Abstract
To assess the hypothesis that the dynamics of plasma angiogenic and inflammatory cytokines after docetaxel chemotherapy with or without the c-kit/abl/platelet-derived growth factor receptor (PDGFR) inhibitor imatinib mesylate for prostate cancer are associated with outcome, the kinetics of 17 plasma cytokines before versus after chemotherapy were assessed and associations with progression-free survival (PFS) examined. After adjusting for multiple tests, significantly different declines in placental growth factor (PIGF), soluble vascular endothelial growth factor receptor-1 (VEGFR1), VEGF, and soluble c-kit were observed with docetaxel plus imatinib (n = 41) compared to docetaxel alone (n = 47). Based on a piecewise linear regression model for change in concentration of each cytokine as a function of the probability of change in p-PDGFR in vivo, only the dynamics of PIGF (P < 0.0001) and soluble c-kit (P < 0.0001) differed with imatinib therapy. In a Bayesian log-normal regression model for PFS, a rise in human matrix metalloproteinase 9 after docetaxel alone associated with a longer PFS. Distinct plasma angiogenic cytokines are modified by imatinib and partitioned by in vivo p-PDGFR dynamics after docetaxel chemotherapy for metastatic prostate cancer. Plasma PIGF and soluble c-kit kinetics are candidate biomarkers of imatinib effect. The predictive value of human matrix metalloproteinase 9 kinetics for docetaxel efficacy requires prospective validation.
Introduction
Improved outcomes with docetaxel chemotherapy for advanced castration-resistant prostate cancer are being sought with novel combinations that target putative mechanisms of disease progression and drug resistance. Preclinical modeling indicated that the platelet-derived growth factor and its receptor (PDGFR) were upregulated in prostate cancer cells proliferating within the bone microenvironment (Uehara and others 2003). The PDGFR was observed to be upregulated in endothelial cells of vasculature specifically associated with PDGF-expressing tumor, and the PDGFR inhibitor imatinib potentiated taxane efficacy via enhanced endothelial apoptosis, an antivascular effect (Uehara and others 2003; Kim and others 2006).
Contrary to preclinical estimates, a randomized controlled study that compared the efficacy of imatinib in combination with docetaxel versus docetaxel alone in men with castration-resistant prostate cancer and bone metastases showed no added benefit with imatinib (Mathew and others 2007). Unexpectedly, in vivo pharmacodynamic monitoring of PDGFR inhibition showed that, within the docetaxel arm, an increased probability of PDGFR activation in peripheral blood leucocytes correlated with improved progression-free survival (PFS) and overall survival (OS) (Mathew and others 2008). Rising plasma PDGF levels were associated with a decreased probability of PDGFR activation and inferior PFS (Mathew and others 2008). While the fundamental biological implications of these observations are yet to be determined, these partitioned outcomes were not equally detected in the docetaxel–imatinib combination arm.
To further explore the dynamic signature of plasma cytokines and their prognostic impact after docetaxel chemotherapy, a panel of 17 additional angiogenic and inflammatory cytokines was constructed. Individual cytokine kinetics between baseline (BL) and after docetaxel exposure, modulation by concurrent PDGF inhibitor therapy, and association with PFS outcomes were studied.
Methods
Patients
One hundred sixteen men were enrolled to a randomized study of docetaxel with placebo or imatinib for metastatic castration-resistant prostate cancer and bone metastases (Mathew and others 2007). Of these, 88 paired plasma samples at BL and 6 weeks later after one cycle of weekly docetaxel-based therapy at cycle 2 day 1 (C2D1) were available.
Multiplex cytokine assay
Plasma levels of all analytes described here were subsequently analyzed in duplicates using a multiplex platform, Meso Scale Discovery (MSD) (Gaithersburg, MD). The analytes were soluble c-kit receptor (c-kit), soluble vascular endothelial growth factor receptor-2 (sVEGFR2, KDR), fibroblast growth factor, VEGF, sVEGFR1, placental growth factor (PIGF), interleukin (IL)2, IL8, IL12p70, IL10, granulocyte macrophage-colony stimulating factor, interferon-γ, IL6, IL10, tumor necrosis factor-α, transforming growth factor-β, and matrix metalloproteinase-(MMP)-9. All reagents were provided with the MSD kits and tests conducted according to the manufacturer's instructions.
Statistical methods
Numerical variables were summarized using means and standard deviations, with association between pairs of variables estimated by Pearson's correlation coefficient (Snedecor and Cochran 1980). The Wilcoxon signed rank test was used for 2 sample comparisons of numerical variables (Hollander and Wolfe 1979), applying the Bonferroni P value correction for multiple tests (Snedecor and Cochran 1980). For each cytokine, the Bayesian regression model and method of Morita and others (2010) were employed to evaluate the effects of change in the cytokine level from BL to C2D1 on PFS time while accounting for the effects of hemoglobin, change in prostate-specific antigen (PSA), and change in p-PDGFR. For each patient, because p-PDGFR was measured in ∼2,000 cells both at BL and at C2D1, the within-patient BL and C2D1 distributions of p-PDGFR could be estimated very reliably. Because both the BL and C2D1 distributions of p-PDGFR were clearly bimodal for all patients, the within-patient change in p-PDGFR could not be summarized usefully as the difference between the C2D1 and BL sample means. Rather, a mixture model accounting for the observed bimodality first was fit and used to estimate the differences between the right modes, denoted by δRi, and the differences between the left modes, denoted by δLi, for the within-patient C2D1-versus-BL distributions of p-PDGFR, for each patient, i = 1, … , 88.
In the Bayesian regression model for PFS (Morita and others 2010), δRi was used as a covariate representing change in p-PDGFR from BL to C2D1. This was done because the values of δRi were much larger than δLi, and moreover δRi was strongly associated with longer PFS. Based on preliminary goodness-of-fit analyses, it was assumed that the logarithm of PFS time was normally distributed, equivalently, that PFS was lognormal. The linear component of the lognormal regression model is the mean of log(PFS time), defined as follows. For patient i and cytokine j = 1, … , 17, denote the (BL, C2D1) cytokine values by (Xij, Yij), the difference between the log-transformed cytokine values by Wij = log(Yij) – log(Xij), Z1i = 1 if treatment was docetaxel+imatinib (DI) and 0 if docetaxel+placebo (DP), Z2i = Hb at BL, and Z3i = change in PSA from BL to C2D1. For cytokine j and patient i, the linear component was assumed to be
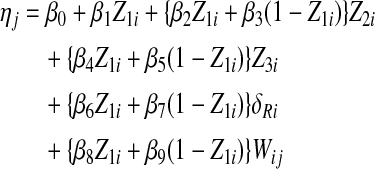 |
In terms of their effects on PFS time, the parameters in the linear term may be interpreted as follows:
β1 = main DI-vs-DP treatment effect
β2 = effect of baseline Hb in the DI arm
β3 = effect of baseline Hb in the DP arm
β4 = effect of change in PSA in the DI arm
β5 = effect of change in PSA in the DP arm
β6 = effect of change in p-PDGFR in the DI arm
β7 = effect of change in p-PDGFR in the DP arm
β8 = effect of change in cytokine value in the DI arm
β9 = effect of change in cytokine value in the DP arm
Using the large (n = ∼2,000 cells) within-patient p-PDGFR samples taken at BL and at C2D1, the probability of decrease in p-PDGFR after treatment, denoted by Pr(Decr), was estimated very reliably for each patient as a standardized Wilcoxon statistic. Specifically, each patient's Pr(Decr) was computed as the mean over all 0/1 indictors that each BL value of p-PDGFR was larger than each C2D1 value. For each cytokine, the following piecewise linear regression model for the BL to C2D1 change in cytokine value, Wij, as a function of the estimated Pr(Decr) was fit.
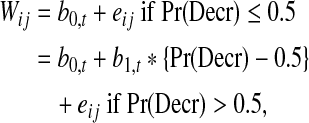 |
for treatment arm t = DI or DP, where eij denotes normally distributed random measurement error. Under this model, in treatment arm t, on average the BL to C2D1 change in the cytokine value equals the constant b0,t if Pr(Decr) ≤0.5 and equals the straight line b0,t + b1,t *{Pr(Decr) – 0.5} if Pr(Decr) > 0.5. The cut-off 0.5 was used because Pr(Decr) = 0.5 corresponds to no change in the cytokine from BL to C2D1, whereas Pr(Decr) ≥ 0.5 and Pr(Decr) < 0.5 correspond, respectively, to the cytokine going down or up, on average. The piecewise linear form was chosen based on preliminary goodness-of-fit plots of each cytokine change as a function of Pr(Decr). Under the null hypothesis (b0,DP, b1,DP) = (b0,DI, b1,DI), the piecewise linear model is the same for the 2 treatment arms. This null hypothesis corresponds to the kinetics of the cytokine, as a function of Pr(Decr), not changing with the addition of imatinib to docetaxel.
Results
The distributions of the 17 plasma angiogenic and inflammatory cytokines at BL and at C2D1 within each treatment arm are summarized in Table 1. These results indicate a significant decline in IL6 and significant increases in PIGF and soluble VEGFR1 in the docetaxel-placebo arm, and a significant decline in soluble c-kit and increase in IL10 in the docetaxel-imatinib arm. Table 2 summarizes changes in cytokine values from BL to C2D1, compared between treatment arms using the Wilcoxon rank sum test. These tests indicate significantly larger declines in PIGF, soluble c-kit, VEGF, and sVEGFR1 in the docetaxel-imatinib arm compared to the docetaxel-placebo arm, on average. The largest individual quantitative difference in cytokines between the arms was the decline in soluble c-kit in the docetaxel-imatinib arm.
Table 1.
Means and Standard Deviations (in Parentheses) of Cytokine Values at Baseline and at Course 2 Day 1 of Chemotherapy
| |
Docetaxel + placebo |
Docetaxel + imatinib |
||||
|---|---|---|---|---|---|---|
| Cytokines | BL | C2D1 | P | BL | C2D1 | P |
| TGFβ | 0.84 (0.22) | 0.90 (0.19) | 0.009 | 0.82 (0.22) | 0.79 (0.18) | 0.586 |
| bFGF | −1.67 (0.24) | −1.67 (0.24) | 0.439 | −1.65 (0.22) | −1.64 (0.21) | 0.881 |
| PIGF | −1.30 (0.09) | −1.20 (0.12) | <0.001a | −1.28 (0.09) | −1.35 (0.11) | 0.002 |
| sVEGFR1 | −0.68 (0.08) | −0.60 (0.10) | <0.001a | −0.65 (0.14) | −0.61 (0.26) | 0.166 |
| VEGF | −0.77 (0.14) | −0.73 (0.17) | 0.05 | −0.80 (0.17) | −0.86 (0.16) | 0.004 |
| c-kit | 0.85 (0.16) | 0.86 (0.15) | 0.508 | 0.83 (0.13) | 0.70 (0.15) | <0.001a |
| sVEGFR2 | 1.23 (0.13) | 1.24 (0.14) | 0.317 | 1.21 (0.15) | 1.19 (0.15) | 0.021 |
| hMMP9 | 1.95 (0.22) | 1.99 (0.29) | 0.354 | 1.91 (0.25) | 1.83 (0.23) | 0.074 |
| GM-CSF | −0.64 (1.14) | −0.68 (1.10) | 0.529 | −0.47 (0.71) | −0.58 (0.80) | 0.05 |
| IFNγ | −0.02 (0.74) | −0.20 (0.77) | 0.071 | 0.13 (0.67) | 0.09 (0.89) | 0.834 |
| IL10 | 0.39 (0.92) | 0.56 (0.79) | 0.019 | 0.64 (0.67) | 0.91 (0.75) | <0.001a |
| IL12p70 | 0.46 (0.72) | 0.50 (0.70) | 0.184 | 0.40 (0.52) | 0.39 (0.55) | 0.167 |
| IL1β | −0.77 (0.75) | −0.84 (0.73) | 0.253 | −0.49 (0.64) | −0.58 (0.72) | 0.265 |
| IL2 | −0.15 (0.55) | −0.27 (0.59) | 0.013 | 0.02 (50) | −0.03 (0.57) | 0.677 |
| IL6 | 0.43 (0.45) | 0.06 (0.59) | <0.001a | 0.57 (0.52) | 0.30 (0.54) | 0.002 |
| IL8 | 0.76 (0.20) | 0.72 (0.24) | 0.068 | 0.76 (0.18) | 0.81 (0.27) | 0.178 |
| TNFα | 0.90 (0.18) | 0.84 (0.19) | 0.012 | 0.97 (0.37) | 0.97 (0.32) | 0.752 |
Comparisons of C2D1-versus-BL for each cytokine within each treatment arm were done using the Wilcoxon rank sum test. Using testwise P value 0.05 and a Bonferroni adjustment for multiple testing, with 34 tests, a P value <0.00147 implies significant change for that cytokine in that treatment arm.
Significant P values.
bFGF, basic fibroblast growth factor; BL, baseline; C2D1, cycle 2 day 1; GM-CSF, granulocyte macrophage-colony stimulating factor; hMMP9, human matrix metalloproteinase; IFNγ, interferon gamma; IL, interleukin; PIGF, placental growth factor; sVEGFR, soluble vascular endothelial growth factor receptor-2; TGFβ, transforming growth factor beta; TNFα, tumor necrosis factor alpha.
Table 2.
Means and Standard Deviations (in Parentheses) of Change from Baseline to Course 2 Day 1 of Chemotherapy for Each Cytokine Variable, Within Each Treatment Arm, Compared Between Arms Using the Wilcoxon Rank Sum Test
| Cytokines | Docetaxel + placebo | Docetaxel + imatinib | P |
|---|---|---|---|
| TGFβ | 0.07 (0.23) | −0.03 (0.22) | 0.020 |
| bFGF | 0.01 (0.28) | 0.03 (0.28) | 0.847 |
| PIGF | 0.12 (0.14) | −0.08 (0.14) | <0.0001a |
| sVEGFR1 | 0.07 (0.12) | 0.04 (0.24) | 0.001a |
| VEGF | 0.04 (0.13) | −0.06 (0.14) | <0.0001a |
| c-kit | <0.01 (0.08) | −0.14 (0.12) | <0.0001a |
| sVEGFR2 | 0.01 (0.07) | −0.03 (0.08) | 0.017 |
| hMMP9 | 0.04 (0.25) | −0.08 (0.26) | 0.049 |
| GM-CSF | −0.04 (0.99) | −0.08 (0.99) | 0.509 |
| IFNγ | −0.20 (0.94) | 0.11 (1.24) | 0.099 |
| IL10 | 0.19 (0.51) | 0.32 (0.52) | 0.137 |
| IL12p70 | 0.04 (0.22) | −0.01 (0.70) | 0.075 |
| IL1β | −0.09 (0.94) | −0.06 (1.07) | 0.913 |
| IL2 | −0.14 (0.66) | 0.01 (0.50) | 0.095 |
| IL6 | −0.39 (0.49) | −0.27 (0.48) | 0.278 |
| IL8 | −0.05 (0.20) | 0.09 (0.46) | 0.053 |
| TNFα | −0.06 (0.18) | 0.02 (0.23) | 0.042 |
Using testwise P value 0.05 and a Bonferroni adjustment for multiple testing, with 17 tests, a P value <0.00294 implies significant change for that cytokine in that treatment arm.
Significant P values.
The fitted piecewise linear regression models are summarized in Table 3. For each cytokine, the test of (b0,DP, b1,DP) = (b0,DI, b1,DI) between the 2 treatment groups was performed using an F statistic with (2, 84) degrees of freedom. The results indicate that, among the 17 cytokines, the kinetics of 2 specific angiogenic cytokines, PIGF and soluble c-kit, differed significantly between the 2 treatment arms in terms of relationship to in vivo p-PDGFR dynamics, as summarized by Pr(Decr). These 2 cytokines were previously identified as among the 4 cytokines decreasing in the docetaxel–imatinib arm compared to the docetaxel–placebo arm (Table 2).
Table 3.
Summaries of 17 Fitted Regression Models, One for Each Cytokine
| |
|
Docetaxel + placebo |
Docetaxel + imatinib |
Test for homogeneity between treatment groups |
||
|---|---|---|---|---|---|---|
| Cytokine | Parameter | Estimate | SE | Estimate | SE | P value |
| TGFβ | 0.013 | |||||
| Intercept | 0.024 | 0.034 | −0.061 | 0.041 | ||
| Slope | 1.825 | 0.661 | 0.555 | 0.379 | ||
| bFGF | 0.431 | |||||
| Intercept | −0.038 | 0.044 | −0.008 | 0.053 | ||
| Slope | 1.826 | 0.858 | 0.504 | 0.492 | ||
| PlGF | <0.001a | |||||
| Intercept | 0.131 | 0.023 | −0.084 | 0.027 | ||
| Slope | −0.336 | 0.442 | 0.037 | 0.253 | ||
| sVEGFR1 | 0.772 | |||||
| Intercept | 0.070 | 0.030 | 0.052 | 0.036 | ||
| Slope | 0.036 | 0.585 | −0.228 | 0.335 | ||
| VEGF | 0.004 | |||||
| Intercept | 0.032 | 0.022 | −0.067 | 0.026 | ||
| Slope | 0.321 | 0.421 | 0.114 | 0.241 | ||
| c-kit | <0.001a | |||||
| Intercept | 0.005 | 0.017 | −0.139 | 0.020 | ||
| Slope | −0.046 | 0.321 | −0.005 | 0.184 | ||
| sVEGFR2 | 0.157 | |||||
| Intercept | 0.01 | 0.012 | −0.018 | 0.015 | ||
| Slope | −0.01 | 0.235 | −0.16 | 0.135 | ||
| hMMP9 | 0.111 | |||||
| Intercept | 0.041 | 0.041 | −0.095 | 0.05 | ||
| Slope | −0.097 | 0.797 | 0.255 | 0.457 | ||
| GM-CSF | 0.122 | |||||
| Intercept | −0.160 | 0.159 | 0.073 | 0.190 | ||
| Slope | 5.201 | 3.057 | −2.396 | 1.752 | ||
| IFNγ | 0.630 | |||||
| Intercept | −0.265 | 0.178 | 0.010 | 0.212 | ||
| Slope | 2.894 | 3.422 | 1.531 | 1.962 | ||
| IL10 | 0.246 | |||||
| Intercept | 0.245 | 0.083 | 0.358 | 0.100 | ||
| Slope | −2.28 | 1.606 | −0.552 | 0.921 | ||
| IL12p70 | 0.474 | |||||
| Intercept | 0.031 | 0.081 | 0.111 | 0.097 | ||
| Slope | 0.505 | 1.558 | −1.782 | 0.893 | ||
| IL1β | 0.922 | |||||
| Intercept | −0.112 | 0.164 | −0.034 | 0.195 | ||
| Slope | 0.986 | 3.148 | −0.463 | 1.805 | ||
| IL2 | 0.475 | |||||
| Intercept | −0.117 | 0.097 | 0.020 | 0.116 | ||
| Slope | −0.865 | 1.865 | −0.097 | 1.069 | ||
| IL6 | 0.169 | |||||
| Intercept | −0.447 | 0.078 | −0.236 | 0.093 | ||
| Slope | 2.393 | 1.499 | −0.464 | 0.860 | ||
| IL8 | 0.156 | |||||
| Intercept | −0.062 | 0.056 | 0.114 | 0.067 | ||
| Slope | 0.638 | 1.077 | −0.36 | 0.618 | ||
| TNFα | 0.129 | |||||
| Intercept | −0.071 | 0.033 | 0.037 | 0.040 | ||
| Slope | 0.348 | 0.632 | −0.227 | 0.362 | ||
In each model, the change in cytokine value from BL to C2D1 is a piecewise linear function of the estimated Pr(Decr) for p-PDGFR, with different parameters for the 2 treatment groups, where Pr(Decr) is the estimated probability that p-PDGFR decreased from BL to C2D1. For each fitted model, the test for identical intercept and slope parameters in the treatment groups, “homogeneity,” is based on an F-statistic with (2, 84) degrees of freedom. Using testwise P value 0.05 and a Bonferroni adjustment for multiple testing, with 17 tests, a P value <0.00294 implies significant heterogeneity between treatment groups, implying different p-PDGFR dynamics with versus without imatinib for that cytokine.
Significant P values.
PDGFR, platelet-derived growth factor and its receptor; SE, standard error.
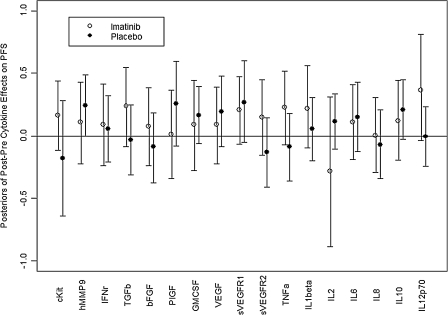
A total of 17 Bayesian log-normal regression models for PFS were fit, one for each cytokine. Because it would be far too cumbersome to tabulate all 17 fitted models, we present only the estimated effects of the C2D1-versus-BL cytokine changes, within each treatment arm, on PFS time. These are the parameters denoted above by β8 and β9 in the model linear component. Because parameters are random quantities under a Bayesian model, each parameter has a posterior distribution under the fitted model. For each combination of cytokine and treatment arm, Fig. 1 summarizes the posterior distribution of the parameter in terms of a 95% credible interval. This interval is represented by a vertical line running from the 2.5th percentile up to the 97.5th percentile of the effect's posterior distribution, with the posterior mean represented by an open circle for the DI arm and by a filled circle for the DP arm. Thus, each vertical line summarizes the middle 95% of the effect's posterior distribution. Under the Bayesian model, a line having lower limit near or above the horizontal line at 0 corresponds to a significant increase in PFS as a function of the C2D1-versus-BL cytokine change. For example, a line for β8 having lower limit 0 would correspond to posterior probability Pr(β8 > 0|data) = 0.975. This would say that, given the observed data, the posterior probability that the effect of the cytokine's change on PFS is positive equals 0.975, a nominally significant effect. A vertical line with mean at 0 would correspond approximately to posterior probability Pr(β8 > 0|data) = 0.50, interpreted as the cytokine change having no effect on PFS. Figure 1 shows that, in the DP arm, human MMP9 (hMMP9) had a significant effect, whereas nearly significant effects on PFS were seen for soluble VEGFR1 and IL-10. In the DI arm, a nearly significant effect on PFS was seen for IL-12p70.
FIG. 1.
Estimated posterior effect of each cycle 2 day 1-to-baseline cytokine change on progression-free survival (PFS) the baseline to cycle 2 day 1 change on PFS time for each cytokine within each treatment arm. Each effect was estimated under a Bayesian log-normal regression model, also accounting for the effects of Hb, change in prostate-specific antigen, and change in p-platelet-derived growth factor and its receptor. The posterior distribution of the parameter quantifying the effect of the in terms of a 95% credible interval. This interval is represented by a vertical line running from the 2.5th percentile to the 97.5th percentile of the effect's posterior distribution, with the posterior mean represented by an open circle for the docetaxel+imatinib arm and a filled circle for the docetaxel+placebo arm.
Discussion
In this study, the kinetics of 17 angiogenic and inflammatory cytokines in men with metastatic castration-resistant prostate cancer receiving docetaxel with or without the c-kit/abl/PDGFR inhibitor imatinib mesylate were examined. Post-treatment cytokines are significantly modified compared to BL in both treatment arms (Table 1), and several differences vary significantly between both treatment arms (Table 2). Our prior observations had indicated that the status of p-PDGFR activation in peripheral blood leucocytes after docetaxel chemotherapy for castration-resistant prostate cancer associated with PFS and OS (Mathew and others 2008). We then studied the differences in cytokine kinetics between the 2 treatment arms when specifically partitioned by post-treatment in vivo p-PDGFR dynamics in peripheral blood leucocytes (Table 3). We find that among these 17 cytokines, PIGF and soluble c-kit dynamics specifically comprise the cytokine signature of imatinib effect after docetaxel chemotherapy.
Decline in soluble c-kit after imatinib therapy has been previously reported in gastrointestinal stromal tumors and has been proposed as a predictive factor for favorable outcome in that disease state (Bono and others 2004, DePrimo and others 2009). In this study, soluble c-kit decline in the imatinib-containing arm was the largest quantitative cytokine difference between the 2 arms. Along with PIGF kinetics, soluble c-kit post-treatment differences retained strong statistical significance when partitioned by p-PDGFR dynamics in peripheral blood leucocytes. These observations may be concordant with the mechanism of action of imatinib as a PDGFR and c-kit inhibitor.
Surprisingly however, in the imatinib arm, increases in soluble c-kit rather than decreases trended toward a favorable PFS profile (Fig. 1) and similarly larger post-treatment values of PIGF and VEGF after docetaxel-alone therapy trended toward an improved PFS. Together, these trends suggest that the cytokine profiles associated with imatinib (c-kit, PIGF, and, to a lesser extent, VEGF declines) compare unfavorably when compared to those generated by docetaxel alone. These findings are also compatible with our previous observations that decreased activation of p-PDGFR in peripheral blood leucocytes after imatinib exposure associated with shorter PFS times (Mathew and others 2008). With the exception of hMMP9 kinetics after docetaxel therapy alone, multivariate analysis of individual cytokine profiles did not yield an independent predictor of outcome. It is conceivable that, with larger numbers of patients, a composite picture of a favorable cytokine signature potentially linked to an in vivo mechanism of action of docetaxel may emerge through such cytokine profiling studies.
Declines in the angiogenic cytokines, PIGF, and VEGF after imatinib therapy have not been reported previously. The altered dynamics of these cytokines together with those previously reported with PDGF (Mathew and others 2008) comprise a candidate cytokine signature of imatinib effect in prostate cancer and bone metastases after docetaxel chemotherapy. Formal mechanistic studies will be required to identify the putative link between the regulation of plasma PIGF and VEGF levels and imatinib therapy. It is conceivable that kinetics of these markers may have predictive value in other disease states, hematological and solid neoplasia, in which imatinib has been established as standard therapy, as these circulating cytokines may not be tumor specific.
Before this report, there have been few studies that demonstrate the profile of changes and/or the predictive value of inflammatory and angiogenic cytokine dynamics after docetaxel therapy in prostate cancer. The wide range of nonhematological toxicities observed with docetaxel, such as peripheral edema or pleural effusions that reflect vascular effects, or fatigue and pneumonitis that reflect proinflammatory effects, are likely to be reflected in plasma cytokine dynamics after treatment. In 2 prior studies, declines in plasma IL6 associated with PSA-declines after docetaxel were reported; however, associations with PFS or OS were not assessed (Domingo-Domenech and others 2006; Ignatoski and others 2009). Our observations do not support a significant association of IL6 decline after docetaxel with PFS (Fig. 1). While significant increases in PIGF and sVEGFR1 and significant decreases in IL6 were observed after docetaxel therapy (Table 1), only an increase in hMMP9 associated with improved PFS (Fig. 1). Elevated hMMP9 expression in prostate cancer has been associated with improved disease-free and OS after prostatectomy for localized prostate cancer (Boxler and others 2010), but a link of plasma MMP9 dynamics with docetaxel efficacy has not been described to our knowledge. These findings suggest the potential predictive value of a cytokine dynamic signature after chemotherapy for prostate cancer, for which larger prospective studies will be required for validation.
Acknowledgments
The authors acknowledge the assistance of Erin Horne and Sherryl Smith (Department of Genitourinary Medical Oncology) for research and administrative support and the M.D. Anderson Cancer Center Immune Monitoring Core Laboratory (IMCL) for assistance with the immunological assays. The IMCL is funded by the M.D. Anderson Cancer Center Support Grant (NCI # CA16672). P.F.T.'s research was partially supported by NCI grant RO1-CA-83932. The authors thank two referees for their detailed and constructive comments.
Author Disclosure Statement
P.M. is on the Speaker Bureau of Sanofi-Aventis. No other disclosures.
References
- Bono P. Krause A. von Mehren M. Serum KIT and KIT ligand levels in patients with gastrointestinal stromal tumors treated with imatinib. Blood. 2004;103:2939–2935. doi: 10.1182/blood-2003-10-3443. others. [DOI] [PubMed] [Google Scholar]
- Boxler S. Djonov V. Kessler TM. Matrix metalloproteinases and angiogenic factors: predictors of survival after radical prostatectomy for clinically organ-confined prostate cancer? Am J Pathol. 2010;177:2216–2224. doi: 10.2353/ajpath.2010.091190. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePrimo SE. Huang X. Blackstein M. Garrett CR. Harmon CS. Circulating levels of Soluble KIT serve as a biomarker for clinical outcome in gastrointestinal stromal tumor patients receiving sunitinib following imatinib failure. Clin Cancer Res. 2009;15:5869–5877. doi: 10.1158/1078-0432.CCR-08-2480. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Domenech J. Oliva C. Rovira A. Interleukin-6, a nuclear factor-kB target, predicts resistance to docetaxel in hormone-independent prostate cancer and nuclear factor-kB inhibition by PS-1145 enhances docetaxel antitumor activity. Clin Cancer Res. 2006;12:5578–5586. doi: 10.1158/1078-0432.CCR-05-2767. others. [DOI] [PubMed] [Google Scholar]
- Hollander M. Wolfe DA. Introduction to the theory of nonparametric statistics. New York: John Wiley; 1979. [Google Scholar]
- Ignatoski KMW. Friedman J. Escara-Wilke J. Change in markers of bone metabolism with chemotherapy for advanced prostate cancer: interleukin-6 response is a potential early indicator of response to therapy. J Interferon Cytokine Res. 2009;29:105–111. doi: 10.1089/jir.2008.0024. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ. Uehara H. Yazici S. Targeting platelet-derived growth factor receptor on endothelial cells of multidrug-resistant prostate cancer. J Natl Cancer Inst. 2006;98:783–793. doi: 10.1093/jnci/djj211. others. [DOI] [PubMed] [Google Scholar]
- Mathew P. Thall P. Bucana CD. Platelet-derived growth factor receptor inhibition and chemotherapy for castration-resistant prostate cancer with bone metastases. Clin Cancer Res. 2007;13:5816–5824. doi: 10.1158/1078-0432.CCR-07-1269. others. [DOI] [PubMed] [Google Scholar]
- Mathew P. Thall PF. Wen S. Dynamic change in phosphorylated platelet-derived growth factor receptor in peripheral blood leucocytes following docetaxel therapy predicts progression-free and overall survival in prostate cancer. Br J Cancer. 2008;99:1426–1432. doi: 10.1038/sj.bjc.6604706. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S. Thall PF. Bekele BN. Mathew P. A Bayesian hierarchical mixture model for platelet derived growth factor receptor phosphorylation to improve estimation of progression-free survival in prostate cancer. J R Stat Soc C. 2010;59:19–34. doi: 10.1111/j.1467-9876.2009.00680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor GW. Cochran WG. Statistical methods. 7th. Ames: Iowa State University Press; 1980. [Google Scholar]
- Uehara H. Kim SJ. Karashima T. Effects of blocking platelet-derived growth factor-receptor signaling in a mouse model of experimental prostate cancer bone metastases. J Natl Cancer Inst. 2003;95:458–570. doi: 10.1093/jnci/95.6.458. others. [DOI] [PubMed] [Google Scholar]