Summary
Gluconeogenesis converts three carbon units into glucose. Here we identify an analogous pathway in Saccharomyces cerevisiae for converting three carbon units into ribose, a component of nucleic acids and nucleotides. This riboneogenic pathway involves the enzyme sedoheptulose-1,7-bisphosphatase (SHB17), whose activity was identified based on accumulation of sedoheptulose-1,7-bisphosphate in the corresponding knockout strain. We determined the crystal structure of Shb17 in complex with sedoheptulose-1,7-bisphosphate, and found that the sugar is bound in the closed furan form in the active site. Like fructose-1,6-bisphosphate, sedoheptulose-1,7-bisphosphate is produced by aldolase, in this case from erythrose 4-phosphate and dihydroxyacetone phosphate. Hydrolysis of sedoheptulose-1,7-bisphosphate by SHB17 provides an energetically favorable input to the non-oxidative pentose phosphate pathway to drive ribose production. Flux through SHB17 is enhanced under conditions when ribose demand is high relative to demand for NADPH, including during ribosome biogenesis in metabolically synchronized yeast cells. Thus, riboneogenesis provides a thermodynamically-driven route of ribose production uncoupled from formation of NADPH.
Introduction
Eukaryotic cells consume glucose through two primary routes, glycolysis and the pentose phosphate pathway. Glycolysis produces ATP, NADH, and trioses for amino acids and glycerolipid biosynthesis. The pentose phosphate pathway generates NADPH, ribose for DNA and RNA synthesis, and erythrose for amino acid biosynthesis. The rate and direction of the glycolytic pathway are controlled by heavily regulated enzymes that catalyze strongly thermodynamically favored reactions, e.g., phosphofructokinase, fructose bisphosphatase, and pyruvate kinase. Glucose enters the pentose phosphate pathway either through glucose-6-phosphate dehydrogenase, the committed step of the oxidative arm of the pathway, or via glycolytic intermediates to the non-oxidative arm of the pathway.
The enzymatic steps in the non-oxidative pentose phosphate pathway are controversial. While textbooks contain a reasonable sequence of mass-balanced steps catalyzed by known enzymes, their ability to account for all isotopic distributions observed has been debated (Berthon et al., 1993; McIntyre et al., 1989; Williams et al., 1987). Moreover, regulation of the pathway flux, including whether net flow is towards or away from ribose, remains poorly understood. These issues are clearly important, as the growth of many tumor cells requires up-regulated activity of the non-oxidative pentose phosphate pathway (Deberardinis et al., 2008).
Advances in mass spectrometry permit measurement of metabolites more sensitively, specifically and conveniently than previously possible, allowing the revisitation of fundamental questions regarding metabolic pathway architectures and regulation (Fendt et al., 2010; Nakahigashi et al., 2009; Walther et al., 2010). Untargeted methods, based on full scan mass spectrometry, can detect both known and unexpected metabolites (Allen et al., 2003; Saito et al., 2006). These untargeted methods have been applied successfully in discovery metabolite profiling, an approach that identifies the endogenous roles of enzymes by searching for metabolites that accumulate upon enzyme inhibition (Saghatelian and Cravatt, 2005). This approach has revealed that the endogenous substrates of enzymes are often not accurately predicted by in vitro activity measurements (Saghatelian et al., 2004).
In the best-characterized eukaryote, S. cerevisiae, more than 900 genes remain uncharacterized according to the Saccharomyces Genome Database. Some of these genes likely encode enzymes catalyzing metabolic reactions, which potentially could be revealed by discovery metabolite profiling. The availability of sequence, transcription, and proteomic data facilitates prioritization of ubiquitously expressed, widely conserved candidate enzymes of unknown function for metabolic analysis. In this study we describe SHB17, an abundant budding yeast protein that links the pentose phosphate pathway and glycolysis. Shb17 is a phosphatase that catalyzes the specific dephosphorylation of the seven- and eight-carbon sugars sedoheptulose-1,7-bisphosphate (SBP) and octulose-1-8-bisphosphate (OBP). Shb17 catalyzes the committed reaction of a sequence that leads ultimately to ribose-5-phosphate production:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Shb17 flux (rxn 3) is altered by the nutrient and growth demands. In accordance with its role in feeding carbon to the pentose phosphate pathway, expression of SHB17 is coordinated with other enzymes in the non-oxidative PPP including transketolase (TKL1 and TKL2, rxns 1 and 4) and ribose phosphate isomerase (RKI1, rnx 5). These riboneogenic enzymes work in concert during times of peak nucleotide demand to convert glycolytic triose and hexose units to ribose.
Results
A yeast shb17 deletion mutant accumulates SBP and OBP
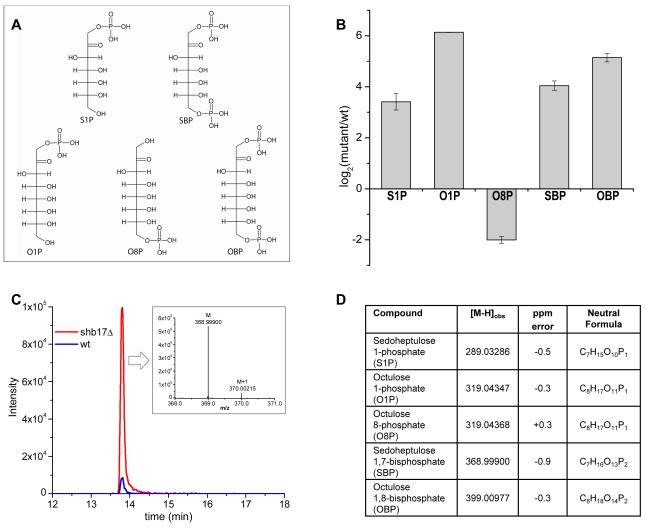
We initiated a metabolomic screen of yeast deletion mutants of genes of unknown function. Candidate genes encoding protein domains similar to enzymes were identified by comparative sequence analysis. Metabolomic phenotypes of mutant strains were measured by reversed phase ion pairing chromatography coupled with high resolution full scan mass spectrometry (Lu et al., 2010). We found that deletion of SHB17, formerly known as the uncharacterized gene YKR043c, consistently led to the accumulation of four metabolites, and the depletion of a fifth (Figure 1).
Figure 1.
Metabolomic phenotype of shb17Δ.
(A) Metabolite structures associated with metabolic phenotype of shb17Δ. For fragmentation data confirming compound structures, see Supplemental Figure 1.
(B) Relative quantitation of metabolites. Data shown is arithmetic mean ± SE of N=4 independent biological replicates.
(C)The negative ionization mode extracted ion chromatogram for SBP in shb17Δ and wild type S. cerevisiae. Inset: Mass spectrum displaying the accurate mass for the parent ion (M) and natural 13C abundance ion (M+1) observed for SBP in negative ionization mode via LC/Exactive Orbitrap MS.
(D) Table of [M-H] ions with altered abundance between shb17Δ and wild type.
Candidate formulae were obtained based on exact masses, and included seven- or eight-carbon mono- or bis-phosphorylated sugars. The formulae were verified by labeling cells with 15N and 13C (Hegeman et al., 2007) and observing no shift for the nitrogen labeling and a shift of +7 or +8 daltons for the carbon labeling (for the putative seven and eight carbon sugars respectively). The presence of phosphate was verified based on mass spectral fragmentation to [H2PO4]−. Collectively, the analytical data revealed that the compounds observed were seven- and eight-carbon mono- and bis-phosphorylated metabolites. We hypothesized that the accumulated compounds were sedoheptulose-1-phosphate (S1P), sedoheptulose-1,7-bisphosphate (SBP), octulose-1-phosphate (O1P), and octulose-1,8-bisphosphate (OBP), and that the depleted compound was octulose-8-phosphate (O8P) (Figure 1). Standards were synthesized for S1P, SBP and OBP, and retention time and fragmentation patterns matched to the accumulating endogenous compounds. Although we did not synthesize O1P and O8P, the location of the phosphate moiety was deduced by comparing fragmentation patterns and retention times to those obtained with S1P and S7P (Supplemental Figure 1). Seven carbon bis-phosphorylated sugars were known in the KEGG database, but not thought to exist in yeast. Eight carbon mono- and bis-phosphorylated sugars were not in metabolite databases but had been proposed in older literature (Bartlett and Bucolo, 1968; Horecker et al., 1982; Paoletti et al., 1979).
Shb17 is a selective sedoheptulose and octulose bisphosphatase
To determine which of the accumulated compounds was the endogenous substrate for Shb17, we performed biochemical assays incubating recombinant Shb17 protein with each of the candidate substrates (S1P, SBP and OBP). Incubation with Shb17 led to depletion of SBP and OBP but not S1P. S7P and O8P accumulated as the products of reactions with SBP and OBP, respectively. S7P is part of the non-oxidative pentose phosphate pathway, which converts ribose-5-phosphate to the glycolytic intermediates fructose-6-phosphate (F6P) and glyceraldehyde-3-phosphate (GAP). The sedoheptulose bisphosphatase activity of Shb17 is inhibited by vanadate (data not shown). Phosphate transfer to AMP or ADP was not observed.
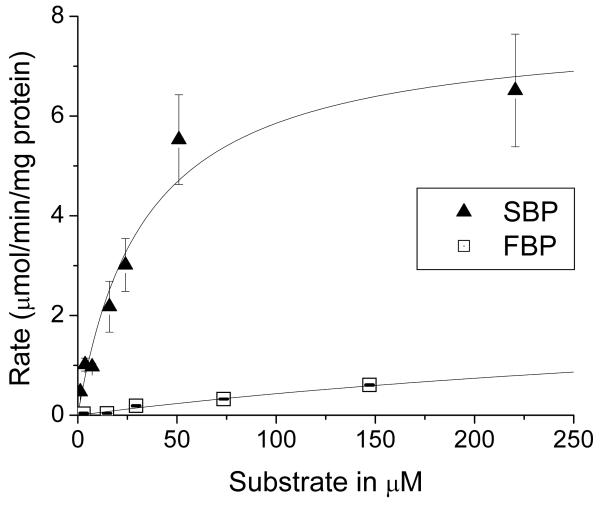
In addition to its observed activity as a sedoheptulose bisphosphatase, Shb17 has previously been shown to exhibit phosphatase activity against the structurally similar metabolite fructose-1,6-bisphosphate (FBP) in vitro (Kuznetsova et al., 2010). FBP does not accumulate in the shb17Δ strain (log2[mutant/wt]=0.048, p=0.6). This suggests that SBP might be the more important in vivo substrate. We measured the kinetic parameters of purified Shb17 using both FBP and SBP as substrates (Figure 2). Shb17 preferentially hydrolyzes SBP, with a lower KM and higher νmax than observed using FBP as a substrate (Table 1). Intracellular concentrations of 19 mM FBP and 0.19 mM SBP were determined by an isotope ratio-based approach (Bennett et al., 2008). The velocity of the enzyme acting on each substrate, assuming that the in vitro measured parameters apply also in vivo, is given by equation (6) for the example of SBP as the substrate (Webb, 1964):
| (6) |
Figure 2.
Hydrolysis of SBP and FBP by purified recombinant Shb17: dependence on substrate concentration. Data are the mean ± standard error for two independent experiments. Full data for FBP are available in Supplemental Figure 2.
Table 1.
Kinetic parameters of Shb17 with FBP and SBP as substrates
| Substrate | KM | ν max | νmax/KM | Intracellular concentration |
Intracellular concentration, 95% confidence interval |
|---|---|---|---|---|---|
| μM | μmoles/min mg protein | L/min mg protein | mM | mM | |
| FBP | 510 ± 50 | 2.6 ± 0.1 | 5.1×10−3 ± 5×10−4 | 19 | 19.9 - 20.5 |
| SBP | 34 ± 8 | 7.8 ± 0.7 | 2.3×10−1 ± 6×10−2 | 0.19 | 0.189 - 0.194 |
Based on equation (1), the estimated dephosphorylation rates for SBP and FBP are similar. Using the observed concentration of 11,500 molecules of Shb17 protein per cell (Ghaemmaghami et al., 2003), we calculate that Shb17 dephosphorylates approximately 300,000 molecules of SBP and 800,000 molecules of FBP per cell per minute. At an average haploid cell volume of 42 fl (Tyson et al., 1979), there are ~ 5 million molecules of SBP and ~ 500 million molecules of FBP per cell. Thus, Shb17 activity substantially affects SBP but not FBP levels. Moreover, typical flux through F6P is typically 10 – 20-fold greater than that through S7P in glucose-grown yeast (Kleijn et al., 2005; Wang and Hatzimanikatis, 2006). Thus, the relative contribution of Shb17 to S7P production is greater than for F6P; this was confirmed based on isotope labeling studies (see below). Thus, while the intrinsic preference of Shb17 for SBP over FBP (i.e., higher νmax/Km), is substantially offset by the higher cellular concentration of FBP than SBP, the primary physiological role of Shb17 appears to be the dephosphorylation of SBP and perhaps also OBP.
Structure of Shb17-SBP complex
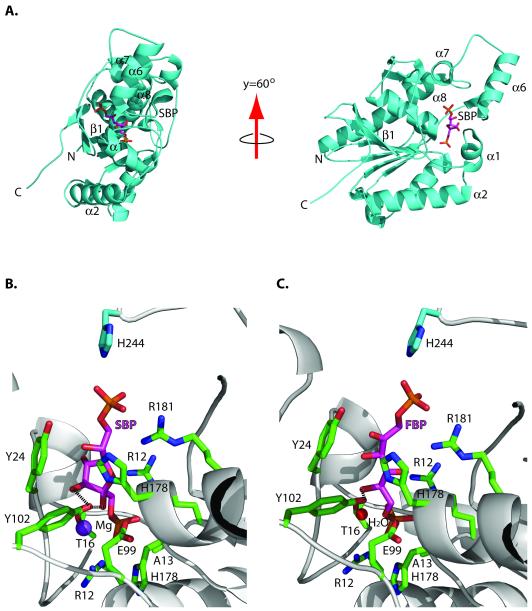
Recently, Kuznetsova et al. (2010) determined the crystal structure of a catalytically inactive mutant of Shb17 (H13A) in complex with FBP bound in the active site. FBP was bound in an extended linear form, unlike the thermodynamically preferred cyclic beta-furanose form observed in complexes with the gluconeogenic enzyme fructose bisphosphatase (Brown et al., 2009, Koerner et al., 1973). To examine the interaction of Shb17 with SBP, we co-crystallized Shb17 (H13A) with SBP and solved the structure of this complex at 2.2 Å resolution (Supplemental Table 1, Supplemental Figure 3). The structure revealed a protein dimer, as in the previous crystal structures of Shb17 (Kuznetsova et al., 2010). Each monomer contains a mixed α-β structure, with a central six-stranded beta sheet flanked by alpha helices on both sides. On one side of this central beta sheet is a substrate binding site formed by the C-terminus of strand β1, the α1 helix and the β1-α1 and α1-α2 loops (residues 14-29), and the α7, α8 and α9 helices and their intervening loops (residues 101-139). Strand β7 (residues 228-234) forms both another boundary for the ligand-binding site, as well as constituting part of the dimer interface, hydrogen-bonding with the major beta-sheet of the interacting monomer. The ligand sedoheptulose 1,7-bisphosphate is best modeled in its beta-furanose form, which is the most common conformer of sedoheptulose and its derivatives in solution (Kuchel et al., 1990).
The Shb17 structure revealed that SBP is bound in the active site in a very similar manner to FBP (Figure 3A). This is possible because SBP is bound in a furanose (cyclic) form, and therefore it has almost the same length as FBP (the distances between the P1 and P2 phosphorus atoms are 10.7 Å and 10.6 Å, respectively). The SBP molecule is coordinated by positively-charged residues on each end of the binding site that position the two terminal phosphate groups. The C1-phosphate group (P1) that is hydrolyzed by the enzyme is highly buried and held in position by hydrogen-bonds and salt bridges from the side chains of R12, R69 and H176 (H13 would presumably also form a salt bridge in the wild-type enzyme) (Figure 3B). The C7-phosphate (P2) is in contact with the side chains of H178 and R181, and the side chain of H244 from the second molecule in the asymmetric unit hydrogen-bonds to the C7-phosphate group. The phenyl ring of Y24 ring lies underneath the ligand to form a platform for the sugar molecule. In the beta-furanose conformation, the C2- and C3-hydroxyl groups point into the protein and away from the solvent, each forming two hydrogen-bonds with the protein. The hydroxyl group of Y102 contributes one hydrogen bond each to the C2- and C3-OH. In addition, the E99 carboxyl group and the T25 backbone amide hydrogen-bond to the 2- and 3-hydroxyl group, respectively. The C4-hydroxyl group points towards the solvent and makes no direct hydrogen-bonding contacts with the protein. In addition, the Shb17-SBP structure revealed the presence of an additional density close to the side chains of the conserved T16 and T25 (2.2 Å and 3.2 Å) and to the P1 oxygen (2.9 Å), which was interpreted as a Mg2+ cation (Figure 3B). This type of Mg2+ coordination is very similar to that found in the regulatory and dynamin-like GTPases/ATPases, where Mg2+ is not required for substrate hydrolysis, but contributes to the phosphate coordination (Daumke et al., 2007; Rutthard et al., 2001; Zhang et al., 2000). There is a similar density at this position in the structure of the Shb17 complex with FBP, which was annotated as a water molecule (Figure 3C), but potentially it might also represent a metal ion. Therefore, although Shb17 and other members of the histidine phosphatase superfamily are known to be metal-independent enzymes, it is possible that some of them can use Mg2+ for the cleavable phosphate coordination and charge neutralization.
Figure 3.
Structure of the Shb17/SBP complex.
(A) Overall fold of the Shb17 (H13A) in complex with SBP (PDB 3OI7, grey ribbon) shown in two orientations with secondary structural elements being labeled. The SBP molecule (magenta carbon atoms) is shown in a stick representation.
(B) Close-up view of the active site of Shb17 in complex with SBP. The side chains of residues in contact with SBP are displayed in a stick representation (green carbon atoms) and labeled. SBP is shown in a stick representation (magenta carbon atoms) and labeled, whereas the Mg2+ ion is shown as a purple sphere and labeled.
(C) Active site of Shb17 in complex with FBP, a similar view as (B). The red sphere denotes a water molecule. Y102 makes two hydrogen bonds with SBP, whereas only one hydrogen bond can be formed between this residue and FBP. These hydrogen bonds are shown by dashed lines in parts B and C.
Overall, the structures of Shb17 complexed with SBP or FBP revealed similar positioning of the active site residues, suggesting that both substrates are hydrolyzed using the same mechanism. Compared to the structure of the Shb17-FBP complex, however, Shb17 makes an additional interaction with SBP: between the C3-OH of SBP and the Y102 of the protein, and another between the 1-phosphate and R12of the protein. These hydrogen bonds, in addition to the more favorable sugar conformation, presumably account for the increased affinity of Shb17 for SBP relative to FBP.
Sedoheptulose 1,7-bisphosphate is synthesized in vivo by fructose bisphosphate aldolase
There are two likely routes for in vivo synthesis of sedoheptulose 1,7-bisphosphate. The first involves formation of SBP by the phosphorylation of S1P, which also accumulates in shb17Δ. The second is via the aldol addition of dihydroxyacetone-phosphate (DHAP) and erythrose-4-phosphate (E4P) catalyzed by the ubiquitous glycolytic enzyme fructose bisphosphate aldolase. Similarly, octulose-1,8-bisphosphate might be synthesized by phosphorylation of O1P or by the aldol addition of DHAP and ribose-5-phosphate.
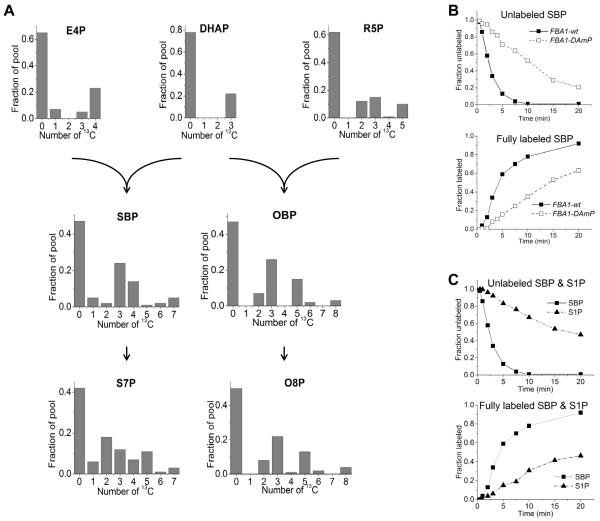
To explore these possibilities, we fed yeast a mixture of 70:30 [U-13C6]-glucose:unlabeled glucose. This mixture yields fully labeled and unlabeled DHAP and E4P, as well as partially labeled forms produced from scrambling reactions of the non-oxidative PPP. From these heterogeneous pools, a labeled subunit may react with an unlabeled subunit, giving rise to a partially labeled product (Szyperskia et al., 1996). In shb17Δ mutant cells, the predominant partially labeled forms of SBP were 13C3 and 13C4, consistent with SBP being synthesized by aldolase-catalyzed condensation of triose and tetrose subunits (Figure 4). Similarly, 13C3 and 13C5 were the predominant partially labeled forms of OBP. The labeling patterns of S7P and O8P were similar to those of SBP and OBP. Partially labeled forms of S1P and O1P could not be measured due to poor signal-to-noise ratio.
Figure 4.
SBP and OBP are synthesized in vivo by C3 + C4 and C3 + C5 subunits via fructose bisphosphate aldolase.
(A) Cells were switched from unlabeled to 70:30 unlabeled glucose:[U-13C]-glucose. Labeling patterns of erythrose-4-phosphate (E4P), dihydroxyacetone-phosphate (DHAP), ribose-5-phosphate (R5P), SBP and OBP were measured in shb17Δ, where SBP and OBP accumulate and hence are more readily quantitated. The reaction products sedoheptulose-7-phosphate (S7P) and octulose 8-phosphate (O8P) were measured in wild type (for data on S7P in shb17Δ see Supplementary Figure 4A). Labeling is reported 20 minutes after nutrient switch for all compounds except OBP, where data is taken at 120 min due to its slower labeling.
(B) Kinetics of labeling of SBP after switching shb17Δ cells with wild type fructose bisphosphate aldolase (FBA1-wt), or the Decreased Abundance by mRNA Perturbation allele (FBA1-DAmP) into [U-13C6]-glucose. For associated pool size and kinetic data, see Supplemental Figure 4 (B-C).
(C) Kinetics of labeling of SBP and S1P after switching shb17Δ cells into [U-13 C6]-glucose.
To examine the role of fructose bisphosphate aldolase (FBA) in the in vivo synthesis of SBP and OBP, we decreased aldolase activity, encoded by the essential gene FBA1, by lowering its expression using a decreased abundance by mRNA perturbation (DAmP) allele (Breslow et al., 2008). The flux of glucose toward SBP and OBP was then measured by kinetic flux profiling (Yuan et al., 2008). Cells were initially grown on unlabeled media, and then switched to fully isotope-labeled [U-13C6]-glucose. Metabolism was quenched at various time points following the switch, enabling us to monitor the rate at which isotopically labeled carbon filled each metabolite pool. We applied this method to four strains: wild type, shb17Δ, FBA1-DAmP, and a FBA1-DAmP / shb17Δ double mutant. Labeling of the direct upstream intermediates of SBP, E4P and DHAP, was unaffected by the FBA1-DAmP allele; however, labeling of SBP and OBP was markedly slowed, verifying their cellular synthesis by aldolase (Figure 4B).
Labeling dynamics provides an additional assessment of the likelihood of SBP and OBP synthesis by aldolase versus by S1P or O1P phosphorylation, which would require that the isotopic labeling of S1P and O1P is faster than labeling of SBP and OBP. Instead, S1P and O1P label more slowly than SBP and OBP. This slow labeling of S1P or O1P is consistent with their formation by hydrolysis of SBP and OBP, perhaps catalyzed by an unidentified phosphatase.
Quantitation of flux through SHB17 into the non-oxidative PPP
S7P can be produced either from SBP by Shb17 or through the canonical pentose phosphate pathway (PPP). Flux from SBP to S7P can be specifically measured based on the pseudo-steady state labeling patterns of SBP and S7P in cells fed glucose singly labeled in its C6 position, [6-13C1]-glucose. Shb17 but not the canonical PPP produces [1,7-13C2]-S7P from [6-13C1]-glucose (Figure 5A).
Figure 5.
Shb17 feeds carbon into the non-oxidative pentose phosphate pathway.
(A) Flux through Shb17 into S7P can be measured using [6-13C1]-glucose. [6-13C1]-glucose leads to [7-13C1]-S7P when S7P is made via the oxidative PPP or the non-oxidative PPP. However, when S7P is produced from SBP via Shb17, a fraction of the S7P pool is doubly labeled: [1,7-13C2]-S7P. Flux is calculated based on the measured isotopic distribution of SBP and S7P.
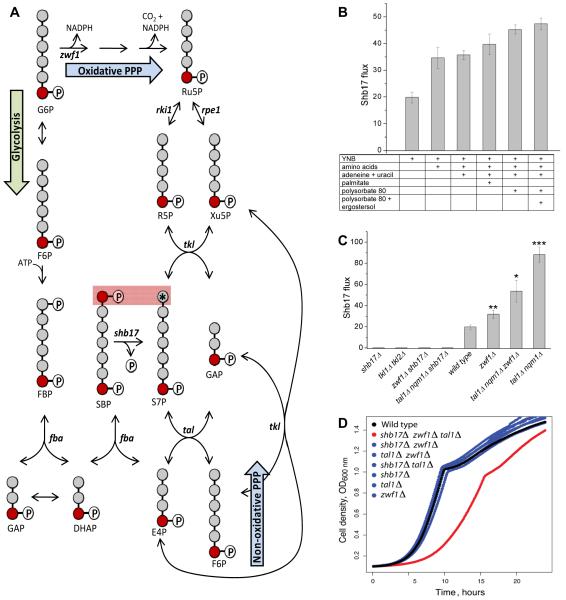
(B) Flux through Shb17 is increased by supplementation with nutrients whose endogenous production requires NADPH, and thus drives oxidative PPP flux. All measurements are performed in wild type yeast. YNB is yeast nitrogen base without amino acids plus 2% glucose. Supplementation with amino acids includes 17 amino acids. Data shown is the arithmetic mean ± SE of N=3 technical replicates.
(C) Effects of PPP gene deletions on Shb17 flux. Deletions are: glucose 6-phosphate dehydrogenase zwf1Δ; transketolase tkl1Δ/ tkl2Δ; transaldolase is tal1Δ/nqm1Δ. Less than 1% doubly labeled S7P was observed in any shb17Δ strain in all measured conditions. All strains were grown in YNB + 2% glucose and supplements as required: methionine for zwf1Δ; synthetic complete media including aromatic amino acids for tkl1Δ/tkl2Δ.
(C) Triple deletion of the sedoheptulose bisphosphatase SHB17, the glucose-6-phosphate dehydrogenase ZWF1, and the transaldolase TAL1, causes a growth defect. Optical density was measured during growth at 30 degrees C in YPD. Growth data are presented in Supplemental Table 3.
The metabolism of [6-13C1]-glucose through the PPP produces only [7-13C1]-S7P. PPP flux in the oxidative direction results in [6-13C1]-glucose-6P being decarboxylated at C1 yielding [5-13C1]-pentose phosphates: ribulose-5P, ribose-5P and xylulose-5P (Xu5P). Transketolase then transfers the C1 and C2 carbons of xylulose-5P to ribose-5P, producing [7-13C1]-S7P and [3-13C1]-glyceraldehyde-3-phosphate (GAP). Non-oxidative PPP flux from glycolytic intermediates towards S7P produces the same labeling pattern. [6-13C1]-fructose-6P combines with [3-13C1]-GAP via transketolase to produce [5-13C1]- Xu5P and [4-13C1]- erythrose-4P (E4P). [4-13C1]- E4P then reacts with another [6-13C1]-fructose 6P, producing via transaldolase [7-13C1]-S7P.
When S7P is made via the Shb17 pathway, however, double labeling from [6-13C1]-glucose can occur. The origin of this double labeling is the aldolase-catalyzed condensation of [3-13C1]-GAP with [4-13C1]- E4P. This yields [1,7-13C2]-SBP, which is dephosphorylated by Shb17 to [1,7-13C2]-S7P. The steady-state fraction of [13C2]-S7P relative to the fraction of [13C2]-SBP equals the fraction of S7P produced through Shb17.
To determine flux through Shb17 by this approach, cells in yeast nitrogen base were first grown on unlabeled glucose, and then switched to otherwise identical media with [6-13C1]-glucose for 90 min, and the ratio of the doubly labeled fractions of S7P and SBP was determined. The labeling duration was selected to allow quasi-steady state labeling of S7P and SBP while minimizing carbon scrambling that can occur when the products of Shb17 undergo further PPP reactions (e.g. formation of doubly labeled pentoses from S7P via transketolase). To validate this strategy, we compared shb17Δ mutants to wild type yeast. In wild-type, 5% of S7P was doubly labeled, while in shb17Δ cells there was < 1% doubly labeled S7P (Figure 5B). The 5% 13C2-labeling of S7P compared with 25% labeling of SBP implies that 20% of S7P is synthesized by dephosphorylation of SBP by Shb17 for wild-type yeast grown in minimal media. Conversely, despite the 10% doubly labeled FBP measured in wild-type yeast, no doubly labeled glucose-6-phosphate was observed, implying that hydrolysis of FBP by Shb17 is not a significant source of hexose phosphates.
Shb17 activity provides a strongly thermodynamically driven route to S7P. Accordingly, we hypothesized that its function might be to drive flux from dihydroxyacetone-phosphate and erythrose-4-phosphate towards ribose-5-phosphate. Such flux would be useful when demand for ribose is not fully met by the oxidative PPP, i.e., when demand for ribose exceeds that for NADPH. In growing yeast, NADPH is consumed substantially for amino acid, nucleic acid base, lipid, and sterol biosynthesis. Accordingly, we hypothesized that providing exogenous amino acids, nucleic acid bases, fatty acids and sterols would increase Shb17 flux. As shown in Figure 5B, such supplementation increased flux through Shb17 from 20% in minimal medium up to almost 50% in media containing the full set of additives.
A more overt way of increasing the requirement for Shb17 flux is genetic elimination of other routes of ribose production. Deletion of glucose 6-phosphate dehydrogenase (ZWF1), the enzyme responsible for the first committed step of the oxidative PPP, caused ~ 50% enhancement of Shb17 flux (Figure 5C). Deletion of the transaldolase genes TAL1 and NQM1, which catalyze a portion of the nonoxidative pentose phosphate pathway, quadrupled flux through Shb17 to S7P. The combination of zwf1Δ mutation with tal1Δ nqm1Δ mutations increased flux over zwf1Δ alone. The overall flux through S7P is higher in tal1Δ nqm1Δ than in zwf1Δ tal1Δ nqm1Δ, but zwf1 mutants grow slowly in the methionine supplemented minimal media used in our flux experiments (data not shown). We anticipate that this growth defect alters overall fluxes in the cell, which is supported by prior evidence that zwf1 mutation reduces flux to the nonoxidative pentose phosphate pathway (Zhao et al., 2004) (Jeppsson et al., 2002). Simultaneous deletion of ZWF1 and the transaldolase TAL1 yielded cells that depend on SHB17 for efficient growth: the triple deletion zwf1Δ tal1Δ shb17Δ grows slowly even on rich medium (Figure 5D). The three single mutants, double mutants with binary combinations of the mutations, or these single and double mutants in combination with mutation of the TAL1 paralog NQM1 (Supplemental Table 3), had no significant effect on growth.
These results are consistent with three effective routes of ribose production from glucose: the oxidative PPP (via Zwf1), the canonical non-oxidative pentose phosphate pathway running in reverse (via Tal1), or riboneogenesis (via Shb17). Both of the latter two also require transketolase activity, which is itself sufficient to make ribose by converting F6P and GAP to Xu5P and E4P, with the disadvantage that Xu5P and E4P are made in stoichiometric amounts, even though cellular demand for R5P exceeds that for E4P. This explains the viability but fitness defect of the zwf1Δ tal1Δ shb17Δ triple deletion mutant. Consistent with the essential role for transketolase in converting S7P formed by Shb17 into ribose, the zwf1Δ tkl1Δ tkl2Δ triple mutation is lethal (Schaaff-Gerstenschlager et al., 1993). Moreover, in double mutants lacking all transketolase activity (tkl1Δ tkl2Δ), there is no measurable flux through Shb17 (Figure 5C).
Shb17 oscillates with ribosomal transcripts in the yeast metabolic cycle
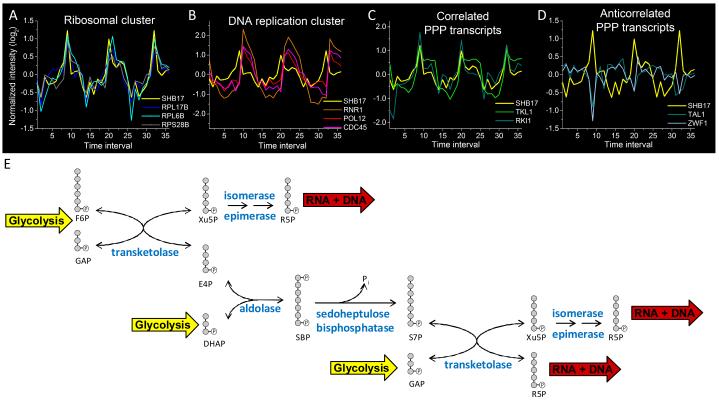
Further evidence supporting the role of Shb17 as a riboneogenic enzyme can be observed through analysis of periodic gene expression during the yeast metabolic cycle (Tu et al., 2005). In nutrient-limited yeast cultures, yeast cells can spontaneously synchronize their metabolism and cell cycle, so that the culture alternates between respiration and fermentation. In gene expression data collected from these metabolically synchronized cells, we observed that levels of the SHB17 transcript are correlated with expression levels of the ribosomal proteins (Figure 6A), which are expressed prior to genes required for DNA synthesis (Figure 6B) (Kudlicki et al., 2007). Thus, periodic SHB17 expression coincides with the peak demand for ribose phosphate that occurs during ribosome biosynthesis.
Figure 6.
SHB17 expression cycles in concert with the yeast metabolic cycle.
(A)-(D) A time series of gene expression during the ~300 minute yeast metabolic cycle is plotted from data presented in (Tu et al., 2005), where each time interval represents ~25 minutes. (A) SHB17 is co-expressed with ribosomal transcripts (shown here: two components of the 60S ribosomal subunit, RPL17B and RPL6B, and one of the 40S subunit, RPS28B). (B) Ribosomal protein transcript expression precedes transcripts associated with DNA replication (shown here: ribonucleotide reductase, RNR1, the B subunit of DNA polymerase, POL12, and a DNA replication initiation factor, CDC45). (C) SHB17 expression correlates with selected PPP transcripts including transketolase (TKL1) and ribose 5-phosphate ketol-isomerase (RKI1). (D) SHB17 expression is anticorrelated with other PPP transcripts including transaldolase (TAL1) and glucose 6-phosphate dehydrogenase (ZWF1). Y-axis displays log2 transformed intensity, with each gene median centered at 0.
(E) Riboneogenic pathway in yeast. The expression data in (A)-(D) suggest a coordinated role of the transketolase TKL1, the ribose ketoisomerase RKI1, and the sedoheptulose bisphosphatase SHB17 in riboneogenesis. The aldolase FBA1 is constituively expressed, consistent with its central role in both glycolysis and gluconeogenesis. The ribulose epimerase RPE1 is also continually expressed. Together, the enzymes work to shunt glycolytic intermediates to ribose. The overall scheme converts one hexose-P and three triose-P to three pentose-P units.
Shb17 facilitates the conversion of glycolytic intermediates to pentose phosphate units. Together with aldolase and transketolase, it can convert one mole of F6P plus three moles of triose phosphate to three moles of pentose phosphate. These reactions define the previously undescribed pathway of riboneogenesis (Figure 6E). TKL1, the primary source of transketolase activity (Schaaff-Gerstenschlager et al., 1993), cycles concurrently with SHB17. The interconversion of the pentose phosphates Xu5P and R5P is achieved in two steps through the intermediate ribulose-5P (Ru5P). In the first step, Xu5P is converted to Ru5P via ribulose 5-phosphate epimerase (Rpe1). Ru5P is then converted to R5P via ribose-5-phosphate ketol-isomerase (Rki1). RKI1 expression oscillates during the metabolic cycle in concert with TKL1 and SHB17, while expression levels of RPE1 are constant (Figure 6C).
Some of the transcripts of the PPP are anti-correlated with SHB17 expression, including glucose 6-phosphate dehydrogenase (ZWF1), the first committed step of the oxidative branch of the PPP (Figure 6D). The anti-correlation of SHB17 and TKL1 with ZWF1 expression suggests that riboneogenesis and the oxidative branch of the PPP are active at distinct phases of the yeast metabolic cycle.
In the canonical non-oxidative PPP, transketolase and transaldolase are considered to convert two moles of F6P and one mole of triose phosphate to three moles of pentose phosphate. However, TAL1 expression is anticorrelated with TKL1, SHB17 and RKI1 across the yeast metabolic cycle (Figure 6D). Because TAL1 and TKL1 are transcribed at different points in the yeast metabolic cycle, Tal1 may not act in concert with Tkl1 for the production of ribose-5-phosphate. Instead, Shb17, Tkl1, and Rki1 presumably act in concert to produce ribose.
Discussion
We report a previously unidentified enzymatic activity encoded by SHB17 that hydrolyzes sedoheptulose-1,7-bisphosphate to sedoheptulose-7-phosphate. In combination with transketolase, ribulose-5-phosphate epimerase, and ribose-5-phosphate isomerase, the activity of SHB17 provides a thermodynamically driven pathway from trioses produced by glycolysis to the synthesis of ribose. The flux through the Shb17 pathway is regulated in response to biosynthetic and redox demands on the cell. This alternative pathway may explain the observed increase in S7P levels when oxidative pentose phosphate activity is inhibited, where previous models had predicted a decrease in S7P (Ralser et al., 2007).
In a recent study, (Kuznetsova et al., 2010) showed the ability of Shb17 to hydrolyze the homologous substrate FBP and determined the crystal structure of Shb17 in complex with it. SHB17 deletion, however, does not alter the cellular levels of FBP, whereas it elevates those of SBP. Motivated by this metabolomic data, we demonstrate that purified Shb17 exhibits higher activity and affinity for SBP. The structure of the Shb17-SBP complex reveals that this higher affinity results from binding of SBP in the preferred beta-furanose sugar conformation and from additional hydrogen bonds between Shb17 and SBP. The current study accordingly is an example of the power of combining metabolomics with structural biology to assign enzyme function, and argues for similar integrated metabolomic-structural biology analysis of other enzymes of unknown function.
Sedoheptulose- and octulose-bisphosphates are absent from most reported pathways of microbial and animal metabolism, but metabolomic analysis in multiple species reveals these compounds are not only present but abundant. In the distantly related fission yeast S. pombe, we observe the formation of doubly labeled S7P in cells fed [6-13C1]-glucose, suggesting a similar flux from SBP to S7P (data not shown). In E. coli, transaldolase mutants fed xylose employ transketolase to convert xylose to S7P, which builds up to sufficient levels to result in its phosphorylation by phosphofructokinase. The resulting SBP is then cleaved by aldolase to generate E4P and DHAP (Nakahigashi et al., 2009). Sedoheptulose and octulose compounds have also been observed in human tissues (Bartlett and Bucolo, 1960; Bucolo and Bartlett, 1960). In macrophages stimulated by endotoxin, a sedoheptulose kinase that produces S7P from sedoheptulose must be downregulated for proper activation (A. Haschemi, personal communication). Sedoheptulose-1,7-bisphosphate is elevated in tumor material (Meijer and Elias, 1984) and in oncogene-transformed cultured mouse cells (Fan and Rabinowitz, unpublished data).
Previous description of SBP and OBP in mammalian metabolism came from studies of an alternative form of the pentose phosphate pathway from the canonical reaction sequence shown in textbooks. The alternative pathway, known sometimes as the L-type (in contrast to the canonical F-type), is purported to be active in liver (Williams et al., 1987). Similar to the pathway described here, the L-type PPP involves interconversion of DHAP and E4P with SBP catalyzed by aldolase. The L-type PPP, however, lacks sedoheptulose-1,7-bisphosphatase activity. Instead, it relies on OBP-S7P phosphotransferase activity, which has never been purified to homogeneity or cloned genetically. Moreover, the net products and reactants of the L-type pathway are identical to that of the canonical F-type. In contrast, in the pathway described here, there is net loss of one high energy phosphate bond, which serves to provide thermodynamic driving force for ribose formation. This, in turn, conveys a physiological function, riboneogenesis.
The riboneogenic pathway has substantial similarity to the Calvin Cycle, the light-independent phase of photosynthesis in which CO2 is condensed with ribulose-1,5,-bisphosphate. In the Calvin Cycle, as in riboneogenesis, sedoheptulose-1,7-bisphosphate is formed when aldolase catalyzes condensation of erythrose 4-phosphate and dihydroxyacetone phosphate. Then, a sedoheptulose-bisphosphatase dephosphorylates sedoheptulose-1,7-bisphosphate to yield sedoheptulose-7-phosphate, which is converted to ribulose-1,5-bisphosphate, the substrate for addition of carbon dioxide. The plant sedoheptulose-bisphosphatases are members of the phosphoglycerate mutase family distantly related to not only Shb17 but also to the fructose bisphosphatase Fbp1 and various phosphoglycerate mutases. The plant enzymes are regulated by light and are localized to the stroma of the chloroplast where they participate in carbon fixation. In contrast, Shb17 is localized to the cytoplasm to coordinate glycolysis with the pentose phosphate pathway.
The demand for the products of the pentose phosphate pathway varies depending on cell growth rate, redox stress, and nutrient availability. It also varies during the cell cycle, as ribose is ultimately the source of ribo- and deoxyribonucleotides. Riboneogenesis allows cells to balance the demands of redox homeostasis and biosynthesis.
Experimental Procedures
Strains and Reagents
S. cerevisiae strains were derived from the synthetic genetic analysis (SGA) deletion set (BY4743 background) (Tong et al., 2001) with the genotype MATa ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 met15Δ0 can1Δ::LEU2+-MFA1pr-HIS3/CAN1 ykr043cΔ::kanMX and a yar047cΔ::kanMX control in the same background. Prototrophic deletions were also created by homologous recombination using the YKR043CΔ::ClonNAT allele amplified by PCR from the above deletion. Prototrophic controls were wild type FY4 (Mat a) and FY5 (Mat α)(Winston et al., 1995). Cells were grown in minimal media comprising 6.7 g/L Difco Yeast Nitrogen Base without amino acids plus 2% (w/v) glucose. Additives were added in selected cases as described in the text and Extended Experimental Procedures. Chemistry solvents and reagents were generally the highest purity that is commercially available. Full details of source materials are available in the Extended Experimental Procedures.
Screening for Metabolic Phenotypes
Single colonies of yeast deletion and control strains (N=4 of each) were grown overnight to saturation at 30°C in minimal medium. Cultures were set back to OD600 ~0.1 in 25 mL media and allowed to grow at 30°C to mid-log phase (OD600 between 0.4 to 0.6). Cells were harvested by vacuum filtering the cultures onto 47 mm diameter 0.45 μm nylon filters. Metabolism was quenched by quickly placing the filter cell side down in 0.8 mL of −20°C extraction solvent (40/40/20 acetonitrile/methanol/water). The cells were allowed to sit in the extraction solvent at −20°C for 15 min, at which time cells were washed off of the filter with an additional 200 μL solvent. The solvent-cell mixture was then centrifuged. The supernatant was removed and placed on ice. Pellets were resuspended in 200 μL fresh solvent, and a second extraction was performed for fifteen minutes at 5°C. The resulting mixture was then centrifuged, and the two supernatants were combined. Aliquots were dried under N2(g) and resuspended in HPLC-grade water prior to LC-MS analysis.
LC-MS Analysis
Full scan LC-MS analysis, without the capability for MS/MS, was performed on an ultra high performance LC system coupled by negative ion mode electrospray ionization to a stand-alone orbitrap mass spectrometer (Lu et al., 2010). The LC method involves reversed phase ion-pairing chromatography on a C18 column with tributylamine as the ion pairing agent (Luo et al., 2007). The mass spectrometer was run at 1 Hz scan speed with 100,000 resolving power. Metabolites differing between wild-type and knock-out strains were determined using MAVEN, an open-source LC-MS data analysis package developed in-house (Melamud et al., 2010). Instrumentation and run parameters are described in detail in Extended Experimental Procedures. LC-MS/MS data was collected in selected cases on a hybrid ion trap-orbitrap instrument.
General Synthesis of Mono and Diphosphosaccaarides
A series of coupled enzymatic reactions were used to produce sedoheptulose-1,7-bisphosphate (SBP), sedoheptulose-1-phosphate (S1P), and d-glycero-d-altro-octulose-1,8-bisphosphate (OBP). In Reaction I, aldolase (4.1.2.13) cleaves fructose-1,6-bisphosphate (FBP) into glyceraldehyde-3-phosphate (GAP) and dihydroxyacetone-phosphate (DHAP). Reaction II uses aldolase (4.1.2.13) to couple glyceraldehyde-3-phosphate to either erythrose-4-phosphate, erythrose, or ribose-5-phosphate to form the products SBP, S1P, or OBP respectively. In the case of SBP, erythrose-4-phosphate is derived from the cleavage of fructose-6-phosphate by transketolase (2.2.1.1) in the presence of GAP which produces xylulose-5-phosphate as a side product. Finally, triose phosphate isomerase (5.3.1.1) is used to facilitate conversion of DHAP to GAP, thus driving the reaction and increasing yield. These methods are modifications of previous work (Smyrniotis, 1956; Valentin, 1993). For detailed syntheses, see Extended Experimental Procedures.
Protein Purification and Enzymatic Assays
Initial enzymatic screens for enzymatic activity were performed using in vitro synthesized, untagged Shb17 (New England Biolabs Inc. PURExpress® In Vitro Protein Synthesis Kit). Subsequent studies were performed using N-terminal His-tagged recombinant protein purified from E. coli. Reactions were carried out at 30°C at pH 7 and quenched using acetic acid. Reactants and products in the quenched solutions were analyzed by LC-MS. For details, see Extended Experimental Procedures.
Cell Growth Assays
Overnight cultures of the indicated mutant strains were diluted back to OD600 nm ~ 0.1 and grown for 4 hours. These cultures were diluted to OD600 nm ~ 0.05 and growth was monitored by measuring optical density at 600 nM in a BioTek plate reader incubated with shaking at 30°C.
Crystallography
Crystals of SHB17(H13A) were grown at room temperature by hanging-drop vapor diffusion. These crystals were soaked for 10 minutes in well solution plus 10 mM sedoheptulose-1,7-bisphosphate, and then cryoprotected and flash-frozen. The structure of SHB17(H13A) with bound sedoheptulose-1,7-bisphosphate was solved by molecular replacement from the protein coordinates of SHB17(H13A) solved with fructose-1,6-bisphosphate (PDB code 3LL4). Additional details regarding structural determination methods are described in Extended Experimental Procedures. Statistics for data collection and structure refinement are summarized in Supplemental Table 1.
Supplementary Material
Highlights.
Riboneogenesis converts three carbon sugars into ribose
Sedoheptulose-1,7-bisphosphate is a key intermediate in riboneogenesis
Shb17 is a sedoheptulose bisphosphatase that thermodynamically drives ribose biosynthesis
Flux through Shb17 is high when demand for ribose exceeds that for reducing power
Acknowledgements
This research was supported by NSF Career Award MCB-0643859 to J.D.R. and NIH Grant GM071508 for the Center of Quantitative Biology at Princeton University. M.F.C. gratefully acknowledges financial support from Merck and Company through a Doctoral Research Fellowship, and Amy Caudy support through the Lewis-Sigler Fellows program. Additional support came from the Beckman Foundation, American Heart Association Grant 0635188N, NSF Career Award MCB-0643859, NIH Grant AI078063, the DOE Biohydrogen program (to J.D.R.), and from the government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405) (to A.F.Y.). We thank Kate Kuznetsova for providing purified proteins, Wenyun Lu and Saw Kyin for assistance with mass spectrometry, and Dannie Durand and David Botstein for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- SGD project [November 16, 2010];Saccharomyces Genome Database. http://www.yeastgenome.org/
- Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- Bartlett GR, Bucolo G. Octulose phosphates from the human red blood cell. Biochem Biophys Res Commun. 1960;3:474–478. doi: 10.1016/0006-291x(60)90158-3. [DOI] [PubMed] [Google Scholar]
- Bartlett GR, Bucolo G. The metabolism of ribonucleoside by the human erythrocyte. Biochim Biophys Acta. 1968;156:240–253. doi: 10.1016/0304-4165(68)90253-5. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon HA, Bubb WA, Kuchel PW. 13C n.m.r. isotopomer and computer-simulation studies of the non-oxidative pentose phosphate pathway of human erythrocytes. Biochem J. 1993;296(Pt 2):379–387. doi: 10.1042/bj2960379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucolo G, Bartlett GR. Sedoheptulose diphosphate formation by the human red blood cell. Biochem Biophys Res Commun. 1960;3:620–624. doi: 10.1016/0006-291x(60)90158-3. [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fendt SM, Buescher JM, Rudroff F, Picotti P, Zamboni N, Sauer U. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol Syst Biol. 2010;6:356. doi: 10.1038/msb.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hegeman AD, Schulte CF, Cui Q, Lewis IA, Huttlin EL, Eghbalnia H, Harms AC, Ulrich EL, Markley JL, Sussman MR. Stable isotope assisted assignment of elemental compositions for metabolomics. Anal Chem. 2007;79:6912–6921. doi: 10.1021/ac070346t. [DOI] [PubMed] [Google Scholar]
- Horecker BL, Paoletti F, Williams JF. Occurrence and significance of octulose phosphates in liver. Ann N Y Acad Sci. 1982;378:215–224. doi: 10.1111/j.1749-6632.1982.tb31198.x. [DOI] [PubMed] [Google Scholar]
- Jeppsson M, Johansson B, Hahn-Hagerdal B, Gorwa-Grauslund M. Reduced Oxidative Pentose Phosphate Pathway Flux in Recombinant Xylose-Utilizing Saccharomyces cerevisiae Strains Improves the Ethanol Yield from Xylose. Appl Environ Microbiol. 2002:1604. doi: 10.1128/AEM.68.4.1604-1609.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijn RJ, van Winden WA, van Gulik WM, Heijnen JJ. Revisiting the 13C-label distribution of the non-oxidative branch of the pentose phosphate pathway based upon kinetic and genetic evidence. FEBS J. 2005:4970–4982. doi: 10.1111/j.1742-4658.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- Kudlicki A, Rowicka M, Otwinowski Z. SCEPTRANS: an online tool for analyzing periodic transcription in yeast. Bioinformatics. 2007;23:1559–1561. doi: 10.1093/bioinformatics/btm126. [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Xu L, Singer A, Brown G, Dong A, Flick R, Cui H, Cuff M, Joachimiak A, Savchenko A, et al. Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae. J Biol Chem. 2010;285:21049–21059. doi: 10.1074/jbc.M110.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Clasquin MF, Melamud E, Amador-Noguez D, Caudy AA, Rabinowitz JD. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- McIntyre LM, Thorburn DR, Bubb WA, Kuchel PW. Comparison of computer simulations of the F-type and L-type non-oxidative hexose monophosphate shunts with 31P-NMR experimental data from human erythrocytes. Eur J Biochem. 1989;180:399–420. doi: 10.1111/j.1432-1033.1989.tb14662.x. [DOI] [PubMed] [Google Scholar]
- Meijer AE, Elias EA. Significance of pentosephosphate cycle capacity increase in malignant tumors for energy metabolism. Acta Histochem Suppl. 1984;29:141–148. [PubMed] [Google Scholar]
- Melamud E, Vastag L, Rabinowitz JD. Metabolomic Analysis and Visualization Engine for LC-MS Data. Anal Chem. 2010 doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Toya Y, Ishii N, Soga T, Hasegawa M, Watanabe H, Takai Y, Honma M, Mori H, Tomita M. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol Syst Biol. 2009;5:306. doi: 10.1038/msb.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti F, Williams JF, Horecker BL. Detection and estimation of sedoheptulose and octulose mono- and bisphosphates in extracts of rat liver. Arch Biochem Biophys. 1979;198:620–626. doi: 10.1016/0003-9861(79)90539-3. [DOI] [PubMed] [Google Scholar]
- Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol. 2007;6:10. doi: 10.1186/jbiol61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutthard H, Banerjee A, Makinen MW. Mg2+ is not catalytically required in the intrinsic and kirromycin-stimulated GTPase action of Thermus thermophilus EF-Tu. J Biol Chem. 2001;276:18728–18733. doi: 10.1074/jbc.M102122200. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Cravatt BF. Discovery metabolite profiling--forging functional connections between the proteome and metabolome. Life Sci. 2005;77:1759–1766. doi: 10.1016/j.lfs.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Saito N, Robert M, Kitamura S, Baran R, Soga T, Mori H, Nishioka T, Tomita M. Metabolomics approach for enzyme discovery. J Proteome Res. 2006;5:1979–1987. doi: 10.1021/pr0600576. [DOI] [PubMed] [Google Scholar]
- Schaaff-Gerstenschlager I, Mannhaupt G, Vetter I, Zimmermann FK, Feldmann H. TKL2, a second transketolase gene of Saccharomyces cerevisiae. Cloning, sequence and deletion analysis of the gene. Eur J Biochem. 1993;217:487–492. doi: 10.1111/j.1432-1033.1993.tb18268.x. [DOI] [PubMed] [Google Scholar]
- Smyrniotis PZ, Horecker BL. The preparation of sedoheptulose diphosphate. J Biol Chem. 1956;218:745–752. [PubMed] [Google Scholar]
- Szyperskia T, Baileyb JE, Wüthrich K. Detecting and dissecting metabolic fluxes using biosynthetic fractional 13C labeling and two-dimensional NMR spectroscopy. Trends in Biotechnology. 1996;14:453–459. [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. Journal of bacteriology. 1979:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin M, Bolte J. Fructose-1,6-diphosphate aldolase from spinach leaves, a challenger for enzymatic synthesis of ketoses. Tet Lett. 1993;34:8103–8106. [Google Scholar]
- Walther T, Novo M, Rossger K, Letisse F, Loret MO, Portais JC, Francois JM. Control of ATP homeostasis during the respiro-fermentative transition in yeast. Mol Syst Biol. 2010;6:344. doi: 10.1038/msb.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hatzimanikatis V. Metabolic engineering under uncertainty--II: analysis of yeast metabolism. Metabolic Engineering. 2006:142–159. doi: 10.1016/j.ymben.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Webb M, Dixon EC. Enzymes. 2nd edn Academic Press; New York: 1964. [Google Scholar]
- Williams JF, Arora KK, Longenecker JP. The pentose pathway: a random harvest. Impediments which oppose acceptance of the classical (F-type) pentose cycle for liver, some neoplasms and photosynthetic tissue. The case for the L-type pentose pathway. Int J Biochem. 1987;19:749–817. doi: 10.1016/0020-711x(87)90239-4. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem. 2000;275:25299–25307. doi: 10.1074/jbc.M001027200. [DOI] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metabolic Engineering. 2004:164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.