Abstract
Abcission, the natural shedding of leaves, flowers and fruits, is a fundamental component of plant development. Abscission is a highly regulated process that occurs at distinct zones of cells that undergo enlargement and subsequent separation. Although some components of abscission, including accumulation of the hormone ethylene and cell wall-degrading enzymes, have been described, the regulatory pathways remain largely unknown. In this paper we describe a critical component required for floral organ abscission in Arabidopsis thaliana, the receptor-like protein kinase HAESA. Histochemical analysis of transgenic plants harboring a HAESA promoter:: β-glucuronidase reporter gene and in situ RNA hybridization experiments show HAESA expression in the abscission zones where the sepals, petals, and stamens attach to the receptacle, at the base of pedicels, and at the base of petioles where leaves attach to the stem. Immunodetection, immunoprecipitation, and protein kinase activity assays reveal HAESA is a plasma membrane serine/threonine protein kinase. The reduction of function of HAESA in transgenic plants harboring an antisense construct results in delayed abscission of floral organs, and the severity of the phenotype is directly correlated with the level of HAESA protein. These results demonstrate that HAESA functions in developmentally regulated floral organ abscission.
Keywords: Arabidopsis, receptor kinase, RLK, HAESA, abscission, HAE
An integral element of plant development is the abscission of senescent tissues, damaged organs, and ripened fruits. Abscission is a complex process that occurs at defined zones of cytoplasmically dense cells, which undergo cell enlargement and accumulation of cell wall and pectin-degrading enzymes. These enzymes dissolve the cementing substances between primary walls of contiguous cells, which subsequently differentiate into scar tissue (Bleecker and Patterson 1997; Gonzalez-Carranza et al. 1998).
The contribution of plant hormones to the abscission process has been the focus of a large body of research. The prevailing view is that auxin delays abscission, whereas ethylene promotes abscission. Although there are conflicting reports in the literature, the effects of other well-characterized plant hormones, such as abscisic acid (ABA), gibberellic acid (GA), and cytokinins, primarily have been attributed to altering levels of or sensitivity to auxin and ethylene in abscising tissue (Sexton and Roberts 1982).
The most widely recognized abscission process is the annual shedding of leaves in deciduous forests. A more genetically tractable system in which to elucidate the mechanisms of abscission is the developmentally governed shedding of Arabidopsis floral organs (i.e., sepals, petals, and stamens). Arabidopsis mutants defective in hormone production or sensing have allowed an assessment of the role of hormone signaling in the floral organ abscission process. The ethylene-insensitive mutants etr1-1 and ein2 exhibit delayed abscission of floral organs (Ecker 1995; Bleecker and Patterson 1997). However, abscission does occur in these mutants, and the molecular markers used to define the abscission process are present, provoking Bleecker and Patterson (1997) to conclude that ethylene may be involved in controlling the timing of floral organ abscission and that ethylene-independent pathways are required. The identity of these proposed pathways is not known but must involve the coordinated response of numerous cells in the abscission zone.
One predominant mechanism of coordinating intercellular responses involves reversible protein phosphorylation mediated by transmembrane receptor protein kinases that are responsible for integrating developmental and environmental cues to the cell's interior (van der Geer et al. 1994). In plants, the receptor-like protein kinases (RLKs) have been implicated in prevention of self-pollination, pathogen response, hormone perception and signaling, and plant development (Becraft 1998; Lease et al. 1998). Many RLKs, whose functions have been deduced from their mutant phenotypes, are involved in plant developmental processes. One class of these, the leucine-rich repeat (LRR) RLKs, have emerged as key developmental regulators. The Arabidopsis gene CLAVATA1, controls the balance between cell proliferation and differentiation at the shoot and floral apical meristems (Clark et al. 1993, 1997), whereas ERECTA controls organ shape (Torii et al. 1996). Perception and signal transduction of brassinosteroids depends on the action of another LRR–RLK, Bri1 (Li and Chory 1997; Altmann 1998).
In this work we show that the Arabidopsis LRR–RLK HAESA (formerly named RLK5) controls floral organ abscission. HAESA is plasma membrane-associated and has serine/threonine protein kinase activity. HAESA is expressed at the base of the petioles and pedicels, as well as in abscission zones of the floral organs, as assessed by both a HAESA promoter::β-glucuronidase (GUS) reporter gene in transgenic plants and by in situ RNA hybridization. To assign a function for HAESA in abscission zones, transgenic plants expressing a constitutive antisense HAESA construct were generated, and abscission of floral organs was scored. Antisense lines showed varying levels of HAESA protein, and the amount of HAESA protein is inversely correlated with defective floral organ abscission. Failure to abscise floral organs is due to the presence of the antisense transgene, as individuals from segregating populations that do not inherit the transgene exhibit normal floral organ abscission. These results demonstrate a role for HAESA receptor kinase in floral organ abscission and provide insights into how plant cells regulate cellular processes.
Results
The Arabidopsis RLK5 gene encodes a RLK (Walker 1993). We implemented multiple strategies to determine the function of RLK5. The expression pattern was especially illuminating, and generation of transgenic reduction-of-function plants established a role for RLK5 in floral organ abscission. To reflect the reduction of RLK5 function phenotype, inability to precisely abscise floral organs, RLK5 has been renamed HAESA (HAE), a Latin word meaning to stick to, adhere to, or cling to.
HAESA expression is tissue-specific and developmental stage-dependent
To establish HAESA function in Arabidopsis we first determined its expression pattern using two different approaches: (1) examination of transgenic plants harboring a HAESA::GUS reporter gene fusion; and (2) in situ RNA hybrizidation with a HAESA antisense probe. In flowers, HAESA promoter activity is observed in the abscission zones, in which the sepals, petals, and stamens attach to the receptacle (Fig. 1A), and weak expression is observed at the base of pedicels (the stalks of individual flowers in an inflorescence) at their attachment points (data not shown). In situ RNA hybridization experiments demonstrate that HAESA is expressed in the floral organ abscission zones (Fig. 1B), consistent with the reporter gene data (Fig. 1A). HAESA expression is dependent on floral stage, with expression in maturing flowers coinciding with competence to self-pollinate (Fig. 1C). This expression corresponds to stage 14–15 (Smyth et al. 1990), around the time when the abscission zones first begin to differentiate (Bleecker and Patterson 1997). However, this inflorescence expression is not dependent on pollination, as removal of anthers to prevent self-pollination without disturbing other floral organs did not interfere with HAESA::GUS expression (Fig. 1D).
Figure 1.
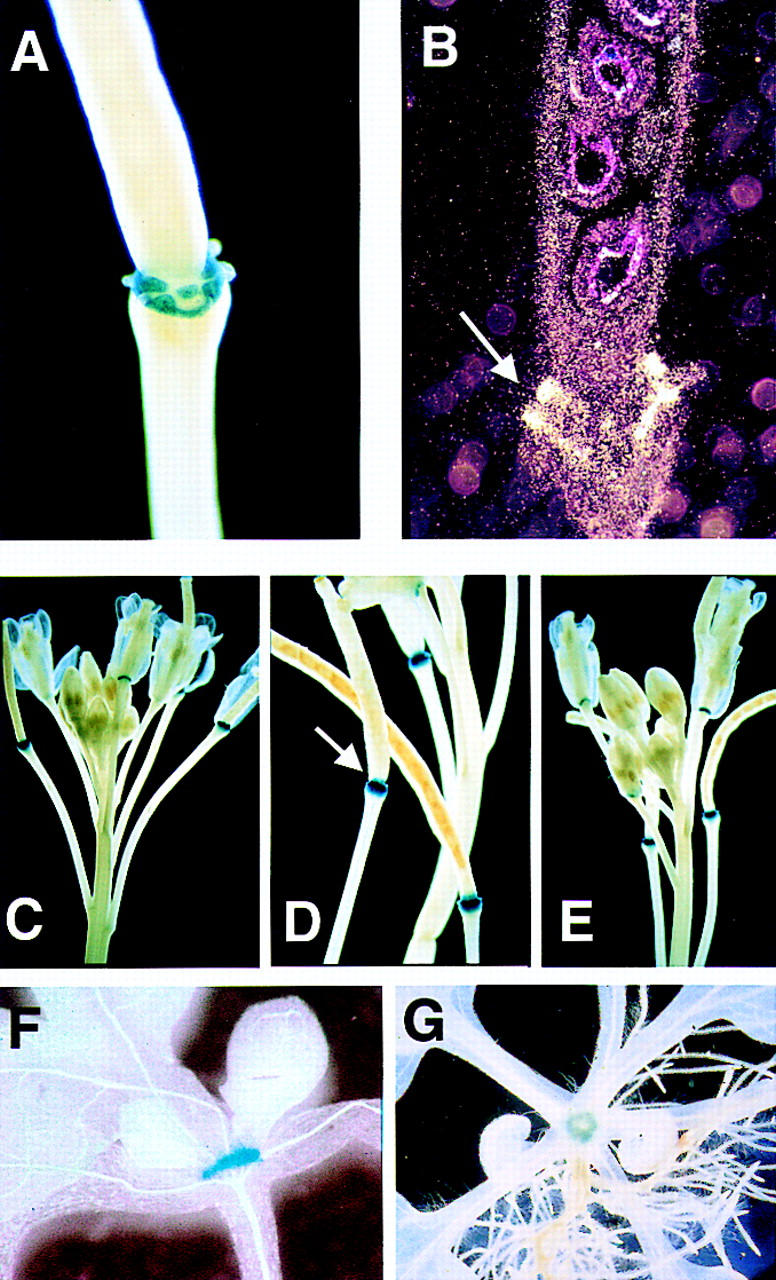
HAESA expression pattern. (A) Transgenic plants harboring the HAESA promoter fused to the GUS reporter gene were histochemically stained for GUS activity using the chromogenic substrate 5-bromo-4-chloro-3 indolyl glucuronide. GUS activity (indicated by blue color) at the base of the flowers shows that HAESA expression is restricted to the abscission zones, in which the sepals, petals, and stamens have detached. (B) In situ RNA hybridization confirms the results of the HAESA::GUS expression in reproductive tissues. Longitudinal sections of a mature silique were hybridized with a HAESA antisense probe. Silver grains deposited after development of the photographic film (indicated by the small white-colored dots) correspond to HAESA mRNA and can be seen at the base of the silique (arrow). No detectable signal was observed in similar sections hybridized with a HAESA sense probe. (C) Developmental stage-specific HAESA expression, in which the first flower competent for pollination shows GUS activity at the base of the flower. (D) Pollination-independent HAESA::GUS expression. An emasculated flower in which the anthers were removed prior to dehiscence still shows GUS activity (arrow, unpollinated silique without seeds). (E) HAESA::GUS expression in the F1 hybrid derived from etr1-1 and the HAESA::GUS transgenic, showing that HAESA expression is independent of ethylene signal transduction. HAESA::GUS expression was also observed in vegetative tissues. (F) A side view of a seedling at the four-leaf stage; (G) a view from the top of a seedling at the six- to eight-leaf stage. GUS activity is restricted to a donut-shaped region at the base of the petioles where the leaves attach to the stem.
HAESA::GUS expression pattern and the in situ RNA hybridization data show that HAESA is strongly expressed in floral organ abscission zones (Fig. 1A–D). Therefore, we proposed that HAESA may function in the abscission of floral organs. Ethylene perception is important for abscission in many plants, and mutations in ETR1, which encodes an ethylene receptor, result in significant delay of floral organ abscission (Bleecker and Patterson 1997). HAESA::GUS expression in the F1 hybrid derived from a cross between the dominant etr1-1 mutant and the HAESA::GUS transgenic (Fig. 1E) is indistinguishable from the transgenic in a wild-type background (Fig. 1C), suggesting that HAESA::GUS expression is independent of ethylene signal transduction.
HAESA::GUS expression is not restricted to floral organs, in vegetative tissues HAESA::GUS expression is first observed in seedlings that have reached the two-leaf stage and continued through maturity. Seedlings at the four-leaf stage (Fig. 1F) and in plants at the six- to eight-leaf stage (Fig. 1G) show HAESA specifically expressed in a donut shape at the region where the petioles attach to the stem. In situ hybridization experiments with an anti-HAESA-specific riboprobe showed similar HAESA expression patterns as the reporter gene data (data not shown). These data demonstrate that HAESA expression is restricted to distinct regions in both floral and vegetative tissues and is independent of ethylene signal transduction.
HAESA is a plasma membrane-localized protein
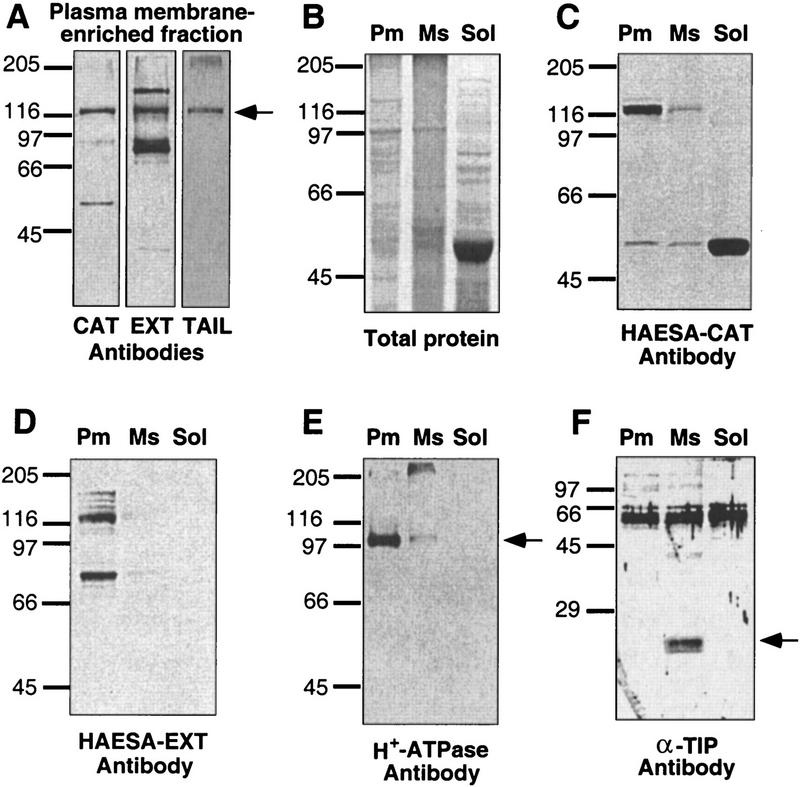
To determine the subcellular location of HAESA and generate tools for assessing the level of HAESA protein in antisense-suppressed transgenic plants, we developed antibodies that recognize the native HAESA protein. Because of the prevalence of LRR RLKs in the Arabidopsis genome, it was necessary to generate multiple antibodies against HAESA to ensure specificity. We raised polyclonal antibodies against recombinant fusion proteins containing the HAESA extracellular domain (EXT) and the HAESA catalytic domain (CAT). Sequence alignments between several different RLK protein kinase catalytic domains revealed that the carboxy-terminal 36 amino acids of RLK5 are unique (Braun et al. 1997). This region was chosen for generation of the catalytic-tail (TAIL) antibody. The use of three different antibodies directed against different portions of the recombinant HAESA protein allowed us to assign an antigenic polypeptide from plasma membrane-enriched fractions to the native HAESA protein.
The cDNA sequence predicts HAESA encodes a 999-amino-acid polypeptide with a molecular mass of 109 kD (107 kD assuming cleavage of the signal peptide). An amino-terminal signal sequence and the apparent absence of any organelle retention signals suggest that the protein is plasma membrane-localized (Walker 1993). Using a two-phase partition system for plasma membrane purification (Yoshida et al. 1983) followed by Western analyses, all three affinity-purified antibodies recognize a 120-kD protein found in the plasma membrane-enriched fraction (Fig. 2A–D). As is evident from Figure 2A, the TAIL antibody showed the highest degree of specificity. Antibodies against plasma membrane H+–ATPase (ATPase; 100 kD) (DeWitt et al. 1996) and tonoplast intrinsic protein (α-TIP; 27 kD), which is found in the microsomal fraction (Johnson et al. 1989), were used to depict the quality of the purified plasma membrane fraction (Fig. 2, E and F, respectively). The size of the native HAESA protein corresponds reasonably well with the predicted mass (120 vs. 109 kD). The discrepancy in the apparent molecular masses of the native protein and the predicted protein may be due to glycosylation. There are six putative N-linked glycosylation sites (NXS/T) in the HAESA LRR extracellular domain (Walker 1993). Alternatively, the mobility shift could be due to autophosphorylation of HAESA (Horn and Walker 1994).
Figure 2.
Subcellular immunolocalization of HAESA. Three different polyclonal antibodies were raised against portions of recombinant HAESA protein (CAT, EXT, and TAIL) and affinity purified for Western blot analyses. Protein extracts from mature rosettes were fractionated using a PEG/dextran two-phase partition system to obtain a plasma membrane-enriched (Pm), a total microsomal membrane (Ms) and a soluble protein (Sol) fraction. (A) A comparison of Western blot analyses using the three different antibodies against plasma membrane-enriched fractions (the arrow indicates the ∼120-kD protein recognized by all three HAESA antibodies); (B) total protein profiles assessed by Coommassie blue staining; and Western blot analyses of Pm, Ms, and Sol fractions using CAT (C) and EXT (D) antibodies. To illustrate the purity of the two-phase partitioned fractions, Western blot analyses with various fractions were performed with antibodies to the plasma membrane-specific H+–ATPase (E; the arrow indicates the ∼100-kD protein) and the tonoplast-specific α-TIP (F; the arrow indicates the ∼27-kD protein). Sizes of the molecular mass markers (kD) are indicated at left.
HAESA is a serine/threonine protein kinase
Recombinant HAESA has serine/threonine protein kinase activity in vitro (Horn and Walker 1994). To determine whether native HAESA also exhibits kinase activity, protein solubilized from Arabidopsis plasma membrane-enriched fractions was immunoprecipitated with preimmune or affinity-purified CAT, EXT and TAIL antibodies. The precipitated immunocomplexes were assayed in the presence of [γ-32P] ATP and analyzed by SDS-PAGE. These three different HAESA antibodies could immunoprecipitate an active protein kinase with an apparent molecular mass of 120 kD (Fig. 3A–C). To assess the amino acid specificity of HAESA, the 120-kD autophosphorylated protein was isolated, acid-hydrolyzed, and separated by two-dimensional thin layer electrophoresis. Incompletely digested peptides are evident near the origin (+), and radioactively labeled phosphoserine and phosphothreonine, but not phosphotyrosine, were observed (Fig. 3D). These results demonstrate that native HAESA protein is an active protein kinase that autophosphorylates on serine and threonine residues, consistent with results obtained with recombinant fusion protein containing the catalytic domain of HAESA (Horn and Walker 1994).
Figure 3.
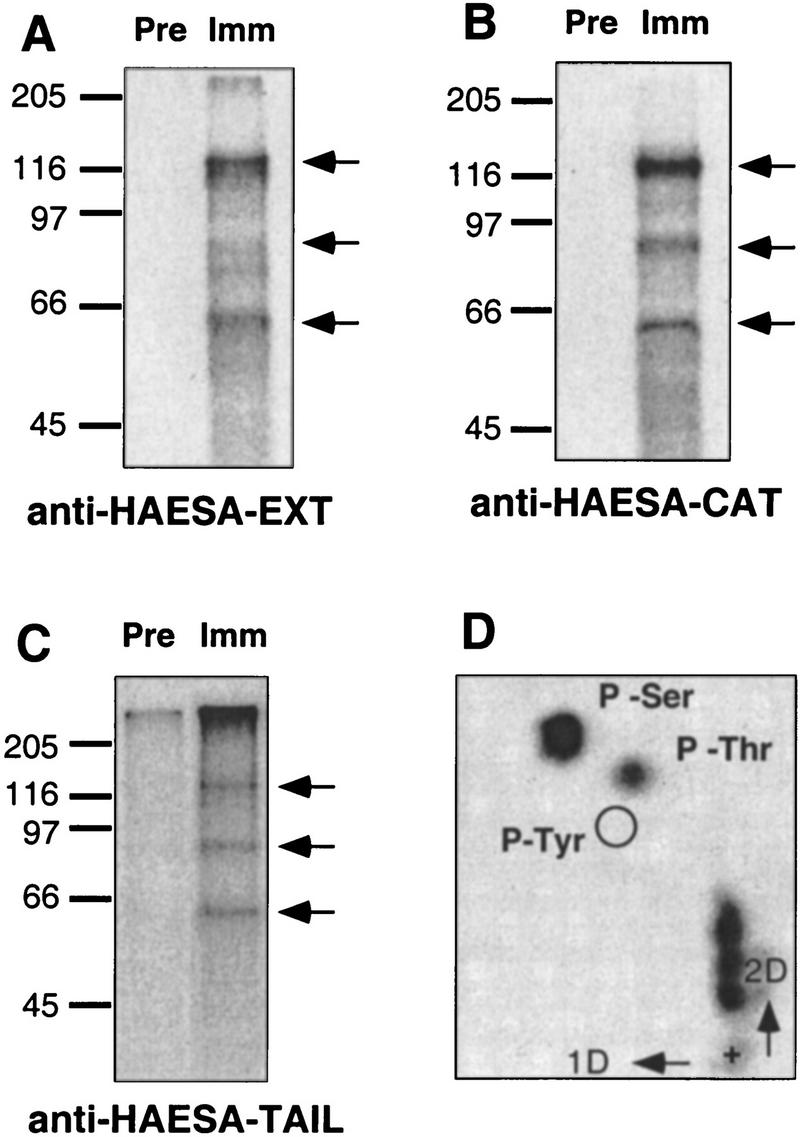
Immunoprecipitation and autophosphorylation of HAESA. Solubilized proteins from plasma membrane-enriched fractions obtained by two-phase partitioning were immunoprecipitated with affinity-purified antibodies from preimmune (Pre) or immune (Imm) sera raised against three different portions of the HAESA protein (A, EXT; B, CAT; and C, TAIL). Immunocomplexes were autophosphorylated in the presence of [γ-32P]ATP, resolved by SDS-PAGE, and exposed to autoradiographic film. Sizes of the molecular mass markers are indicated at left; the HAESA 120-kD protein and ∼85- and ∼65-kD unknown polypeptides are indicated with arrows. Phosphoamino acid content was determined by isolation of the 120-kD phosphoprotein, acid-hydrolysis, and separation by two-dimensional thin-layer electrophoresis (D). The sample origin is indicated by +, and positions of phosphoamino acid standards visualized by staining with ninhydrin are circled. Incompletely hydrolyzed peptides are evident above the origin, and radioactive phosphoserine (P-Ser) and phosphothreonine (P-Thr) were detected, but phosphotyrosine (P-Tyr) was not.
Phosphoproteins with apparent molecular masses of 85 and 65 kD coimmunoprecipitated with HAESA (Fig. 3A–C). Phosphoamino acid analysis revealed phosphorylation on both serine and threonine residues (data not shown). These polypeptides may correspond to HAESA proteolytic products produced during immunoprecipitation or endogenous substrates that interact with the HAESA receptor kinase.
HAESA reduction of function using an antisense construct
To ascertain a physiological role for HAESA, we generated HAESA reduction-of-function transgenic plants by antisense suppression. Multiple transgenic lines were selected by use of a kanamycin resistance gene, and seed from 11 independent primary transformants that contained the 35S CaMV::HAESA antisense transgene inserted at a single locus were characterized further. Transgenic seedlings were transplanted into soil, and a membrane fraction was prepared from plants at the six- to eight-leaf stage. HAESA protein expression was determined by Western blot analysis using the TAIL antibody (Fig. 4A) and quantitated by densitometry. Three lines (C, E, and K) have <10% of HAESA wild-type levels, whereas line H is only reduced to 88% of wild type and other lines exhibited intermediate levels (Fig. 4A).
Figure 4.
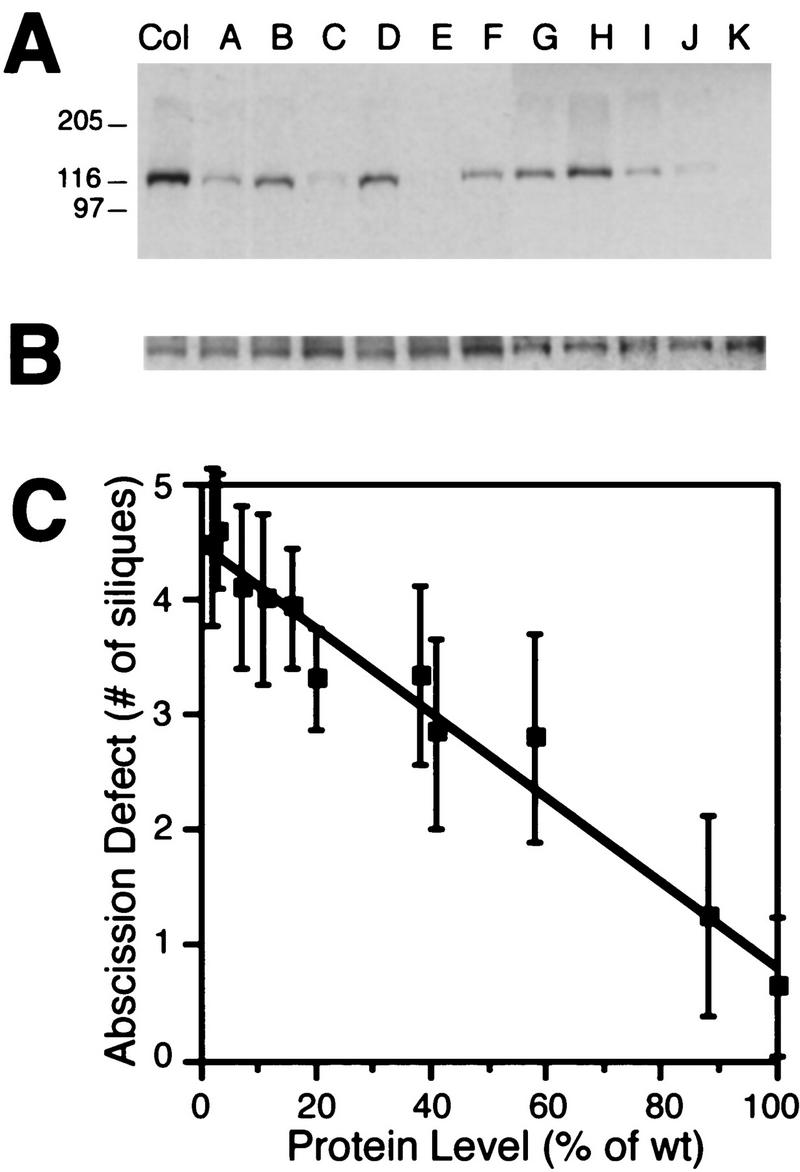
Antisense suppression of HAESA and floral organ abscission phenotype. (A) Transgenic lines containing single copies of the strong, constitutive 35S cauliflower mosaic virus (CaMV) promoter driving an antisense version of HAESA cDNA were generated, and the levels of HAESA protein determined by Western blot analysis using the TAIL antibody. The parental wild-type (Col) is compared with 11 independent lines (indicated above each lane). Sizes of molecular mass markers (kD) are indicated at left. (B) An antibody raised against the plasma membrane-specific H+–ATPase (DeWitt et al. 1996) was used as a protein loading control. Densitometric quantitation indicates that the transgenic lines have suppressed levels of HAESA protein ranging from <10% of wild type (lines C, E, and K) to 88% of wild type (line H). (C) Floral organ abscission in the five most mature siliques were scored. Those that had failed to detach their floral organs were scored as defective. The number of siliques that failed to abscise floral organs (±s.d.) was plotted against the level of HAESA protein in wild type (Col), and the various antisense-suppressed transgenic lines determined by densitometry. A direct, inverse correlation between the amount of HAESA protein and the number of siliques in which floral organs failed to abscise was observed (r2 = 0.964). Col n = 150 flowers; antisense transgenic lines n = 45 flowers each.
Observation of HAESA expression in floral organ abscission zones (Fig. 1A–E) prompted us to investigate whether HAESA reduction-of-function transgenic plants displayed floral organ abscission defects. Eight-week-old HAESA antisense transgenic lines, wild-type plants, and the well-characterized ethylene-insensitive mutant etr1-1 were scored for floral organ abscission in the first five most mature siliques. The transgenic lines with the least amount of HAESA protein (C,E, and K) exhibited a strong phenotype similar to the etr1-1 mutant, in which the first five mature siliques failed to abscise their floral organs. In these strong reduction-of-function lines, floral organs never abscise; abscission is not simply delayed. Line H, which has near wild-type levels of HAESA protein (88% of wild type), is able to abscise the floral organs during maturation, similar to wild type. The lines expressing intermediate levels of HAESA had differing degrees of the floral organ abscission defect. By plotting the amount of HAESA protein (determined by densitometry) in the various transgenic lines against the number of siliques in which floral organs failed to detach, a direct, inverse correlation (r2 = 0.964) between HAESA protein level and the floral organ abscission defect was found (Fig. 4C).
To confirm that the observed abscission defect phenotype is due to the HAESA antisense transgene, segregating progeny from primary transformants E and K were investigated for both the presence of the transgene and the floral organ abscission defect. The presence of the transgene in segregating siblings was discerned by PCR using specific primers that hybridize within the 35S CaMV promoter and the HAESA gene and was further confirmed by scoring the kanamycin sensitivity of their progeny (data not shown). Inflorescence total membrane fractions from wild type (Columbia), siblings from lines E and K that retained the HAESA antisense construct, and siblings that had lost the transgene (E* and K*) were used for Western blot analysis and densitometry (Fig. 5A). As predicted, those plants that no longer contained the HAESA antisense transgene recover normal levels of the HAESA gene product. Equal protein loading is depicted by probing the Western blot with an antibody to the plasma membrane H+–ATPase (Fig. 5B). Those plants with wild-type levels of HAESA protein possess typical wild-type phenotypes with normal floral organ abscission during maturation (Fig. 5C). A close-up view of the base of siliques from wild-type (Col), transgenic line E, and a sibling, E*, without the transgene are shown in Figure 5D.
Figure 5.
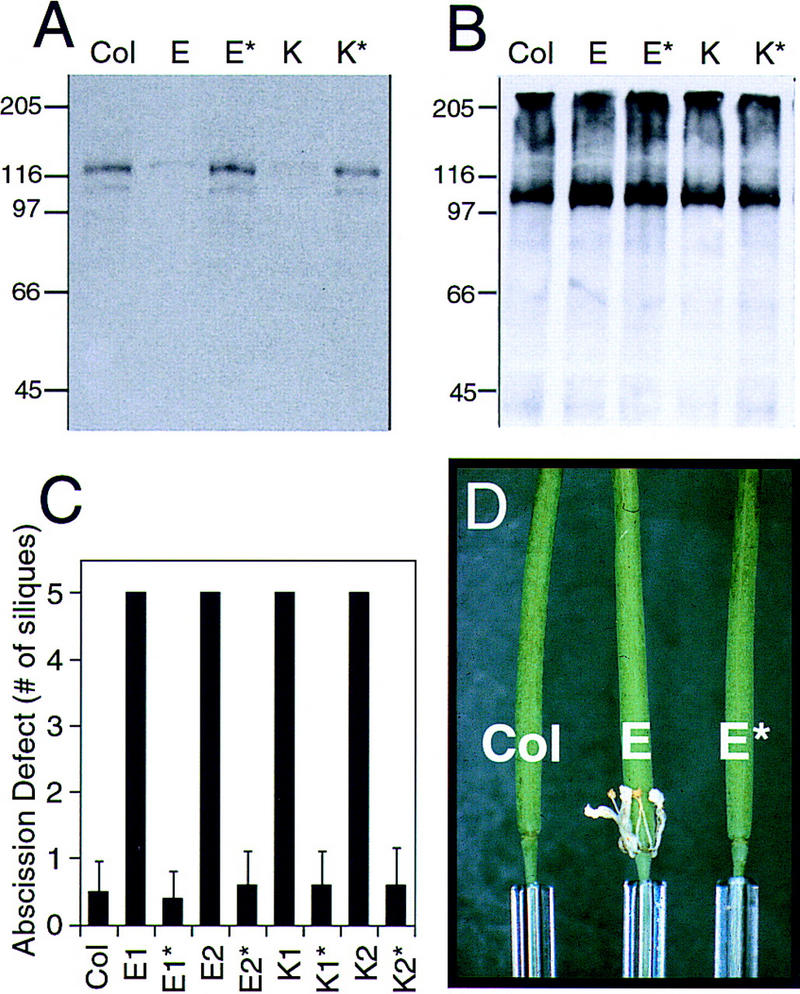
Loss of the transgene leads to loss of the floral organ abscission phenotype. To confirm that the floral organ abscission defect observed in antisense-suppressed transgenic lines is due to the presence of the transgene, HAESA protein levels in wild-type (Col) and siblings that retain the transgene (E and K) or have lost the transgene (E* and K*) were determined by Western blot analysis using the TAIL antibody (A). (B) A duplicate Western blot probed with the plasma membrane H+–ATPase antibody ensures equal loading. Sizes of molecular mass markers (kD) are indicated at left. (C) Floral organ abscission (no. of siliques ±s.d.) of these segregating plants. Those plants that no longer carry the transgene (indicated by * ) have normal floral organ abscission. Col n = 100 flowers; lines derived from primary transformants E and K n = 60 flowers each. (D) A representative example of siliques from wild-type Col (left), antisense trangenic line E (middle; for clarity, the front sepal and petal are removed), and a sibling that had lost the transgene (E*; right).
Discussion
All multicellular organisms must respond to intrinsic developmental and externally generated cues to survive and successfully reproduce. In plants, the RLKs have been proposed to serve the function of detecting and transducing some of these signals. Although many genes encoding RLKs have been identified, their functions have been somewhat elusive. Of those with known functions, most have been deciphered from the phenotypes associated with RLK mutants (Becraft 1998; Lease et al. 1998). In the absence of available rlk5 mutants, we used a reverse genetic approach to determine RLK5's function in Arabidopsis. Multiple independent antisense-suppressed trangenic lines with varying levels of protein were generated that displayed a phenotype characterized by an inability to precisely abscise floral organs (sepals, petals, and stamens). Furthermore, there is a direct, inverse correlation between RLK5 protein levels in these transgenic lines and severity of the floral organ abscission defect. To reflect this phenotype we have renamed RLK5 and now refer to it as HAESA.
Our first clues to HAESA's function were provided by the expression patterns observed in transgenic plants harboring HAESA::GUS and in situ RNA hybridization using a HAESA antisense probe. HAESA is expressed in the tissues in which we detected the floral organ abscission defect (i.e., abscission zones of sepals, petals, and stamens) in the HAESA antisense-suppressed transgenic lines. Weak HAESA expression was also observed at the base of the pedicels and petioles in which we failed to detect a phenotype. No phenotypes were observed other than the floral organ abscission defect; therefore, it is likely that the antisense suppression is specific to HAESA. Many LRR RLKs are present in the Arabidopsis genome, but the most closely related genes share only 60%–70% identity within the protein kinase catalytic domain and less in the extracellular domain. One of the most closely related genes is CLAVATA1, and if our antisense lines were down-regulating other RLKs, we would have expected a clv1 phenotype.
From its cDNA sequence, HAESA is predicted to encode a plasma membrane-associated protein with intrinsic serine/threonine-specific protein kinase activity (Walker 1993; Horn and Walker 1994). Subcellular fractionation, Western blot analyses, and immunoprecipitation and protein kinase assays, confirmed that HAESA is localized in the plasma membrane-enriched fraction and has serine/threonine protein kinase activity. On the basis of the predicted structure of the HAESA protein, its subcellular location, and biochemical activity, we conclude that HAESA functions as a transmembrane receptor. If this conclusion is valid, there must exist a ligand(s) and downstream effectors that participate in a HAESA signal transduction pathway controlling floral organ abscission. The extracellular LRR region presumably binds a ligand, and this information is transmitted through the transmembrane domain to affect HAESA's cytoplasmic protein kinase activity. No ligands for the plant RLKs have yet been identified conclusively, but recent reports suggest that a small secreted peptide encoded by CLAVATA3 might be the ligand for the CLAVATA1 LRR RLK (Fletcher et al. 1999). This is supported by both genetic (Clark et al. 1995) and biochemical analyses (Trotochaud et al. 1999). Because of the similarities in the extracellular domains of HAESA and CLAVATA1 and the fact that LRRs commonly mediate protein–protein interactions (Kobe and Deisenhofer 1994), it is likely that the ligand(s) that activates HAESA is also a polypeptide.
Potential downstream effectors of plant RLKs include those that have been shown to interact directly with RLKs, such as thioredoxins (Bower et al. 1996) a protein with a region of homology to β-catenin (Gu et al. 1998), kinase-associated protein phosphatase (KAPP) (Stone et al. 1994, 1998; Braun et al. 1997; Williams et al. 1997), and a Rho GTPase-related protein (Trotochaud et al. 1999). Of these, only KAPP has been demonstrated to mediate RLK signaling. Overexpression of KAPP results in a weak clv1 mutant phenotype (Williams et al. 1997), characterized by enlarged stems and excess floral organs (Clark et al. 1993), whereas sense suppression of KAPP in a clv1 mutant background restores a wild-type phenotype (Stone et al. 1998). These results indicate that KAPP functionally participates as a negative regulator of the CLAVATA1 signal transduction pathway, perhaps by dephosphorylating the activated receptor. KAPP has been found in a CLAVATA1-containing high molecular mass complex, which is presumably the ligand-activated receptor complex (Trotochaud et al. 1999). However, KAPP is also found in high molecular mass complexes lacking CLAVATA1 (Trotochaud et al. 1999), and additional phenotypes were observed in KAPP sense-suppressed lines (Stone et al. 1998), supporting the notion that KAPP may act in multiple signaling pathways. Whether one of the polypeptides that coimmunoprecipitated with HAESA represents KAPP, or other effectors, remains to be determined.
Studies describing the molecular events contributing to abscission point to other possible downstream components of HAESA signal transduction. Accumulation of enzymes involved in degradation of the middle lamella between cell layers in the abscission zone has been widely studied (Gonzalez-Bosch et al. 1997). However, the role of these different enzymes in the abscission process has been difficult to ascertain. The separation zone is only a few layers thick, making biochemical analyses difficult, and molecular approaches are confounded by the fact that most of these enzymes exist in multigene families with only one or a few members contributing to abscission (Lashbrook et al. 1998). Although the current knowledge of genes expressed in Arabidopsis floral organ abscission is limited, it will be interesting to test whether expression of these hydrolytic enzymes is defective in HAESA antisense-suppressed lines.
The interplay between hormonally dependent and independent processes in the control of abscission are also revealed in these studies. Both ethylene and auxin perception are known to be important for the timing of abscission, as the abscission process can be accelerated by exogenous application of ethylene, whereas auxin seems to act antagonistically to delay the response (Brown 1997; Gonzalez-Carranza et al. 1998). Moreover, mutations in the tomato NR and Arabidopsis ETR1 ethylene receptors confer dominant ethylene insensitivity and delay flower abscission (Lanahan et al. 1994; Wilkinson et al. 1995) and floral organ abscission (Bleecker and Patterson 1997), respectively. Many genes expressed primarily in abscission zones are transcriptionally up-regulated by ethylene (Tucker et al. 1988; Ferrarese et al. 1995; Sessa et al. 1996), but examples exist of genes that are ethylene-independent (Coupe et al. 1997). HAESA falls into the second category, and its expression appears to be independent of ethylene signal transduction for several reasons: (1) Expression of HAESA is unaltered in response to treatment with the ethylene precursor, 1-aminocyclopropane-1-carboxylic acid (ACC; data not shown); (2) no established consensus ethylene-responsive elements, such as an ERE or G box (Salinas et al. 1992; Eyal et al. 1993), are found in the HAESA genomic sequence; and (3) when HAESA::GUS plants were crossed with the dominant etr1-1 mutant, expression in the F1 was temporally and spatially indistinguishable from the HAESA::GUS parent. HAESA and ETR1 may act in parallel to control expression of genes that contribute to abscission, with ETR1-dependent signaling directing the subset of ethylene-regulated genes and HAESA-dependent signaling governing expression of ethylene-independent genes.
HAESA clearly participates in floral organ abscission, but HAESA expression was also observed at the base of pedicels and petioles. In many plant species, discrete abscission zones are found at these regions (Sexton and Roberts 1982; Gonzalez-Carranza et al. 1998). Because analogous mechanisms may operate in the formation of abscission zones in different tissues, a cognate LRR RLK may control leaf and flower abscission in these plant species. However, neither Arabidopsis flowers nor leaves abscise naturally. One possible explanation for this is the full complement of components necessary for formation of an effective abscission zone are absent in these tissues. HAESA may simply be unable to direct abscission at the base of pedicels and petioles in Arabidopsis if, for example, the HAESA ligand or downstream effectors are only expressed in the floral organ abscission zones. Perhaps HAESA expression is an evolutionary relic from ancestral plants that undergo leaf and flower abscission. Alternatively, and not mutually exclusively, particular environmental conditions might be required to observe a phenotype in nonfloral tissues of HAESA antisense-suppressed transgenic lines. For example, pathogen attack is known to accelerate leaf abscission in some plants (Hashim et al. 1997), but there are no reports of this occurring in Arabidopsis. Under the appropriate assay conditions, a phenotype in these tissues might have been observed.
The identification of HAESA as a protein regulating floral organ abscission may have practical applications. Premature abscission in agricultural crops, including fruit, results in significant yield losses. Formation of dehiscence zones in pod-bearing plants share many of the features of abscission zones, and early dehiscence (or pod shatter) in crops such as Brassica napus (oilseed rape/canola) and Glycine max (soybean) leads to a major loss of revenue and contributes to soil contamination of future crops (Spence et al. 1996). The information gained from the study of HAESA may also contribute to the floriculture industry, in which a concerted effort to delay senescence and abscission of petals has gained a great deal of attention (van Doorn and Stead 1997).
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia was the wild type used for transformation, and the etr1-1 mutant was obtained from the Arabidopsis Biological Resource Center (CS237, Ohio State University, Columbus). Seeds were imbibed 4–6 days at 4°C before being sown in soil (Pro-mix, Premier, Canada). Plants were grown under constant light at 25°C. Alternatively, seeds were surface-sterilized and germinated on agar-containing MS media plates (Murashige and Skoog 1962) supplemented with kanamycin (50 μg/ml) to select for transformants.
Expression vector construction and fusion protein purification
HAESA (GenBank accession no. M84660) extracellular domain (amino acids 27–621), catalytic domain (amino acids 649–999), and catalytic-tail domain (amino acids 965–999) were amplified by PCR with engineered restriction site primers to facilitate cloning. The PCR products were cloned into pGEX-2T (Pharmacia, Piscataway, NJ) or pMalcRI (New England Biolabs, Beverly, MA) expression vectors. Recombinant plasmids were confirmed by restriction endonuclease mapping and DNA sequencing. Glutathione S-transferase (GST) fusion protein and maltose binding protein (MBP) fusion protein were expressed in Eschericha coli SG1611 or DH5α strains and purified with glutathione–agarose (Sigma, St. Louis, MO) and amylose–agarose (Pharmacia, Piscataway, NJ) affinity columns, respectively (Horn and Walker 1994).
Affinity purification of antibodies
The affinity-purified MBP::HAESA EXT, CAT, and TAIL domain fusion proteins were used to immunize rabbits. The consequent three different antisera were affinity purified with glutathione–agarose columns cross-linked with GST::HAESA–EXT, HAESA–CAT, and HAESA–TAIL fusion proteins (Koff et al. 1992). Antibodies were eluted with 100 mm glycine (pH 2.5), neutralized immediately with 0.1 volume of 1 m Tris-HCl (pH 8.0), dialyzed against 10 mm Na-phosphate (pH 7.5), 135 mm NaCl, and concentrated with Centricon-10 concentrators (Amicon, Beverly, MA).
Plasma membrane protein isolation
All manipulations were conducted in a 4°C cold room or on ice. Plant tissues were homogenized with 10 mm Tris-HCl buffer (pH 7.5), containing 250 mm sucrose, 5 mm EDTA, 1 mm PMSF, protease inhibitor cocktail (10 μg/ml leupeptin, 5 μg/ml chymostatin, and 10 μg/ml aprotinin), and centrifuged at 10,000g for 15 min. The resultant supernatant was centrifuged at 100,000g for 30 min to yield the soluble protein fraction (supernatant) and the total membrane fraction (pellet), which were resuspended in 5 mm potassium phosphate buffer (pH 7.8), with 250 mm sucrose, 4 mm KCl, 1 mm PMSF, and protease inhibitor cocktail. The membrane fraction was then partitioned into a microsomal fraction (L1) and a plasma membrane-enriched fraction (U2) by two-phase partioning (Yoshida et al. 1983; Mito et al. 1988). Protein complexes from the pellets were solubilized with 50 mm bis-HCl buffer (pH 7.0) containing 750 mm 6-aminocaproic acid, 0.25 volume of 10% laurylmaltoside, 1 mm PMSF, and protease inhibitor cocktail, centrifuged at 100,000g for 30 min and the supernatant stored at −80°C until use (Schaegger and von Jagow 1991).
Immunoprecipitation, kinase activity, and phosphoamino acid assay
Proteins from the plasma membrane-enriched fraction were immunoprecipitated with preimmune or purified antibodies as described previously (Borgese and Gaetani 1980). The immunoprecipitated complexes were recovered using protein A–agarose beads (Pierce) and immobilized for protein kinase activity assays. The reaction contained 50 mm HEPES buffer (pH 7.4), 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, and 25 μCi [γ-32P] ATP at room temperature for 1 hr (Horn and Walker 1994). The reaction was stopped by adding Laemmli SDS–polyacrylamide sample buffer, separated by 7.5% SDS-PAGE (Laemmli 1970), Coomassie blue-stained, dried, and exposed to X-ray film (Kodak). The phosphoamino acids (PAA) content of immunoprecipitated complexes was determined essentially as reported (Boyle et al. 1991). Samples were electrophoresed at 1.5 kV for 30 min in pH 1.9 buffer in the first dimension followed by electrophoresis in pH 3.5 buffer (5% acetic acid, 0.5% pyridine) at 1.3 kV for 25 min in the second dimension using a Hunter Thin Layer Electrophoresis System (HTLE 7000; CBS Scientific Company, Del Mar, CA). PAA standards were visualized by spraying with ninhydrin (0.25% in acetone). Plates were analyzed with a Fuji Bas-IIIS phosphoimage analyzer (Fuji PhotoFilm Co., Japan) to detect the radioactive PAA.
Western blot analyses
Samples were resolved by SDS-PAGE (Laemmli 1970) and electroblotted to nitrocellulose membranes (Gelman Sciences, Ann Arbor, MI) as described (Towbin et al. 1979). Membranes were blocked for 3 hr with PBS containing 0.2% Tween 20 and 5% nonfat milk. Protein bands that cross-reacted with HAESA antibodies were identified by reaction with horseradish peroxidase conjugated to goat anti-rabbit IgG, and bound antibodies were visualized with ECL chemiluminescent reagents (Amersham, Arlington Heights, IL) according to the manufacturer's instructions.
In situ hybridization
A 286-bp PCR product from HAESA spanning the last 36 codons of the ORF and 178 bp of 3′ UTR with EcoRI and XhoI engineered restriction sites was cloned into Bluescript SK+ (Stratagene, La Jolla, CA). Antisense RNA probes were synthesized with T7 RNA polymerase after linearization with EcoRI, and sense RNA controls were synthesized using T3 RNA polymerase after linearization with XhoI. Tissues were fixed, embedded, sectioned, hybridized, and exposed as described (Cox and Goldberg 1988).
Construction of reporter gene fusion, plant transformation, and histochemical analyses
The HAESA promoter (1.6 kb) was amplified by PCR, cloned into a GUS expression vector, pGN100 (J.C. Walker, unpubl.), and subcloned into binary vector pGA482 (An et al. 1988). The chimeric construct was transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell 1986). Transformation of Arabidopsis was via vacuum infiltration (Bechtold and Bouchez 1994). Transgenic plants were selected on agar plates by virtue of the kanamycin resistance marker. Transgene copy number was assessed by kanamycin resistance segregation. Histochemical staining for GUS activity was performed with chromogenic substrate 5-bromo-4-chloro-3 indolyl glucuronide (X-gluc) as detailed (Craig 1992). To prepare tissue for histological analysis, the stained tissue was dehydrated with ethanol, treated with xylene, and embedded in Paraplast (Oxford, St. Louis MO), and sections (10 μm) were visualized using dark-field illumination (Sieburth and Meyerowitz 1997).
Antisense suppression of HAESA
The full-length HAESA cDNA was cloned into a 35S CaMV expression vector (J.C. Walker, unpubl.), subcloned into the binary vector pGA482 (An et al. 1988), and used for plant transformation described above.
Acknowledgments
We thank Drs. Steve Alexander, Cathy Gunther, and Pete Yorgey for critical comments on the manuscript. Drs. Michael Sussman and John Rogers provided the antibodies to H+–ATPase and α-TIP, respectively. This research was supported by a National Science Foundation grant (MCB9809884) to J.C.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL WalkerJ@missouri.edu; FAX (573) 882-0123.
References
- Altmann T. Recent advances in brassinosteroid molecular genetics. Curr Opin Plant Biol. 1998;1:378–383. doi: 10.1016/s1369-5266(98)80259-8. [DOI] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB. Binary vectors. Plant Mol Biol Manual. 1988;A3:1–19. [Google Scholar]
- Bechtold N, Bouchez D. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In: Potrykus I, Spangenberg G, editors. Gene transfer to plants. New York, NY: Springer Verlag; 1994. p. 361. [DOI] [PubMed] [Google Scholar]
- Becraft PW. Receptor kinases in plant development. Trends Plant Sci. 1998;3:384–388. [Google Scholar]
- Bleecker AB, Patterson SE. Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Gaetani S. Site of synthesis of rat liver NADH–cytochrome b5 reductase, an integral membrane protein. FEBS Lett. 1980;112:216–220. doi: 10.1016/0014-5793(80)80183-9. [DOI] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR. Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell. 1996;8:1641–1650. doi: 10.1105/tpc.8.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Braun DM, Stone JM, Walker JC. Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: implications for transmembrane signaling in plants. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- Brown KM. Ethylene and abscission. Physiol Plantarum. 1997;100:567–576. [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–2067. [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Roberts JA. Temporal and spatial expression of mRNAs encoding pathogenesis-related proteins during ethylene-promoted leaflet abscission in Sambucus nigra. Plant Cell Environ. 1997;20:1517–1524. [Google Scholar]
- Cox HK, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant molecular biology: A practical approach. Oxford, UK: IRL Press, Oxford; 1988. pp. 1–34. [Google Scholar]
- Craig S. The GUS reporter gene. Application to light and transmission electron microscopy. In: Gallagher SR, editor. GUS protocols. Using the GUS gene as a reporter of gene expression. New York, NY: Academic Press; 1992. pp. 115–124. [Google Scholar]
- DeWitt ND, Hong B, Sussman MR, Harper JF. Targeting of two Arabidopsis H+–ATPase isoforms to the plasma membrane. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Lev-Yaden S, Fluhr R. A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J. 1993;4:225–234. doi: 10.1046/j.1365-313x.1993.04020225.x. [DOI] [PubMed] [Google Scholar]
- Ferrarese L, Trainotti L, Moretto P, Polverino de Laureto P, Rascio N, Casadoro G. Differential ethylene-inducible expression of cellulase in pepper plants. Plant Mol Biol. 1995;29:735–747. doi: 10.1007/BF00041164. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Bosch C, del Campillo E, Bennett AB. Immunodetection and characterization of tomato endo-β-1,4-glucanase Cel1 protein in flower abscission zones. Plant Physiol. 1997;114:1541–1546. doi: 10.1104/pp.114.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Carranza ZH, Lozoya-Gloria E, Roberts JA. Recent developments in abscission—Shedding light on the shedding process. Trends Plant Sci. 1998;3:10–14. [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim M, Roberts JA, Rossall S, Dickinson MJ. Leaflet abscission and phytoalexin production during the response of two faba bean breeding lines to Botrytis infection. Plant Pathol. 1997;46:989–996. [Google Scholar]
- Horn MA, Walker JC. Biochemical properties of the autophosphorylation of RLK5, a receptor-like protein kinase from Arabidopsis thaliana. Biochim Biophys Acta. 1994;1208:65–74. doi: 10.1016/0167-4838(94)90160-0. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Herman EM, Chrispeels MJ. An abundant, highly conserved tonoplast protein in seeds. Plant Physiol. 1989;91:1006–1013. doi: 10.1104/pp.91.3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: A versatile binding motif. Trends Biochem Sci. 1994;19:415–420. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamahita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol & Gen Genet. 1986;204:383–396. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. The never ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Giovannoni JJ, Hall BD, Fischer RL, Bennett AB. Transgenic analysis of tomato endo-beta-1,4-glucanase gene function—Role of CEL1 in floral abscission. Plant J. 1998;13:303–310. [Google Scholar]
- Lease K, Ingham E, Walker JC. Challenges in understanding RLK function. Curr Opin Plant Biol. 1998;1:388–392. doi: 10.1016/s1369-5266(98)80261-6. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Mito N, Kimura T, Asahi T. Partial purification and characterization of an ATPase in mung bean hypocotyl plasma membrane: suggestion for a new type of higher plant plasma membrane ATPase. Plant Cell Physiol. 1988;29:875–882. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Salinas J, Oeda K, Chua N-H. Two G-box-related sequences confer different expression patterns in transgenic tobacco. Plant Cell. 1992;4:1485–1493. doi: 10.1105/tpc.4.12.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaegger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Sessa G, Raz V, Savaldi S, Fluhr R. PK12, a plant dual-specificity protein kinase of the LAMMER family, is regulated by the hormone ethylene. Plant Cell. 1996;8:2223–2234. doi: 10.1105/tpc.8.12.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. Cell biology of abscission. Annu Rev Plant Physiol. 1982;33:133–162. [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Vercher Y, Gates P, Harris N. “Pod shatter” in Arabidopsis thaliana, Brassica napus, and B. juncea. J Microsc. 1996;181:195–203. [Google Scholar]
- Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE. Control of meristem development by CLAVATA1 receptor kinase and KAPP protein phosphatase interactions. Plant Physiol. 1998;117:1217–1225. doi: 10.1104/pp.117.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci. 1979;76:4350–4356. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a rho-related protein. Plant Cell. 1999;11:393–405. doi: 10.1105/tpc.11.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ML, Sexton R, del Campillo E, Lewis LN. Bean abscission cellulase. Characterization of a cDNA clone and regulation of gene expression by ethylene and auxin. Plant Physiol. 1988;88:1257–1262. doi: 10.1104/pp.88.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Stead AD. Abscission of flowers and floral parts. J Exp Bot. 1997;48:821–837. [Google Scholar]
- Walker JC. Receptor-like protein kinase genes of Arabidopsis thaliana. Plant J. 1993;3:451–456. doi: 10.1111/j.1365-313x.1993.tb00164.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Uemura M, Niki T, Sakai A, Gusta LV. Partition of membrane particles in aqueous two-polymer phase system and its practical use for purification of plasma membranes from plants. Plant Physiol. 1983;72:105–114. doi: 10.1104/pp.72.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]