, Hu FB, Liu L. Association between heme oxygenase-1 gene promoter polymorphisms and type 2 diabetes in a chinese population. Am J Epidemiol. 2009;170:747–756. doi: 10.1093/aje/kwp196. [DOI] [PubMed] [Google Scholar]
- 51.Stocker R, Perrella MA. Heme oxygenase-1: A novel drug target for atherosclerotic diseases? Circulation. 2006;114:2178–2189. doi: 10.1161/CIRCULATIONAHA.105.598698. [DOI] [PubMed] [Google Scholar]
- 52.Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochemical Pharmacology. 2010;80:1895–1903. doi: 10.1016/j.bcp.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 53.Juan SH, Lee TS, Tseng KW, Liou JY, Shyue SK, Wu KK, Chau LY. Adenovirus-mediated heme oxygenase-1 gene transfer inhibits the development of atherosclerosis in apolipoprotein e-deficient mice. Circulation. 2001;104:1519–1525. doi: 10.1161/hc3801.095663. [DOI] [PubMed] [Google Scholar]
- 54.Orozco LD, Kapturczak MH, Barajas B, Wang X, Weinstein MM, Wong J, Deshane J, Bolisetty S, Shaposhnik Z, Shih DM, Agarwal A, Lusis AJ, Araujo JA. Heme oxygenase-1 expression in macrophages plays a beneficial role in atherosclerosis. Circ Res. 2007;100:1703–1711. doi: 10.1161/CIRCRESAHA.107.151720. [DOI] [PubMed] [Google Scholar]
- 55.Melo LG, Agrawal R, Zhang L, Rezvani M, Mangi AA, Ehsan A, Griese DP, Dell'Acqua G, Mann MJ, Oyama J, Yet SF, Layne MD, Perrella MA, Dzau VJ. Gene therapy strategy for long-term myocardial protection using adeno-associated virus-mediated delivery of heme oxygenase gene. Circulation. 2002;105:602–607. doi: 10.1161/hc0502.103363. [DOI] [PubMed] [Google Scholar]
- 56.Duckers HJ, Boehm M, True AL, Yet SF, San H, Park JL, Clinton Webb R, Lee ME, Nabel GJ, Nabel EG. Heme oxygenase-1 protects against vascular constriction and proliferation. Nat Med. 2001;7:693–698. doi: 10.1038/89068. [DOI] [PubMed] [Google Scholar]
- 57.Omura S, Suzuki H, Toyofuku M, Ozono R, Kohno N, Igarashi K. Effects of genetic ablation of bach1 upon smooth muscle cell proliferation and atherosclerosis after cuff injury. Genes Cells. 2005;10:277–285. doi: 10.1111/j.1365-2443.2005.00832.x. [DOI] [PubMed] [Google Scholar]
- 58.Popovics, Petra, Stewart JA. Gpr39: A zn[2+]-activated g protein-coupled receptor that regulates pancreatic, gastrointestinal and neuronal functions. Springer; Heidelberg, ALLEMAGNE: 2011. #160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dittmer S, Sahin M, Pantlen A, Saxena A, Toutzaris D, Pina A-L, Geerts A, Golz S, Methner A. The constitutively active orphan g-protein-coupled receptor gpr39 protects from cell death by increasing secretion of pigment epithelium-derived growth factor. Journal of Biological Chemistry. 2008;283:7074–7081. doi: 10.1074/jbc.M704323200. [DOI] [PubMed] [Google Scholar]
- 60.Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.