Summary
Aging research has developed rapidly over the past decade, identifying individual genes and molecular mechanisms of the aging process through the use of model organisms and high throughput technologies. Calorie Restriction (CR) is the most widely researched environmental manipulation that extends lifespan. Activation of the NAD+-dependent protein deacetylase Sir2 (Silent Information Regulator 2) has been proposed to mediate the beneficial effects of CR in the budding yeast S. cerevisiae, as well as other organisms. Here we show that in contrast to previous reports, Sir2 is not stimulated by CR to strengthen silencing of multiple reporter genes in the rDNA of S. cerevisiae. CR does modestly reduce the frequency of rDNA recombination, although in a SIR2-independent manner. CR-mediated repression of rDNA recombination also does not correlate with the silencing of Pol II-transcribed non-coding RNAs derived from the rDNA intergenic spacer, suggesting that additional silencing-independent pathways function in lifespan regulation.
Keywords: Sir2, aging, calorie restriction, silencing, recombination, rDNA, yeast
Introduction
Calorie restriction (CR) extends both average and maximal life span in most organisms tested, suggesting that the fundamental processes of aging are being broadly delayed as opposed to adjusting only specific causes of early death. Although this effect of age delay has been known for over 70 years (McCay & Crowell 1934; McCay et al. 1935), the molecular basis for CR’s effects on aging has only recently been studied and remains largely unclear. Much of the research in understanding the mechanisms of CR has been performed in model organisms, including the budding yeast S. cerevisiae. Reduction of calories by decreasing the level of glucose, the primary carbon source in yeast growth media, results in an increase in both replicative life span (RLS) and chronological life span (CLS) (Jiang et al. 2000; Lin et al. 2000; Kaeberlein et al. 2002; Smith et al. 2007). Previous research has shown that deletion of the SIR2 gene shortens RLS, while SIR2 over-expression extends RLS (Kaeberlein et al. 1999). Sir2 is an NAD+-dependent histone deacetylase required for heterochromatic gene silencing in yeast (Imai et al. 2000; Landry et al. 2000b), and is the founding member of a large family of proteins called the Sirtuins (Brachmann et al. 1995; Frye 1999). Deleting SIR2 causes genomic instability in the ribosomal DNA (rDNA) array of yeast where homologous recombination is normally suppressed by the enzymatic activity of Sir2 (Gottlieb & Esposito 1989). In the absence of Sir2, increased recombination leads to high levels of extra-chromosomal rDNA circles (ERCs) (Kaeberlein et al. 1999), which are causative in the senescence of old mother cells (Sinclair & Guarente 1997).
Early experiments in yeast showed that SIR2 was required for CR-induced RLS extension (Lin et al. 2000), suggesting that CR stimulates Sir2 activity. Supporting this model, transcriptional silencing assays with the repressible MET15 reporter gene integrated within the rDNA locus indicated that CR strengthened the silencing in a SIR2-dependent manner (Lin et al. 2002). This type of silencing in the rDNA (rDNA silencing) is highly responsive to increased SIR2 gene dosage (Fritze et al. 1997; Smith et al. 1998). One model for the stimulation of Sir2 activity by CR proposes that the cellular NAD+/NADH ratio is altered by a CR-induced shift from fermentation toward respiration, causing a relative increased abundance of NAD+ (Lin et al. 2002; Lin et al. 2004). CR does increase the NAD+/NADH ratio in yeast cells, but not through an increase in NAD+. Rather, there is a decrease in the NADH concentration (Lin et al. 2004). Sir2 was reported to be inhibited by NADH (Lin et al. 2004), so the stimulation in Sir2 activity could actually be a relief of inhibition. A different model suggests that CR induces expression of the nicotinamidase protein Pnc1 (Anderson et al. 2003), which converts nicotinamide (NAM) to nicotinic acid as part of the NAD+ salvage pathway (Ghislain et al. 2002; Anderson et al. 2003; Gallo et al. 2004). NAM is a strong non-competitive inhibitor of Sirtuins that is also a byproduct of the NAD+-dependent deacetylase reaction (Landry et al. 2000a; Tanny & Moazed 2001; Bitterman et al. 2002). Therefore, increased Pnc1 would prevent the accumulation of excess NAM, thus promoting Sir2 activity (Anderson et al. 2003; Gallo et al. 2004). While different, these two models are not necessarily mutually exclusive.
SIR2 homologues have also been suggested to function in regulating lifespan and mediating the CR response in C. elegans and D. melanogaster (Tissenbaum & Guarente 2001; Rogina & Helfand 2004). While the role of specific Sirtuins in modulating lifespan appears to be conserved from yeast to metazoans, the role of Sir2 in mediating the extension of lifespan caused by CR in yeast has been controversial. Experiments from some labs have challenged the requirement of SIR2 in mediating the effect of CR in yeast RLS extension (Jiang et al. 2002; Kaeberlein et al. 2004), as well as in worms (Kaeberlein et al. 2006b; Lee et al. 2006; Hansen et al. 2007; Schulz et al. 2007). Additional studies have shown that in CLS, the presence or absence of SIR2 has little effect on CLS during “moderate” CR (Fabrizio et al. 2005; Smith et al. 2007), and sir2Δ mutants actually have a very long lifespan during “extreme” CR (Fabrizio et al. 2005). Thus, it appears that in yeast CLS and RLS, there exists a SIR2-independent/ERC-independent aging pathway that is sensitive to calorie restriction by glucose limitation. To further shed light on the mechanism of CR modulation of lifespan, we have investigated the ability of CR to increase Sir2 activity in yeast using rDNA silencing and recombination as indicators.
Results
Effects of CR on rDNA silencing of a MET15 reporter gene
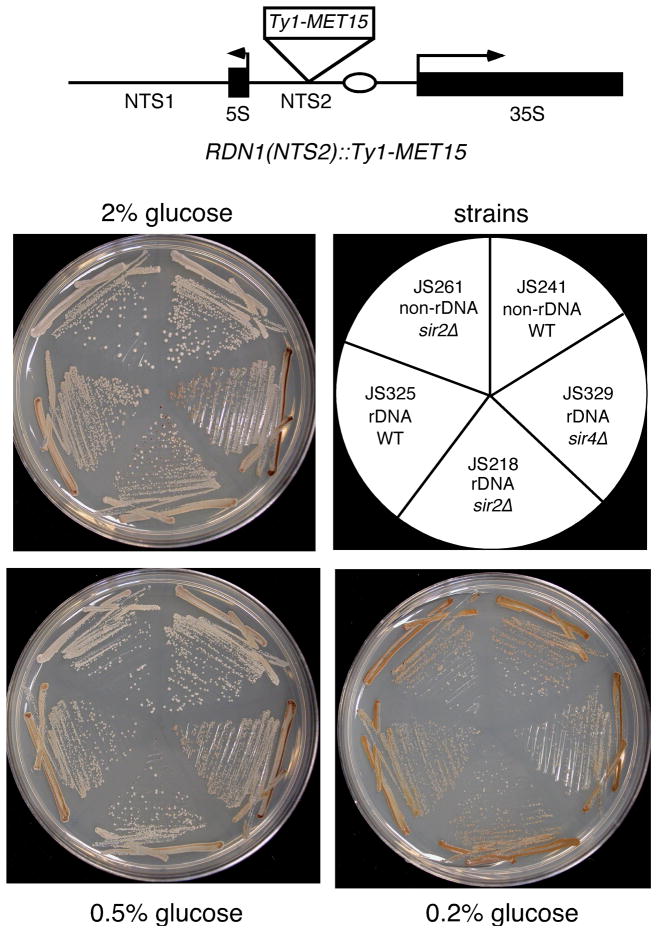
The exquisite sensitivity of rDNA silencing to Sir2 levels implies that it should be an excellent tool for monitoring the effect of CR on Sir2 activity. We first analyzed the effect of lowering the glucose concentration on the silencing of a Ty1-MET15 reporter gene integrated into the rDNA non-transcribed spacer (NTS2) (Smith & Boeke 1997). Ty1 is a retrotransposon that carried MET15 into the rDNA of this strain (see schematic in Fig. 1). MET15 encodes an O-acetyl homoserine-O-acetyl serine sulfhydrylase required for sulfur amino acid synthesis. In the presence of Pb2+ ions, met15Δ strains produce a characteristic dark brown pigment, and MET15+ strains do not produce the pigment (Cost & Boeke 1996). Silencing of MET15 in the rDNA results in an intermediate tan colony color (Smith & Boeke 1997). Eliminating silencing by deletion of SIR2 changes the colony color to white, whereas strengthening silencing by deletion of SIR4 results in a darker shade of tan (Smith & Boeke 1997). As expected (Smith & Boeke 1997), deletion of SIR4 caused a darker tan color than WT, indicating that the plates were sensitive to improved silencing of MET15 on both 2% and 0.5% glucose (Fig. 1). Surprisingly, reducing the glucose concentration from 2% to 0.5% (the typical CR condition) had no measurable effect on the colony color when MET15 was positioned in the rDNA (Fig. 1). The typical CR condition of 0.5% glucose, therefore, does not appear to have any effect on silencing of MET15. This is in contrast to an earlier report that detected a darker colony color with this reporter at 0.5% glucose (Lin et al. 2002).
Fig. 1.
CR effects on silencing of a MET15 reporter integrated into the NTS2 region of an rDNA repeat. The indicated strains were streaked for single colonies onto 0.5xYPD plates containing 0.1% lead nitrate and either 2%, 0.5%, or 0.2% glucose. Darker colony color indicates better rDNA silencing of MET15. Colonies were grown for 5 days.
In some yeast replicative lifespan studies, it was reported that glucose concentrations lower than 0.5% were needed to reliably detect CR-mediated extension of lifespan (Jiang et al. 2000; Kaeberlein et al. 2004). In the case of MET15, reducing the glucose to 0.2% clearly resulted in a darker colony color for the WT JS325 strain, suggestive of stronger silencing (Fig. 1). However, the darker color was not dependent on SIR2, and also developed when MET15 was located at a non-rDNA locus that is not subject to SIR2-dependent silencing (Fig. 1). This result suggested that the MET15 reporter gene was potentially transcriptionally repressed by very low glucose concentrations, but the repression was not specific to the rDNA locus. Therefore, the MET15 reporter system may not be reliable for measuring silencing under CR conditions.
Effects of CR on the rDNA silencing reporter gene mURA3
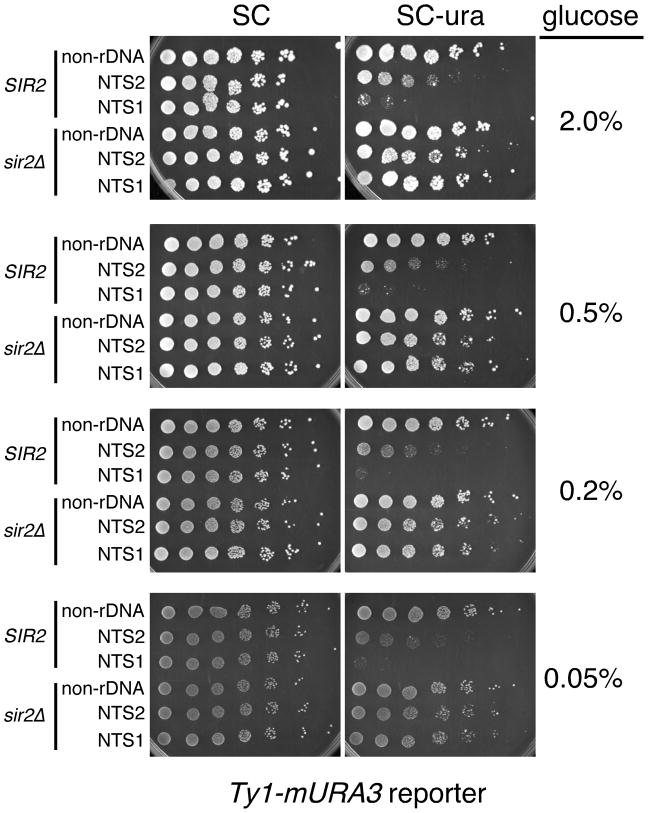
The development of a dark brown colony color at a 0.2% glucose concentration for all strains in Fig. 1 could also indicate an indirect metabolic effect on pigment formation, rather than a transcriptional effect. To test this possibility, we utilized a different rDNA silencing reporter gene called Ty1-mURA3 that is also highly sensitive to changes in Sir2 dosage or activity (Smith et al. 1998; Sauve et al. 2005). The mURA3 reporter is a modified URA3 gene in which the promoter has been replaced with a TRP1 promoter (Smith & Boeke 1997). Silencing in these strains can be easily measured by spotting 5-fold serial dilutions of cells onto SC-ura plates. Improved silencing of the Ty1-mURA3 reporter results in diminished growth on SC-ura. Strains with Ty1-mURA3 integrated within an rDNA gene at NTS1 or NTS2 were compared to a control strain with Ty1-mURA3 integrated at a non-rDNA chromosomal position, and to congenic strains where SIR2 was deleted (Fig. 2). The strains were all spotted onto SC and SC-ura plates containing 2%, 0.5%, 0.2%, or 0.05% glucose. It should be noted that each of the reduced glucose concentrations tested have previously shown some benefit to RLS (Jiang et al. 2000; Kaeberlein et al. 2002), as well as to CLS (Smith et al. 2007). Slight decreases in growth were observed for the NTS2 and NTS1 positions on SC-ura plates when the glucose concentration was lowered (Fig. 2). However, this difference was accounted for by a progressive reduction in colony growth on the SC control plates (Fig. 2), suggesting that silencing was not actually being strengthened. Similar results were observed with the Ty1-mURA3 reporter using a different strain background (BY384) related to BY4741/42 (data not shown), indicating that this was not a strain specificity issue. This result also suggests that the effect of CR on MET15 that caused a darker colony color in Fig. 1 was most likely not transcriptional.
Fig. 2.
CR effects on silencing of a Ty1-mURA3 marker within the tandem array. Strains with Ty1-mURA3 integrated within the tandem array at NTS2 (JS125) and NTS1 (JS128), or at a non-rDNA genomic location (JS122) were spotted as 5-fold serial dilutions onto SC and SC-ura plates containing 2%, 0.5%, 0.2%, or 0.05% glucose. The sir2Δ control strains were JS151 (non-rDNA), JS155 (NTS2), and JS163 (NTS1). Plates were incubated for 3 days prior to photography.
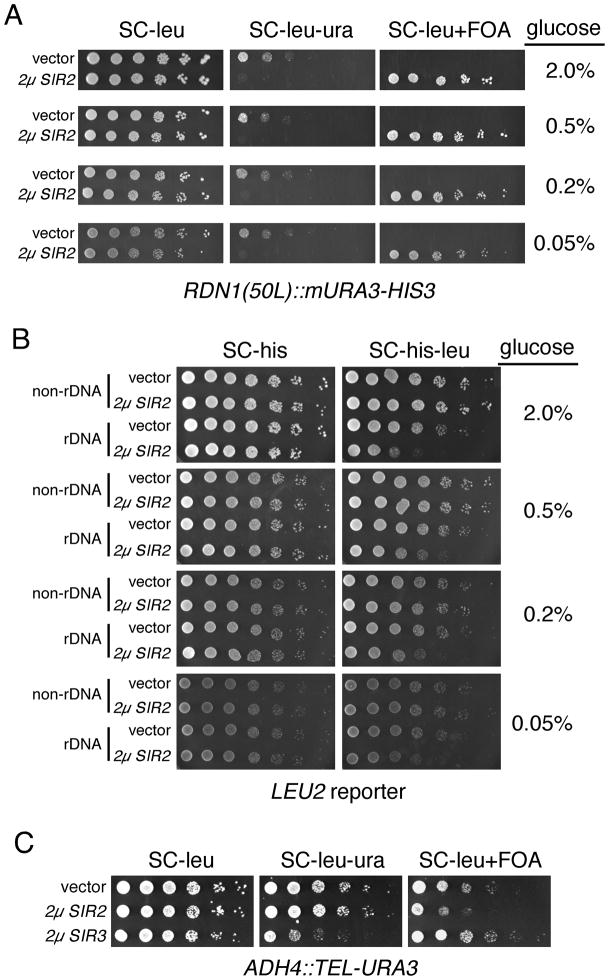
We next tested whether CR induces the spreading of rDNA silencing through the use of a mURA3-HIS3 cassette integrated 50bp left (50L) of the rDNA tandem array, within unique chromosome XII sequence (Buck et al. 2002). Unlike genes integrated within the tandem array, the RDN1(50L)::mURA3-HIS3 cassette is not subject to high recombination frequencies (Buck et al. 2002), allowing the reliable use of 5-FOA to detect improved silencing of mURA3 as in indicator of spreading (McClure et al. 2008). 5-FOA is a uracil analog that kills cells expressing the Ura3 protein (Boeke et al. 1984). The induction of spreading by SIR2 overexpression strengthens mURA3 silencing such that growth is permitted on FOA (McClure et al. 2008). As expected, introduction of a 2μ SIR2/LEU2 plasmid (pSB766) resulted in strong growth on FOA with this reporter, serving as a positive control for the spreading phenotype (Fig. 3A, 2μ SIR2 rows). However, reducing the glucose concentration had no effect on either the SC-leu-ura, or SC-leu FOA plates (Fig. 3A, vector rows), indicating that CR did not induce the spreading of rDNA silencing. The polyphenolic compound, resveratrol, has been reported as a small molecule activator of Sir2 and other Sirtuins (Howitz et al. 2003). We therefore tested whether the addition of resveratrol would strengthen rDNA silencing in combination with CR. Using the RDN1(50L)::mURA3-HIS3 reporter cassette again, 100 μM resveratrol had no effect on rDNA silencing regardless of the glucose concentration. (Supplemental Fig. 1A).
Fig. 3.
SIR2 overexpression strengthens rDNA silencing, but CR does not. A) YSB348 was transformed with either an empty 2μ LEU2 vector (pRS425) or a SIR2 2μ LEU2 plasmid (pSB766). YSB348 contains a mURA3-HIS3 silencing reporter cassette integrated within unique chromosome XII sequence just 50 bp to the left (centromere proximal) of the tandem array (Buck et al. 2002). All plates lacked leucine to select for the plasmids, and contained the indicated glucose concentration. B) JS210 and JS215 were transformed with either an empty 2μ HIS3 vector (pRS423) or a SIR2 2μ HIS3 plasmid (pJSS71-13). JS215 contains LEU2 integrated within the rDNA, and JS210 contains LEU2 outside the rDNA as a control (Smith & Boeke 1997). The SC plates lacked histidine to select for the plasmids. Silencing of LEU2 is indicated by poor growth on SC-his-leu. C) Telomere silencing assay showing the effects of overexpressing SIR2 (strain JS717) or SIR3 (strain JS718) compared to an empty vector (strain JS716). These strains contain a URA3 marker integrated at the ADH4 gene, and flanked by a telomere. The plates lacked leucine to select for the LEU2-containing plasmids.
In yeast, there is a limiting pool of Sir2 shared between the rDNA, telomeres and silent mating loci (Buck & Shore 1995; Smith et al. 1998), raising the possibility that the Sir2 protein pool is normally fully activated, making any potential positive effect by CR on silencing undetectable. Therefore, we hypothesized that increased SIR2 dosage would promote the enhancement of silencing by CR. However, as shown in Fig. 3A, the addition of a 2μ SIR2 plasmid strengthened mURA3 silencing at the 50L position so much that no growth was observed on the SC-leu-ura plates. Thus, although there were no differences in silencing with reduced glucose concentrations, the assay was already at the detection limit even at the 2% glucose level. Therefore, we also measured the effect of a 2μ SIR2 plasmid on silencing of a LEU2 reporter integrated within the tandem array. This marker is not silenced as well as mURA3 (Smith & Boeke 1997), allowing for added strengthening of repression by CR. SIR2 overexpression improved silencing of LEU2 as expected, but reducing the glucose concentration did not improve the silencing any further (Fig. 3B). If anything, there could be slight depression. Together, these results show that in our assays we can easily detect Sir2-mediated improvements in rDNA silencing, but never CR-mediated improvements.
Evaluating the utility of TPE as an indicator of Sir2 activity
The effect of SIR2 overexpression on telomere position effect (TPE) has been less clear than with rDNA silencing, with one study reporting no effect (Renauld et al. 1993), and at least two reporting improved silencing (Cockell et al. 2000; Kaeberlein et al. 2005a). To evaluate the utility of TPE as an indicator of Sir2 activity levels, we transformed high copy LEU2/SIR2 (pSIR2μ) or LEU2/SIR3 (pLP304) plasmids into a reporter strain that contains a telomeric URA3 gene. SIR3 overexpression was already known to strengthen TPE (Renauld et al. 1993), and was used as a positive control in the assay. Improved TPE was indicated by less colony growth on SC plates lacking uracil (SC-leu-ura) and better growth on SC containing 5-FOA). As predicted, the SIR3 control plasmid strengthened TPE (Fig. 3C). However, the SIR2 plasmid actually weakened TPE, as indicated by less growth on FOA compared to the empty vector control (Fig. 3C). As a result of this finding, we concluded that TPE is not a consistent indicator of Sir2 activity. Similarly, Alan Morgan and coworkers have also determined that CR does not strengthen TPE (personal communication).
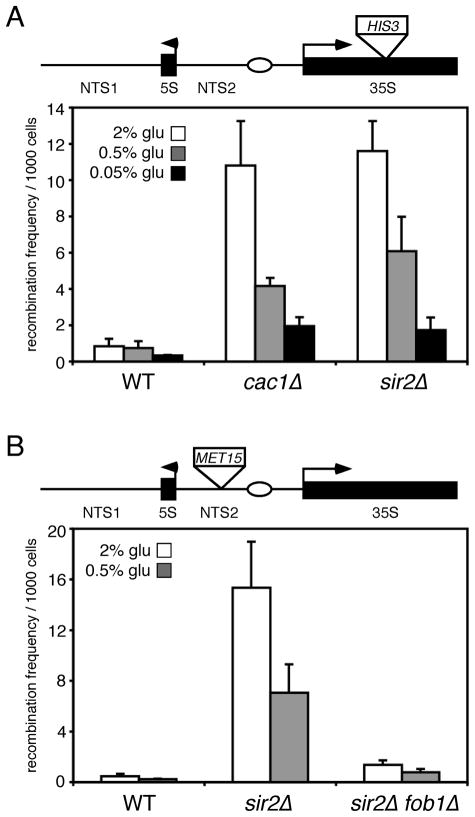
CR represses rDNA recombination independently of SIR2
The above results with rDNA silencing do not support a model for Sir2 activation by CR growth conditions. However, Sir2 is also involved in repressing recombination within the rDNA (Gottlieb & Esposito 1989), leaving open the possibility that stimulation of Sir2 activity by CR could improve the repression of rDNA recombination without strengthening rDNA silencing. Such a model is supported by the previous identification of genes that differentially affect rDNA silencing and recombination when mutated, including SET1, FOB1, and HHO1 (Bryk et al. 2002; Huang & Moazed 2003; Li et al. 2008). To evaluate the effect of CR on rDNA recombination, WT, cac1Δ, and sir2Δ strains containing the mURA3-HIS3 cassette within the 35S transcribed region of the rDNA were used to measure the frequency of HIS3 marker loss on various glucose concentrations by a replica-plating assay for ½ sectored colonies on SC-his media. The frequency of rDNA recombination in a WT strain is quite low, so the cac1Δ mutation was chosen to increase the recombination frequency (Smith et al. 1999), thus making it easier to detect any decrease in the frequency caused by CR. As shown in Fig. 4A, the cac1Δ and sir2Δ mutations both increased the frequency of recombination compared to WT when grown on 2% glucose. The typical CR condition (0.5% glucose) modestly reduced the recombination frequency for each strain, including the sir2Δ mutant, by ~2-fold (Fig. 4A). Stronger CR (0.05% glucose) further reduced the recombination frequency in both mutants by ~5-fold (Fig. 4A). To confirm these results we also measured recombination frequency with an independent strain set that contained the Ty1-MET15 reporter within NTS2. In this case, the frequency of ½ sectored colonies (white/brown) grown on Pb2+-containing plates was calculated for WT, sir2Δ, and sir2Δ fob1Δ strains (Fig. 4B). Again, the recombination frequency was greatly elevated on 2% glucose by the sir2Δ mutation and repressed by ~2-fold on 0.5% glucose. The 0.05% glucose concentration blocked colony growth on Pb2+ plates, so this recombination data from this condition was not obtained. Similar to the silencing results, resveratrol did not suppress rDNA recombination using this assay (Supplemental Fig. 1B). Deletion of FOB1 was already known to partially block the hyper recombination caused by a sir2Δ mutation (Kaeberlein et al. 1999), which we also observed in the sir2Δ fob1Δ double mutant. These results support a model in which CR suppresses rDNA recombination without affecting the transcriptional silencing of Pol II-transcribed reporter genes, although the suppression does not depend on SIR2.
Fig. 4.
CR suppresses rDNA recombination independently of SIR2. A) Recombinational loss of HIS3 from the rDNA was measured by calculating the frequency of half-sectored His+/His− colonies per 1000 cells. Strains tested were JS306 (WT), JS400 (cac1Δ), and JS576 (sir2Δ). Strains were grown in SC media containing either 2%, 0.5%, or 0.05% glucose. The schematic diagram indicates the position of HIS3 (as part of the mURA3-HIS3 cassette) in the 35S transcribed region of an rDNA repeat. The number of colonies for each replicate ranged from ~2000 to 6000. B) Recombinational loss of MET15 from the rDNA was measured by calculating the frequency of half-sectored brown/white colonies per 1000 cells. Strains tested were DSY380 (WT), DSY394 (sir2Δ), and DSY390 (sir2Δ fob1Δ). Lead plates contained 2% or 0.5% glucose. The schematic diagram indicates the position of MET15 (as part of the Ty1-MET15 cassette) in the NTS2 region of an rDNA repeat. The number of colonies for each replicate ranged from ~4000 to 12000. Error bars in both panels represent the standard deviation between biological replicates.
Effects of CR on endogenous transcripts produced from the rDNA NTS
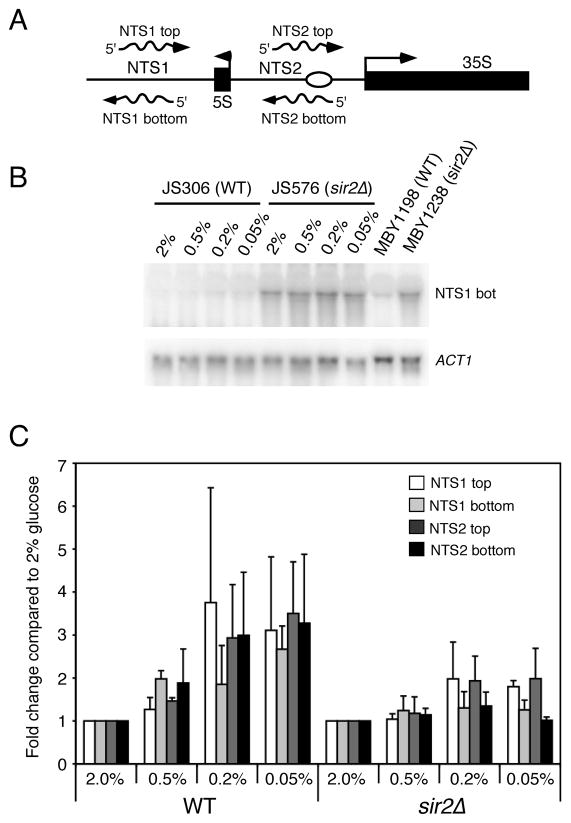
One of the current models for Sir2-mediated suppression of rDNA recombination involves the silencing of a bidirectional Pol II promoter within NTS1 called E-pro (Kobayashi & Ganley 2005). Deletion of SIR2 results in the transcription of E-pro, and the transcription process results in the displacement of cohesin from the NTS (Kobayashi et al. 2004; Kobayashi & Ganley 2005). The loss of cohesin stimulates unequal crossing over between the sister chromatids and a subsequent change in rDNA copy number. It was therefore possible that CR suppressed rDNA recombination in our assays by improving the silencing of transcription derived from E-pro or other Pol II-derived transcripts recently discovered from NTS1 and NTS2 (Li et al. 2006). To test this possibility, we utilized Northern blotting to measure the steady state level of each NTS transcript from WT and sir2Δ strains grown in 2%, 0.5%, 0.2%, and 0.05% glucose. These RNAs are transcribed from both the top and bottom strands of NTS1 and NTS2 (Fig. 5A). As shown in Fig. 5B for the NTS1-top RNA species, deletion of SIR2 greatly increases the expression due to a loss of silencing. Similar results were observed for the other three transcripts (data not shown). Although silencing of these transcripts depends on Sir2, CR did not strengthen their repression. Instead, for each of the four RNAs, CR conditions either had no effect or may have slightly elevated their expression (Fig. 5C). CR did not further increase the expression in the sir2Δ background compared to 2% glucose (Fig. 5C). This data strongly suggests that CR does not suppress rDNA recombination by improving the silencing of non-coding transcripts from the NTS.
Fig. 5.
Analyzing CR effects on the silencing of non-coding RNAs derived from the NTS1 and NTS2 regions of the rDNA. A) Schematic diagram depicting the Pol II-driven expression of non-coding RNAs from both strands of NTS1 and NTS2. B) Representative Northern blot showing silencing of the NTS1-bottom strand RNA by Sir2. Note the large increase in expression in the sir2Δ mutants JS576 and MBY1238. Expression level of ACT1 was used as a loading control. C) Quantitation of CR effects on the expression of the non-coding RNAs. Since the sir2Δ mutant JS576) caused a large increase in expression for each RNA, we normalized the expression level at 2% glucose to 1.0 for both WT and sir2Δ. Changes in expression relative to 2% glucose are shown for both WT (JS306) and sir2Δ (JS576). Standard deviations from 3 independent experiments are shown.
Discussion
Understanding the molecular mechanism of CR could provide a great benefit in combating age-related degeneration and disease. The highly researched SIR2 pathway has been proposed to mediate the lifespan effects of CR in yeast, worms, flies and even rodents (Lin et al. 2000; Tissenbaum & Guarente 2001; Lin et al. 2002; Anderson et al. 2003; Rogina & Helfand 2004; Chen et al. 2005; Bordone et al. 2007; Boily et al. 2008). Sir2 and other Sirtuins have clearly been implicated in longevity control of yeast and the other model organisms, but the data presented here challenge the initial interpretation that Sir2 activity at the rDNA locus is increased by CR in yeast. By tracking commonly used rDNA reporter genes, as well as endogenous non-coding RNAs from the “non-transcribed spacer”, we were unable to detect improved Sir2-dependent silencing as a result of CR growth conditions. These findings are consistent with previous RLS and CLS studies showing lifespan extension by CR even in the absence of SIR2 (Kaeberlein et al. 2004; Fabrizio et al. 2005; Smith et al. 2007). It has been proposed that the Sir2 homolog, Hst2, can partially substitute for Sir2 in suppressing recombination to extend lifespan in response to CR (Lamming et al. 2005), although a separate study showed that CR of varying glucose concentrations extends RLS even in a fob1Δ sir2Δ hst1Δ hst2Δ quadruple mutant (Kaeberlein et al. 2006a). While we have not tested the effects of the other sirtuins on recombination in this study, it is clear that CR can suppress recombination independently of Sir2 and in the absence of enhanced Pol II silencing within the rDNA. If CR does stimulate Sir2 in yeast cells, then a novel mechanism would have to be invoked in which the pool of Sir2 utilized for rDNA silencing is not subject to the activation. This seems unlikely because activation of Sir2 in vivo by the addition of isonicotinamide generally strengthens HM, telomeric, and rDNA silencing (Sauve et al. 2005). Alternatively, one or more of the other Sirtuins could become engaged at the rDNA without the need to modulate Sir2 activity.
At least three previously published experiments have used silencing as a read-out of Sir2 activation. The first study used a MET15 reporter to show that CR strengthened rDNA silencing to produce a darker colony color (Lin et al. 2002). We did not observe a darker colony color with “moderate” CR of 0.5% glucose, but found that “extreme” CR conditions below 0.5% glucose result in a dark colony color that does not depend on SIR2 or location in the rDNA (Fig. 1). CR either causes metabolic effects that affect colony color development with this system that are variable according to differences in media formulation, or causes a general transcriptional repression of the MET15 promoter regardless of its genomic location. The other two studies measured telomeric silencing of ADE2 or URA3 reporters to show that neither resveratrol nor CR improved Sir2 activity in TPE (Kaeberlein et al. 2005a; Kaeberlein et al. 2005b). Overexpression of SIR2 strengthened repression of the telomeric ADE2 reporter, but did not extend RLS in the PSY316 strain background (Kaeberlein et al. 2005a). Our SIR2 overexpression data in Fig. 3C using a strain background related to BY4741/42 and S288C did not demonstrate improved TPE, which may suggest that the sensitivity of TPE to Sir2 activity is specific to particular reporter genes, strain backgrounds, or even specific levels of SIR2 overexpression (Cockell et al. 2000). In contrast, SIR2 overexpression has universally resulted in stronger silencing at the rDNA regardless of the strain background, reporter gene, or level of SIR2 overexpression (Fritze et al. 1997; Smith et al. 1998; Smith et al. 2007). We propose that rDNA silencing is better suited than TPE in detecting improved Sir2 activity. Despite using rDNA silencing as the reporter assay, we were also unable to detect any effect of resveratrol, even in combination with CR growth conditions.
One of the potential limitations of using reporter genes such as mURA3 or LEU2 as a readout for Sir2 stimulation is that small differences might be difficult to detect due to the qualitative nature of the 5-fold dilution spot assays. However, previous studies have shown that even one extra copy of SIR2 produces an easily visible enhancement of rDNA silencing using the 5-fold spot assays (Smith et al. 1998; Smith et al. 2007). Using 2-fold serial dilutions have not resulted in any improved resolution because colony size also contributes to the readout (data not shown). Additionally, quantitative measures of silencing under CR conditions have similarly failed to demonstrate Sir2 activation (personal communication A. Morgan). Finally, in addition to the spot assays for measuring silencing, northern blotting for NTS-derived non-coding RNAs showed that CR does not improve Sir2-mediated repression of these transcripts.
If CR does not stimulate Sir2 for silencing or recombination suppression in yeast cells, then what alternative pathways could be involved in the suppression of recombination by CR and in the extension of lifespan? Inhibition of the TOR signaling pathway has been shown to extend both RLS and CLS in yeast (Kaeberlein et al. 2005b; Powers et al. 2006; Medvedik et al. 2007), and to extend the lifespan of Drosophila and C. elegans (Vellai et al. 2003; Kapahi et al. 2004). When nutrients are high, TOR activity is high, and when nutrients are reduced, TOR activity becomes decreased. Accordingly, genetic epistasis experiments in yeast have indicated that TOR acts downstream of CR in the extension of lifespan (Kaeberlein et al. 2005b; Powers et al. 2006; Medvedik et al. 2007). As with CR, there is some debate whether SIR2 and HST2 are required for the TOR inhibition-mediated extension of yeast RLS (Kaeberlein et al. 2005b; Medvedik et al. 2007). At least for CLS, it has been reported that the effect of reducing TOR signaling is through increased respiration and upregulation of mitochondrial gene expression (Bonawitz et al. 2007). Alternatively, the effects could be mediated by a decrease in translation capacity (Hansen et al. 2007; Steffen et al. 2008). Interestingly, deletion of TOR pathway genes or the addition of low concentrations of rapamycin both cause a reduction in the frequency of rDNA recombination ((Medvedik et al. 2007); A. Morgan, personal communication), suggesting that even if Sir2 is not mediating the effect, the control of rDNA circles could still be involved in the extension of RLS in a WT strain background. Of particular interest is the recent finding that positioning of the rDNA array near the inner nuclear membrane (INM) is required for maintaining the stability of the rDNA repeats, but not for rDNA silencing of reporter genes (Mekhail et al. 2008). CR could potentially enhance this association with the INM. In conclusion, CR does not appear to activate Sir2 at the rDNA or telomeres in Saccharomyces cerevisiae. Additional effort will be needed to uncover the molecular details of CR-mediated replicative and chronological longevity.
Experimental Procedures
Yeast strains and media conditions
All strains used for rDNA silencing were derived from the GRF167/JB740 background commonly used for Ty1 transposition and rDNA silencing assays (Smith & Boeke 1997), and listed in Table 1. Synthetic Complete (SC) and Yeast Extract/Peptone/Dextrose (YPD) media were formulated as previously described (Burke et al. 2000). Strains were transformed with plasmids where indicated, including the empty 2μ LEU2 plasmid (pRS425), 2μ LEU2 SIR2 plasmids (pSIR2μ or pSB766), and 2μ LEU2 SIR3 plasmid (pLP304). In Fig. 3B, an empty 2μ HIS3 plasmid (pRS423) and 2μ HIS3 SIR2 plasmid (pJSS71-13) were used because LEU2 was the silencing reporter.
Table 1.
Yeast strains used in the study.
| Strain | Genotype |
|---|---|
| DSY380 | MATa his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(18S)::mURA3-HIS3 RDN1(NTS2)::Ty1-MET15 |
| DSY390 | DSY390 sir2Δ::kanMX4 |
| DSY394 | DSY380 sir2Δ::kanMX4 fob1Δ::kanMX4 |
| JS122a | MATα his3Δ200 leu2Δ1 ura3-167 ????::Ty1-mURA3b |
| JS125a | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(NTS2)::Ty1-mURA3 |
| JS128a | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(NTS1)::Ty1-mURA3 |
| JS151a | JS122 sir2Δ::HIS3 |
| JS155a | JS125 sir2Δ::HIS3 |
| JS163a | JS128 sir2Δ::HIS3 |
| JS210a | MATα his3Δ200 trp1 Δ63 ura3-167 leu2Δ1::pJSS60-2c |
| JS215a | MATα his3Δ200 trp1 Δ63 ura3-167 leu2Δ1 RDN1(35S)::pJSS60-2c |
| JS218a | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(NTS2)::Ty1-MET15 sir2Δ::HIS3 |
| JS241a | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 ????::Ty1-MET15b |
| JS261 | JS241 sir2Δ::HIS3 |
| JS306d | MARa his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(18S)::mURA3-HIS3 RDN1(NTS2)::Ty1-MET15 |
| JS325d | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(NTS2)::Ty1-MET15 |
| JS329d | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(NTS2)::Ty1-MET15 sir4Δ::HIS3 |
| JS400d | JS306 cac1Δ::kanMX4 |
| JS576d | JS306 sir2Δ::kanMX4 |
| JS716 | MATa his3Δ200 lys2Δ202 trp1Δ63 leu2Δ1::TRP1 ura3-52 ADH4::URA3-TEL +pRS425 (2μ LEU2 empty) |
| JS717 | MATa his3Δ200 lys2Δ202 trp1Δ63 leu2Δ1::TRP1 ura3-52 ADH4::URA3-TEL +pSIR2μ (2μ LEU2 SIR2) |
| JS718 | MATa his3Δ200 lys2Δ202 trp1Δ63 leu2Δ1::TRP1 ura3-52 ADH4::URA3-TEL +pLP304 (2μ LEU2 SIR3) |
| MBY1198e | MATα, his3Δ200, ade2Δ::hisG, leu2Δ0, ura2Δ0, met15Δ0, trp1Δ63, Ty1his3AI-236r, Ty1ade2AI-515 |
| MBY1238e | MBY1198 sir2Δ |
| YSB348f | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)::mURA3-HIS3 |
Strains described in reference (Smith & Boeke 1997).
Ty1-mURA3 is integrated at a non-rDNA location in the genome.
Plasmid pJSS60-2, containing a LEU2 marker, was integrated into the genome at the indicated locus.
Strains described in reference (Smith et al. 1999).
Strains described in reference (Li et al. 2006).
Strain described in reference (Buck et al. 2002).
Silencing and recombination assays
For the MET15 reporter strains, cells were patched onto YPD plates and allowed to grow overnight at 30°C. Cells were then streaked onto YPD plates containing ½ the normal concentration of Bacto peptone and Bacto yeast extract, and lead nitrate at 0.1% lead nitrate. Using ½ concentrated YPD (0.5xYPD) improved color development compared to typical YPD (data not shown). Glucose concentrations ranged from 2% to 0.05%. The lead plates were then incubated at 30°C for 5 days before photographs were taken. For spot test silencing assays, cells were patched onto the appropriate SC plates containing the indicated glucose concentration and/or missing the indicated amino acids or uracil. After overnight growth at 30°C, the cells were serially diluted in 5-fold increments in 96-well plates and then 5 μl spotted onto the indicated plates. For recombination assays, cells were pre-grown overnight in SC media containing either 2% or 0.5% glucose. JS306, JS400, and JS576 were diluted in water and plated onto YPD containing 2% or 0.5% glucose, and allowed to grow into colonies for 2 days (average of ~200 colonies per plate). The colonies were then replica plated onto SC and SC-his to identify ½ sectored colonies where only half the colony grew on SC-his. The number of half-sectored colonies was divided by the total number of His+ or sectored colonies, and multiplied by 1000 to give the recombination frequency per 1000 cells. For DSY380, DSY394, and DSY390, the culture dilutions were plated directly onto the 0.1% lead nitrate plates (2% or 0.5% glucose) and allowed to grow for 5 days before the frequency of half sectored (brown/white) colonies was calculated. Resveratrol (Sigma) was added to the media were indicated at a concentration of 100 μM. The recombination assays were performed in triplicate starting from three independent colonies for each strain.
Northern blotting
Yeast strains were grown to saturation in YPD containing 2% glucose and supplemented with tryptophan (0.4 mM) and adenine (40 μg/mL) at 30°C. Cultures were then diluted to a final density of 2.5×106 cells/ml in 50 ml of fresh media with 2%, 0.5%, 0.2% or 0.05% glucose. The diluted cultures were grown at 30°C to 2-3×107 cells/ml. Total RNA was isolated as described previously (Bryk et al. 1997). Northern analyses were performed as described in (Swanson et al. 1991). Strand-specific 32P-labeled RNA probes used to detect NTS transcripts are described in (Li et al. 2006). An ACT1 (+564 to +1200) probe was made by PCR amplification of yeast genomic DNA, purified from an agarose gel and labeled with [α-32P]-dATP by random priming (Ausubel et al. 2000). Northern blots were quantified on a GE Healthcare Storm 860 PhosphorImager by using ImageQuant software.
Supplementary Material
Acknowledgments
We thank Alan Morgan for communicating results prior to publication. We also thank members of the Smith and Bryk labs for helpful discussions. This work was supported in part by a Cellular and Molecular Biology training grant from the NIH to D.L.S., and by NIH grants GM075240 and AG027340 to J.S.S. and GM070930 to M.B.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc; 2000. [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Buck SW, Sandmeier JJ, Smith JS. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell. 2002;111:1003–1014. doi: 10.1016/s0092-8674(02)01193-5. [DOI] [PubMed] [Google Scholar]
- Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Cockell MM, Perrod S, Gasser SM. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics. 2000;154:1069–1083. doi: 10.1093/genetics/154.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast. 1996;12:939–941. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C939::AID-YEA988%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Esposito RE. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005a;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006a;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006b;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000a;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000b;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Mueller JE, Bryk M. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol Biol Cell. 2006;17:3848–3859. doi: 10.1091/mbc.E06-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Mueller JE, Elfline M, Bryk M. Linker histone H1 represses recombination at the ribosomal DNA locus in the budding yeast Saccharomyces cerevisiae. Mol Microbiol. 2008;67:906–919. doi: 10.1111/j.1365-2958.2007.06101.x. [DOI] [PubMed] [Google Scholar]
- Lin S-J, Defossez P-A, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- McCay CM, Crowell MF. Prolonging the life span. Sci Mon. 1934;39:405–414. [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- McClure JM, Gallo CM, Smith DL, Jr, Matecic M, Hontz RD, Buck SW, Racette FG, Smith JS. Pnc1p-mediated nicotinamide clearance modifies the epigenetic properties of rDNA silencing in Saccharomyces cerevisiae. Genetics. 2008;180:797–810. doi: 10.1534/genetics.108.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.